ABSTRACT
The fragile X syndrome (FXS), the most common form of inherited intellectual disability, is due to the absence of FMRP, a protein regulating RNA metabolism. Recently, an unexpected function of FMRP in modulating the activity of Adenosine Deaminase Acting on RNA (ADAR) enzymes has been reported both in Drosophila and Zebrafish. ADARs are RNA-binding proteins that increase transcriptional complexity through a post-transcriptional mechanism called RNA editing.
To evaluate the ADAR2-FMRP interaction in mammals we analyzed several RNA editing re-coding sites in the fmr1 knockout (KO) mice. Ex vivo and in vitro analysis revealed that absence of FMRP leads to an increase in the editing levels of brain specific mRNAs, indicating that FMRP might act as an inhibitor of editing activity. Proximity Ligation Assay (PLA) in mouse primary cortical neurons and in non-neuronal cells revealed that ADAR2 and FMRP co-localize in the nucleus. The ADAR2-FMRP co-localization was further observed by double-immunogold Electron Microscopy (EM) in the hippocampus. Moreover, ADAR2-FMRP interaction appeared to be RNA independent.
Because changes in the editing pattern are associated with neuropsychiatric and neurodevelopmental disorders, we propose that the increased editing observed in the fmr1-KO mice might contribute to the FXS molecular phenotypes.
KEYWORDS: ADAR2, FMRP, Fragile X syndrome, RNA editing
Introduction
Fragile X syndrome (FXS) is one of the most common heritable form of intellectual impairment.1-5 Estimates report that FXS affects approximately 1 in 2,500–5,000 men and 1 in 4,000 – 6,000 women.6,7 The leading cause of the FXS is the expansion of the number of a polymorphic CGG triplet in the 5’ UTR of fmr1 gene, located on the X chromosome (Xq27.3). Through complex epigenetic modifications it results in hypermethylation of the chromosomal region and subsequent transcriptional silencing of fmr1 gene, preventing expression of Fragile-X- Mental Retardation Protein (FMRP).8-10
FMRP is a RNA binding protein that shuttles between the nucleus and the cytoplasm, where it forms ribonucleoparticles.11-13 FMRP regulates several characteristics of RNA metabolism like splicing, stability, subcellular transport and translation of mRNA encoding for proteins involved in synaptic structure and function.14-17 However, the best known function for FMRP is as translational repressor, since its absence in fmr1 KO mice leads to an increase of protein expression.18-23 Mouse models for FXS have revealed diverse phenotypes characterized by altered neuronal development and circuits formation (spine dysmorphogenesis represents the main marker of FXS) and by impairments in long-term synaptic plasticity underlying learning and memory.24,25
Recently, a new and unexpected nuclear function of FMRP has been reported, enhancing its role as post-transcriptional regulator.26 Bhogal and collaborators reported that in D. melanogaster, dFMRP physically and biochemically interacts with dADAR (Adenosine Deaminase Acting on RNA) enzymes, a class of RNA binding proteins that catalyzes the peculiar post transcriptional mechanism called RNA editing.27,28 RNA editing is a post-transcriptional hydrolytic deamination of an adenosine (A) to an inosine (I) which is read by the ribosomes as a guanosine (G). RNA editing might induce aminoacid substitutions contributing to a diversification of the information that is encoded by the genome.29 RNA editing in mammals has been mainly described for genes expressed in the Central Nervous System (CNS) and changes in editing patterns are frequently found associated with neuropsychiatric and neurodevelopment disorders.30-33 Three ADAR enzymes, ADAR1, ADAR2 and ADAR3 are expressed in mammals but only the first 2 are enzymatically active.28,34 The enzymes are able to recognize specific double stranded RNA structures generated by the hybridization of complementary exon and intron sequences in the pre-mRNA of specific transcripts, and catalyze the nucleotide deamination.28,34
The report of Bhogal and collaborators,26 for the first time, correlated the RNA editing pathway with FMRP (see also35). In Drosophila, dADAR and dFMRP interact in the nucleus, binding on several RNAs. dFMRP has been implicated in the modulation of the RNA editing levels of specific mRNAs encoding for proteins necessary for proper function of Drosophila Neuro-Muscular Junction (NMJ), including transcripts encoding a calcium channel (Caα1D), a potassium channel (Shab). Moreover, several ADAR target mRNAs have been shown to be associated with ADAR and FMRP in a messenger ribonucleoprotein complex.26
A recent study from Shamay-Ramot and collaborators,36 using the zebrafish model for FXS, the fmr1 -/- larvae, showed an interaction of ADAR2a with FMRP, also in vertebrates. Furthermore, a mild increase in RNA editing levels of mRNAs encoding neuronal and synaptic proteins like the Calcium Channel, Voltage-Dependent, L Type, α 1D Subunit (cacna1d), glutamate receptor ionotropic kainate type subunit 2 (grik2), glutamate receptor ionotropic AMPA type subunit 4 (gria4b), glutamate receptor ionotropic AMPA type subunit 3b (gria3b), was reported. These data in zebrafish further support a role of FMRP in inhibiting ADAR activity.
In this work, we wanted to elucidate the interplay between FMRP and ADAR enzymes in mammals, focusing mainly on ADAR2. We observed an increase in the level of RNA editing of specific neuronal transcripts in the cortex and hippocampus of fmr1 KO mice. Furthermore, using in vitro and ex vivo approaches we show that ADAR2 and FMRP physically interact in neuronal and non-neuronal cells in an RNA-independent manner. Consistent with the nuclear localization of ADARs, we could detect FMRP in the nuclei. These data reveal a possible novel nuclear function of mammalian FMRP in modulating ADAR enzymes. Because its absence leads to an increase of RNA editing, we suggest that an upregulation of RNA editing might contribute to the synaptic dysfunctions observed in FXS patients.
Results
Fmr1 KO mice show an increase in RNA editing of neuronal mRNAs
To determine whether FMRP might influence ADAR activity, the editing efficiency of different ADAR substrates was analyzed in frontal cortex (FC) and in the hippocampus (HC) of fmr1 KO mice, using a gene candidate approach. Neuronal mRNAs involved in synaptic plasticity and/or known to be affected in FXS has been analyzed. In particular the re-coding editing sites of AMPA receptors subunits GluA2, GluA3, GluA4, Kainate receptor subunits GluK1–2, 5-hydroxytryptamine receptor 2C (5-HT2c), GABA(A) receptor subunit α3 (GABRA3), Calcium Channel, Voltage-Dependent, L Type, α 1D Subunit (Cav1.3), Potassium Channel, Voltage Gated Shaker Related Subfamily A, Member 1 (KV1.1), Cytoplasmic FMR1-interacting protein 2 (CYFIP2), Calcium-Dependent Activator Protein For Secretion 1 (CAPS1), Neuro-Oncological Ventral Antigen 1 (NOVA-1), ADAR2, Bladder Cancer Associated Protein (BLCAP), Filamin-a (FLN-A) were analyzed. The AMPA receptor R/G editing sites were analyzed in combination with the splicing variants called flip and flop.
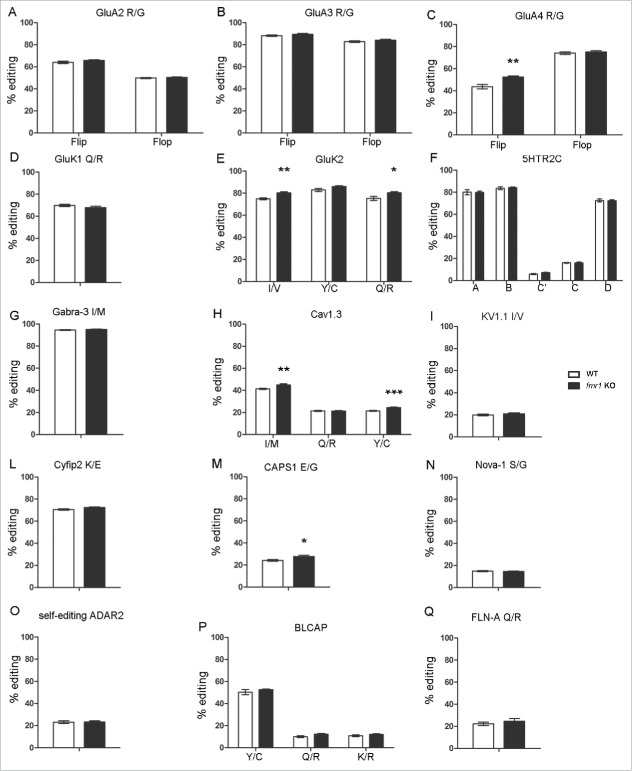
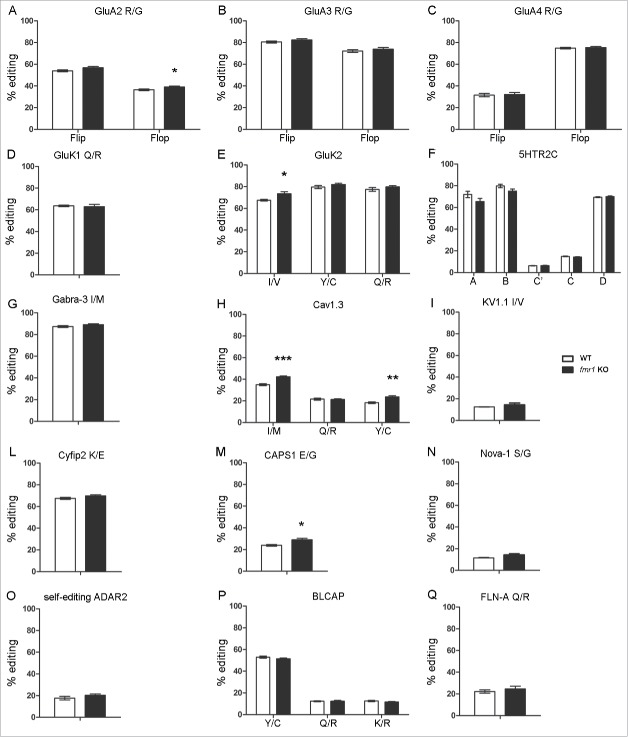
In FC a mild but statistically significant increase of RNA editing level in the fmr1 KO mice compared with the wild type was observed for: the GluA4 R/G site in the flip isoform (WT: 43.6 ± 1.99; KO: 52.4 ± 1.00, p < 0.01 Fig. 1C), the GluK2 I/V site (WT: 74.9 ± 0.88; KO: 80.3 ± 0.99, p < 0.01 Fig. 1E), the GluK2 Q/R site (WT: 75.3 ± 1.83; KO: 80.4 ± 0.93, p < 0.05 Fig. 1E), the Cav 1.3 I/M site (WT: 41.4 ± 0.54; KO: 45 ± 0.8, p < 0.01 Fig. 1H), the Cav 1.3 Y/C site (WT: 21.5 ± 0.4; KO: 24.5 ± 0.5, p < 0.001 Fig. 1H) and CAPS1 E/G (WT: 24.2 ± 0.79; KO: 27.7 ± 1.05, p < 0.05 Fig. 1M). In the HC an upregulation of RNA editing level for the GluA2 R/G site in the flop isoform (WT: 36.4 ± 0.78; KO: 39.0 ± 0.82, p < 0.05 Fig. 2A), the GluK2 I/V site (WT: 67.6 ± 0.80; KO: 73.5 ± 1.82, p < 0.05 Fig. 2E), the Cav 1.3 I/M site (WT: 35 ± 0.87; KO: 42.3 ± 0.79, p < 0.001 Fig. 2H), the Cav 1.3 Y/C site (WT: 18.4 ± 0.71; KO: 23.7 ± 1.16, p < 0.01 Fig. 2H) and CAPS1 E/G (WT: 23.3 ± 0.45; KO: 27.1 ± 1.30, p < 0.05 Fig. 2M) was observed in the fmr1 KO mice compared with control. Notably, while the level of editing is increased for GluK2 and Cav1.3 editing levels in both areas, a cortical specific upregulation was observed for GluA4 R/G and hippocampal upregulation for GluA2 R/G, suggesting a brain specific effect of FMRP. Because absence of FMRP in both in FC and HC leads to an increase of RNA editing, it is tempting to hypothesize that FMRP might function as an inhibitor of ADAR activity at specific editing sites. ADAR2, Bladder Cancer Associated Protein (BLCAP) and Filamin-A editing site (FLNA) were also analyzed, but no statistically significant variations were observed (Fig. 1O, P, Q and 2O, P, Q).
Figure 1.
RNA-editing levels in Frontal Cortex of wild type (white bars) and fmr1 KO (black bars) 3-weeks-old mice. The transcript and the edited site analyzed is reported above the graph. Data are presented as means ± SEM (n = 8). Statistical analysis was performed using unpaired t test with Welch's correction (*p < 0,05; **p <0,01; ***p < 0,001).
Figure 2.
RNA-editing levels in Hippocampus of wild type (white bars) and fmr1 KO (black bars) 3-weeks-old mice. The transcript and the edited site analyzed is reported above the graph. Data are presented as means ± SEM (n = 8). Statistical analysis was performed using unpaired t test with Welch's correction (*p < 0,05; **p <0,01; ***p < 0,001).
To further understand the modifications in RNA editing induced by the absence of FMRP, we focused our attention on Cav1.3 mRNA that showed statistically significant variations in 2 out of 3 editing sites both in HC and in FC regions. Combinations of the edited nucleotides might generate different protein isoforms.37 Interestingly, in HC a downregulation of the unedited IQDY isoforms could be detected with a parallel increase of the double edited MQDC isoforms (Supplemental Figure S1A); in FC the downregulation of IQDY isoforms was detected in parallel with the upregulation of the single edited MQDY and only partially of MQDC (Supplemental Figure S1B). Moreover, we tested also possible modulation in the frequency of 5-HTR2c edited isoform, although no difference were present in the single editing sites. Editing at the 5 5-HTR2c editing sites can generate up to 24 receptor isoforms, ranging from the completely unedited (INI) to the fully edited form (VGV). However, no statistically significant alteration were detected (Supplemental Table 1).
Although FMRP loss induced an upregulation of RNA editing level, this effect is not present on all editing site analyzed and it is slightly area specific. Next, we tested if FMRP absence might modulate the expression level of ADAR1 or ADAR2 mRNAs (Supplemental Figure S2). No statistical significant expression variations were determined for either enzymes, although a trend for increase (up to 20%) could be detected for both enzymes in fmr1 KO mice, both in FC and in HC. We then analyzed the expression pattern of several editing stimulatory factors such as Split Hand/Foot Malformation 1 (SHFM1), and the RNA binding protein hnRNP A2/B1 (hnRNPA2/B1), as well as inhibitory factors such as ribosomal protein S14 (RPS14) and Serine/Arginine-Rich Splicing Factor 9 (SRSF9) (Supplemental Figure S3) which can modulate ADAR activity.38,39 Only the inhibitory factor RPS14 mRNA showed a statistically significant decrease (Supplemental Figures S3A), only in the HC of the fmr1 KO mice. These data might suggest that FMRP loss might influence other members involved in the regulation of RNA editing reaction.
We then tested if a difference in the expression level of FMRP and ADAR2 in HC and FC brain areas might correlate with the specific RNA editing modulation; however no clear variations in the mRNA expression level were reported. (Supplemental Figure S4).
FMRP inhibits ADAR2 activity
To explore a possible role of FMRP in inhibiting ADAR activity, we performed an in vitro editing assay (Tariq et al., 2013) in HEK293T cells stably expressing ADAR2 and transiently expressing FMRP and an editable target. The target is generated by a vector (RNAG) expressing RFP and GFP proteins separated by an editable stop codon (Fig. 3A, see methods for details). Upon editing, the stop codon is converted to a tryptophan codon, allowing the expression of GFP. The ratio between GFP and RFP fluorescence indicated the level of editing (Fig. 3B). HEK293T cells transiently transfected only with the construct RNAG showed a GFP/RFP ratio of 0.06 ± 0.019; this is consistent with the low levels of editing previously observed in this cell line.40 HEK293T cells expressing stably ADAR2 (HEK293T/ADAR2) with showed an increase GFP/RFP ratio (0.32 ± 0.019, p < 0.001). In HEK293T/ADAR2 cells expressing exogenous FMRP, a downregulation of the levels of editing was observed, as shown by a reduction of the GFP/RFP ratio (0.24 ± 0.002, p <0.001 vs only RNAG transfected cells, p <0.01 vs HEK293T/ADAR2 cells). This result supports a role for FMRP as inhibitor of ADAR2 activity.
Figure 3.
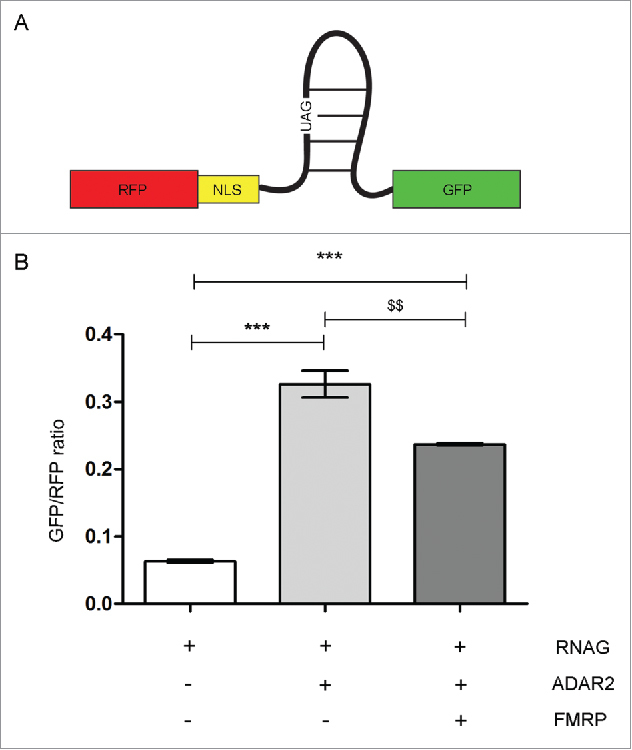
Editing Assay in HEK293T cells. (A) Scheme of a editing reporter construct (RNAG) constitutively expressing RFP and GFP only after editing of an amber-stop codon to a tryptophan codon (UAG->UGG). (B) FACS analysis of transfected HEK293T. Increase of the green to red fluorescence ratio indicates increase in editing level. All cells are transfected with the reported construct RNAG. White bar: HEK293T cells transfected with RNAG vector alone; Gray bar: HEK293T cells transfected with RNAG and ADAR2 vectors; Dark gray bar: HEK293T cells transfected with RNAG, ADAR2 and FMRP vectors. Data are presented as means ± SEM. Statistical analysis was performed using one-way ANOVA (***p <0,001; $$p < 0,01). NLS: nuclear localization signal.
FMRP interacts directly with ADAR2
ADARs are mainly nuclear proteins28 while FMRP localization has been largely described to be cytoplasmic.3,41 To test a possible physical interaction between FMRP and ADAR, we first studied the cytoplasmic and nuclear distribution of FMRP. As reported in Fig. 4, FMRP is present in both cytoplasmic (high levels) and nuclear (low levels) fractions. The enrichment of the 2 fractions is shown by the presence of the RNA binding protein Staufen mainly in the cytoplasmic fraction and Dyskerin and Histone H1 in the nuclear fraction.
Figure 4.
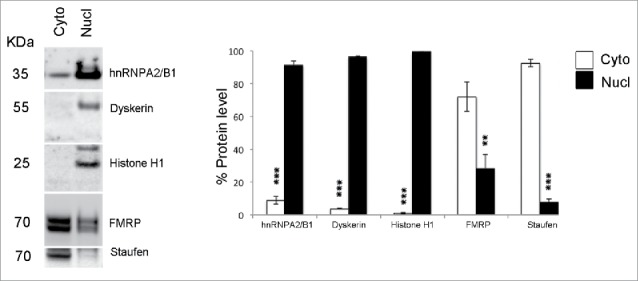
Cytoplasm/nucleus fractionation of FMRP. Cytoplasmic and nuclear fractions were prepared from the P21 mouse cortex. (Left) Western blotting showing the distribution of FMRP in cytoplasmic (Cyto) and nuclear (Nucl) fractions. hnRNPA2/B1, Dyskerin, histone H1 proteins (markers of nucleus) are present only or mainly in nuclear fraction, while the cytoplasmic protein Staufen is present in the cytoplasmic fraction. (Right) The histogram shows the quantification of 4 independent experiments. FMRP is present in both cytoplasmic (high levels) and nuclear (low levels) fractions. Data are presented as means ± SEM. Statistical analysis was performed using Student's t test (**p <0,01; ***p < 0,001).
The ADAR2-FMRP co-localization was at first analyzed ex vivo with double-immunogold Electron Microscopy (EM) (Fig. 5 and Supplemental Figure 5) in hippocampal sections. The co-localization of ADAR2 and FMRP occurs mainly in the nucleus (red boxes in panels A and C and red arrows in panels B and D) and only a few FMRP-ADAR2 dots were detected in the cytoplasm, where the majority of FMRP (smaller dots) is localized.
Figure 5.
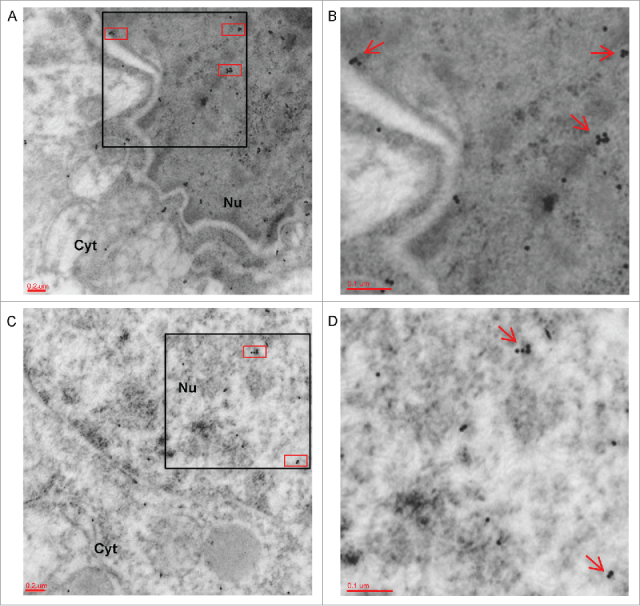
Co-localization of FMRP and ADAR2 in hippocampal sections by electron microscopy. (A-C) Two representative electronic micrographics of a double-immunogold for FMRP (labeled by 10 nm gold particles) and ADAR2 (labeled by 15 nm gold particles) performed on hippocampal ultra-thin sections of adult male mice. The red boxes highlight the co-localization of FMRP and ADAR2 proteins inside the nucleus (Nu). Magnification: 20000 x (B) 30000 x magnification of black box showed in panel A. Red arrows show FMRP/ADAR2 co-localization. (D) 30000 x magnification of black box in panel C. Red arrows show FMRP/ADAR2 co-localization. Nu: Nucleus; Cyt: Citoplasm.
Moreover, the endogenous ADAR2-FMRP interaction was analyzed in primary neurons at DIV 14 using the Proximity Ligation Assay (PLA)42 (Fig. 6). Several dots that represent the interaction of FMRP with ADAR2 were observed mainly in the nuclei (Fig. 6A-B). The PLA for a known FMRP interacting protein FXR1P,43 showed several dots mainly in the cytoplasm (Fig. 6C); without primary ADAR2 antibody processed samples didn't show any dot (Fig. 6D).
Figure 6.
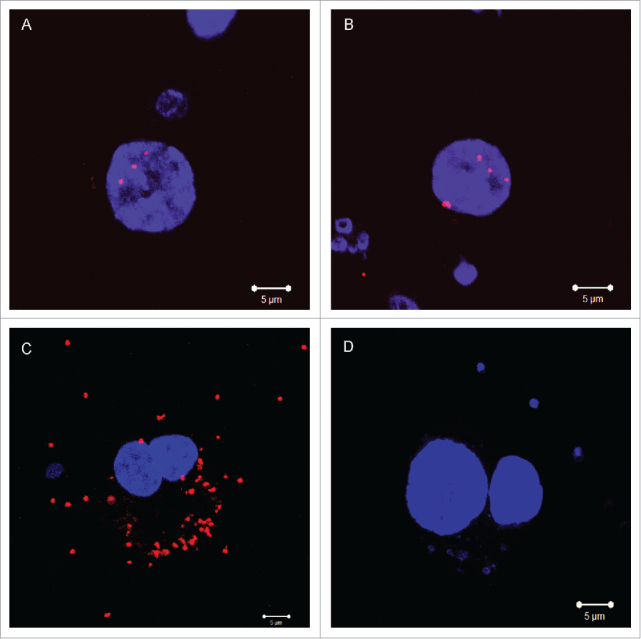
Detection of endogenous ADAR2 and FMRP interaction by Proximity Ligation Assay (PLA) in mouse primary cortical neuron cultures. (A-B) PLA dots for ADAR2 and FMRP show manly a nuclear localization. (C) PLA dots for FMRP and FXR1P interaction are in the cytoplasm; (D) Without-primary-ADAR2 antibody processed samples do not show any dot in PLA experiment. Scale bars 5µm.
These findings were further confirmed in HEK293T non-neuronal cells expressing stably ADAR2-HA and upon the transient exogenous expression of FMRP-Myc. PLA fluorescent signals, using primary antibody against HA and Myc, were detected mainly in the nuclei (Supplemental Figure 6A-B) indicating FMRP-ADAR interaction. A few rare dots were also observed in the cytoplasm possibly due to the overexpression of both proteins. On the other hand, no signals were detected in not-transfected HEK293T cells (Supplemental Figure 6C-D) or in HEK293T/ADAR2 cells overexpressing α-synuclein (Supplemental Figure 6E-F), that does not interact with ADAR2.
To further corroborate the interaction of ADAR2 and FMRP, we performed co-immunoprecipitation experiments from cortical tissues of WT and fmr1 KO mice (Fig. 7). FMRP can be clearly detected in ADAR2 immunoprecipitates (Fig. 7 A) in WT but not in fmr1 KO extracts, confirming the specificity of the interaction. Moreover, the result was not altered by treatment with RNaseA showing that ADAR2 and FMRP interact in a RNA independent manner (Fig. 7 B).
Figure 7.
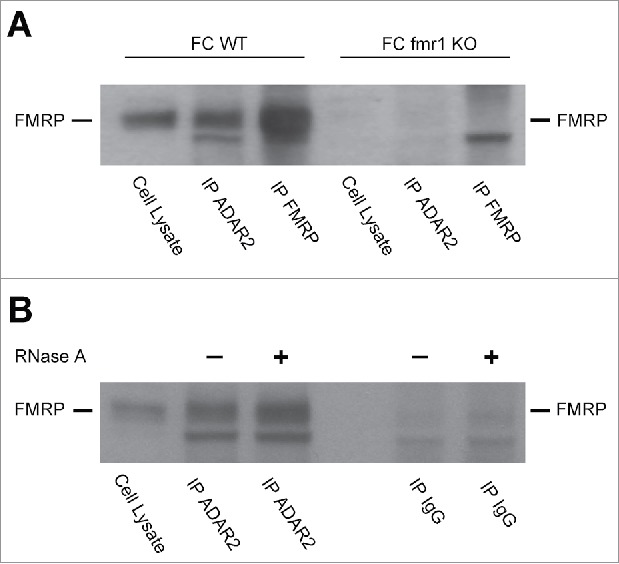
ADAR2-FMRP interaction determined by co-immunoprecipitation experiments. (A) FMRP western blot on frontal cortex total cell lysate prior and after immunoprecipitation with ADAR2 and FMRP antibodies. Both WT and fmr1 KO murine FC were analyzed. (B) ADAR-FMRP interaction is RNA independent. FMRP western blot of frontal cortex total cell lysate from WT mice prior and after immunoprecipitation with ADAR2 and rabbit IgG antibodies treated or not with RNase A.
Discussion
Here we report, for the first time in mammals, that the absence of FMRP increases ADAR enzymes activity both in FC and HC, as measured by enhanced editing level of specific sites. These changes, although of limited magnitude, might be of interest since the edited mRNAs encode for proteins involved in the synaptic structure and function.
Overexpression or downregulation of Drosophila FMRP has been shown to change the editing efficiency of specific dADAR targets involved in synaptic transmission.26 Moreover, zebrafish FMRP was shown to regulate RNA editing, synaptic density and locomotor activity in zebrafish.36 If confirmed also in mammals, these data will clearly indicate the existence of a new and evolutionarily conserved FMRP activity that might be dysregulated in FXS pathogenesis.
We investigated the ADAR-FMRP interaction in mammals and we showed that loss of FMRP leads to a mild increase of ADAR activity both in FC and HC of fmr1 KO mice, resulting in increased editing level. Although the changes in editing are moderate, these could result in considerable functional consequences.32,44,45 In particular, the increased editing levels of the R/G site for GluA2 in flop isoform in HC and for GluA4 in flip isoform in FC, imply a faster recovery rate from desensitization compared with the wild type mice.46 Thus, the increased editing in these sites could enhance synaptic strength.46 Moreover, we found increased editing levels for the I/V site of GluK2 in both tissues. This editing site is located in the first transmembrane domain of the subunit and the editing process seems to be involved in a finely tuned regulation of ion permeability of the channel,47 together with the modulation of editing levels for GluK2 Q/R site, which is increased in both FC of KO mice. Of note, the zebrafish fmr1 -/- shows changes in editing levels of glutamate receptor transcripts, leading to altered synaptic strength and morphology and ultimately defects in locomotor activity. Taken together, these data indicate that loss of mammalian FMRP might alter glutamate receptor excitability via modification of RNA editing efficacy.
Furthermore, we detected increased editing levels for Cav 1.3 subunit, in the I/M and Y/C editing sites both in FC and HC. The editing sites are all ADAR2 specific and they are located in the so called IQ domain of the subunit, a calmodulin-binding site mediating inhibitory Ca2+-feedback (CDI) on channels. The increase of editing levels in these 2 sites leads to downregulation of the IQDY isoform expression, with a parallel increase of the double edited MQDC isoform in HC and of both the single edited MQDY and MQDC edited isoforms in FC. Edited channels exhibit a strong reduction of the CDI,37 leading to a prolonged activation of the channel itself and increased load of calcium. This feature might be linked to the imbalance of the excitatory and inhibitory synaptic pathways described in patients with FXS.3
In addition, E/G editing level of CAPS1 was found to be increased both in FC and HC; the encoded protein is a cytosolic and peripheral membrane protein required for Ca2+ regulated exocytosis of dense-core vesicles carrying neurotransmitters and neuropeptides.48 A recent paper49 for the first time, investigated the functional role of RNA editing at this site, generating mutant mice expressing solely the edited CAPS1. The mutant mice were leaner and exhibited increased physical activity due to augmented neurotransmitters release, most probably dopamine.49 It is intriguing to speculate that FXS patient hyperactivity behavior might be in part due to increased RNA editing at CAPS1 site. Moreover a recent paper50 demonstrates a reduced BDNF release following exocytosis in dendrites of CAPS1-deficient neurons; accordingly, it is possible to speculate that dysregulation in CAPS1 editing could also be linked to the neuronal spine dysmorphogenesis found on postmortem brains from patients lacking FMRP.51
Our data indicate a role of FMRP in the inhibition of the editing process, a mechanism that participate in the physiologic modulation of synapses in the CNS. The way in which ADAR enzymes and FMRP might interact in modulating RNA editing process is not known in mammals. dFMRP and dADAR interact in the nucleus possibly to regulate RNA editing, and dADAR acts downstream FMRP to modulate synaptic morphology of Drosophila neuromuscular junction.26 Similarly, in zebrafish ADAR and FMRP seem to biochemically interact as shown by co-immunoprecipitation experiments, but it is not known if this interaction occurs in the nucleus or in the cytoplasm.36
Using in vitro PLA experiments and double-immunogold Electron Microscopy in brain slices we showed that mammalian FMRP and ADAR2 interact in the nucleus. Moreover, a functional interaction has resulted from the in vitro RNA Editing Assay, confirming an inhibiting role of FMRP on ADAR2 activity. In addition, co-immunoprecipitation experiment and RNase treatment, showed that ADAR2 and FMRP interact in an RNA-independent manner.
It is tempting to hypothesize that FMRP might inhibit the recognition of specific RNA secondary structures by ADAR. Furthermore, ADAR2 mRNA has been reported as a FMRP target14,52 indicating that FMRP might modulate also ADAR activity by regulating its RNA metabolism; however, the global mRNA expression level of ADARs enzymes in fmr1 KO mice is not altered.
Because ADAR2-FMRP interaction is RNA-independent, RNA editing inhibition might occur through protein-protein interaction.
We speculate that the ADAR2- FMRP complex might be assembled on specific editing sites repressing the editing reaction or that FMRP might sequester ADAR2 inhibiting its capability to bind and edit target RNAs, as already proposed for other ADAR co-factors.39 Further studies are needed to address these possibilities.
ADAR enzymes have a main role in the maintenance of the central nervous system homeostasis, given that their absence results in severe neurologic defects or it could be lethal, as shown by animal models.53,54 Although the changes in RNA editing reported in the absence of FMRP in Drosophila, zebrafish and, in the present work, in the mouse are moderated, they occur in evolutionally conserved transcripts that are stringently regulated. Accordingly, only mild alterations in RNA editing level have been reported in patients suffering of neurologic disorders such as ALS, epilepsy, schizophrenia and depression.55
In conclusion, our data support the presence of a nuclear function of FMRP in the regulation of RNA editing and suggest that a dysregulation of this mechanism might contribute to the FXS pathogenesis.
Materials and methods
Animal care
Animal care was conducted conforming to the institutional guidelines that are in compliance with Italian national (DL N116, GU, suppl 40, 18–2–1992) and international laws and policies (European Community Council Directive 86/609, Oja L 358, 1, December 12, 1987; National Institutes of Health Guide for the Care and Use of Laboratory Animals, US National Research Council, 1996). The fmr1-FBV KO and FVB wild type mice were used.
RNA extraction and retro-transcription reaction
Total RNA was extracted from frontal cortex (FC) and hippocampus (HC) of P21 mice (n = 8 per group) using the TRIzol® Reagent (Thermo Fisher Scientific), according to the manufacturer instructions. Reverse transcription was performed using Moloney Murine Leukemia Virus-Reverse Transcriptase (MMLV-RT) (Thermo Fisher Scientific). Briefly, 2 µg of total RNA was mixed with 2.2 µl of 0.2 ng/µl random hexamer (Thermo Fisher Scientific), 10 µl of 5 × buffer (Thermo Fisher Scientific), 10 µl of 2 mM dNTPs, 1 µl of 1 mM DTT (Thermo Fisher Scientific), 0.4 µl of 33 U/µl RNasin (Promega, Madison, WI, USA) and 2 µl MMLV-RT (200 u/µl) in a final volume of 50 µl. The reaction mixture was incubated at 37°C for 2 h, and then the enzyme was inactivated at 75°C for 10 min.
RNA editing quantification
The levels of editing in re-coding sites of AMPA receptors subunits GluA2, GluA3, GluA4, Kainate receptor subunits GluK1–2, 5-hydroxytryptamine receptor 2C (5-HTR2c), GABA(A) receptor subunit α3 (GABRA3), Calcium Channel, Voltage-Dependent, L Type, α 1D Subunit (Cav1.3), Potassium Channel, Voltage Gated Shaker Related Subfamily A, Member 1 (KV1.1), Cytoplasmic FMR1-interacting protein 2 (CYFIP2), Calcium-Dependent Activator Protein For Secretion 1 (CAPS1), Neuro-Oncological Ventral Antigen 1 (NOVA-1), ADAR2 self editing site, Bladder Cancer Associated Protein (BLCAP), Filamin-a (FLN-A) were analyzed. The quantification was performed by sequence analysis as described previously56 using Discovery Studio (DS) Gene 1.5 (Accelrys Inc., San Diego, CA, USA).
In vitro RNA editing assay
For RNA editing assay a vector expressing RFP and GFP proteins separated by an editable stop codon, called RNAG, was used (gift from Prof. Jantsch). Briefly, the stem-loop containing the R/G editing site of glutamate receptor subunit B was modified to contain an amber stop codon at the editing site.39,38 The substrate stem-loop was cloned between the red fluorescent protein (RFP) and green fluorescent protein (GFP) ORFs. The transient transfection of this construct induces the constitutive expression of RFP. The stop codon in the loop prevents GFP translation without editing process; otherwise, the increase of editing levels leads to a conversion of the stop codon to a tryptophan codon inducing the production of GFP. The ratio between the GFP and RFP fluorescence indicates the editing levels in the cell population: if the editing activity increases, the GFP expression increases as well. After 24h from the transfection of the HEK293T cell lines with the RFP/GFP vector, the samples were collected and maintained in PBS EDTA 2 mM until the FACS analysis was performed.
The samples were read on a MACSQuant flow cytometer (Miltenyi Biotec) and analyzed with FlowJo (Tree Star Inc., Ashland, USA). Editing efficiencies were determined by calculating the ratio of green to red arithmetic mean fluorescence of cells with solid RFP expression as described previously.57 At least 10000 events were collected for each sample. Statistical analysis has been performed on triplicate experiments.
Nuclear and cytoplasmic fractionation
Cortices from P21 mice were resuspended in fractionation buffer (75 mg in 600 µl). Cytoplasmic and nuclear fractions were prepared using the Paris kit (Ambion).
Western blotting
For Western blot analysis standard methodologies were used. Protein samples were separated by SDS–PAGE electrophoresis and blotted on a PVDF membrane (Millipore). Membranes were incubated using specific antibodies: mouse anti-hnRNP A2B1 antibody (1:1000 Abcam), Dyskerin (1:1000, kindly provided by Yves Henry, University Paul Sabatier Toulouse), Mouse Anti-Histone H1 (1:1000 SIGMA), Rabbit anti-Staufen (1:1000 Abcam), rabbit anti-FMRP (1:1000, Ferrari et al. 2007).
HRP-conjugated anti-rabbit, anti-mouse antibodies (1:10000) were purchased from Promega or Chemicon.Proteins were revealed using an enhanced chemiluminescence kit (GE Healthcare) and the imaging system LAS-4000 mini (GE).Quantification was performed using the IQ ImageQuant TL software (GE Healthcare). The amount of analyzed proteins was normalized by Coomassie blue staining.
Electron microscopy
Transverse hippocampal sections from 3-month-old mice were fixed 3 hours at 4°C in a mixture of 2% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.6, dehydrated in alcohol at progressively higher concentrations and embedded in Bioacryl resin (British Biocell, Cardiff, United Kingdom), followed by UV polymerization, according to standard procedures.58 Consecutive thin and ultrathin sections were cut using a diamond knife (Diatome, Biel, Switzerland) on a Reichert ultramicrotome (Depew, NY, USA). Ultrathin sections (80–90 nm) were collected on 300 mesh nickel grids. To block non-specific binding sites, these grids were treated with a blocking buffer made of phosphate buffer saline supplemented with 0.1% Tween-20, 0.1% bovine serum albumin and 4% normal goat serum and incubated overnight at 4°C with a rabbit polyclonal anti-FMRP59 and a rabbit anti-ADAR2 (Abcam; cod. ab64830) primary antibodies. The grids were then incubated for 1 hour with goat anti-rabbit IgG conjugated with 10 and 15 nm colloidal-gold particles (British Biocell, Cardiff, United Kingdom), counterstained for 5 min in 4% uranyl acetate (in 70% ethanol) to evidentiate the cell morphology, and observed with EM 109 Zeiss (Oberkochen, Germany). A without-primary-antibody negative control was processed in parallel. Photographs were taken with GATAN Orius SC200 TEM CCD camera.
Primary neuronal and HEK293T cell cultures
Mouse primary cortical cultures were prepared as described previously.60 Briefly, mouse cerebral cortices from day 13.5 mouse embryos were mechanically dissociated in cold HBSS containing 10mM HEPES (Invitrogen); the cell suspension was re-suspended in serum-free Neurobasal medium (Invitrogen) supplemented with B-27 (Invitrogen), 30 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA), 30 µg/ml streptomycin (Sigma-Aldrich) and 0.5 mM Glutamax (Invitrogen). The neurons were then plated at a density of 30,000 cells/cm2 on a poly-D-lysine coating (Sigma-Aldrich) in multi-well plates. Three days after plating, 50% of the medium was replaced with fresh medium; subsequently, half of the medium was replaced once a week for a maximum of 4 weeks.
Stable HEK293T expressing HA tagged ADAR2 enzyme, generated after viral infection with modified pRRLSIN.cPPT.PGK-GFP.WPRE vector (addgene: 12252), were obtained after cloning selection of HA-positive colonies.
HEK293T cell lines were cultured at 37°C and 5% CO2 in DMEM medium (Invitrogen) supplemented with 10% of heat-inactivated fetal bovine serum (FBS), 30 U/ml penicillin (Sigma-Aldrich), 30 µg/ml streptomycin (Sigma-Aldrich), 1% minimum Eagle's medium nonessential amino acids, 1 mM sodium pyruvate.
Transient Transfection
HEK293T cells are plated 24h before transfection at a density of 30’000 cell/cm2 in 6-well plates; the medium was changed 2h before transfection. 2.5µg of the plasmid of interest (pcDNA3.1-FMR1-myc-his43; pcDNA3.1-α-synuclein61; RNAG39) were mixed with 0.1 × TrisEDTA (TE 0.1 ×)/dH2O (2:1) and 2.5M CaCl2; the mixture is maintained 5 min at RT. The precipitate is formed by adding dropwise 2 × HBS solution to the mixture, then the suspension should be added immediately to the cells. The calcium-phosphate plasmid DNA mixture should be allowed to stay on the cells for 14–16h, after which the medium should be replaced with fresh medium.
Proximity Ligation Assay (PLA)
The Duo-link® using PLA Technology® kit (Sigma-Aldrich) was used for the proximity ligation assay, accordingly to the manufacturer instructions with minor modifications. Briefly, the cells were fixed with paraformaldehyde 4% (PFA); in particular neurons were fixed at DIV14. Each sample was permeabilized with PBS-Triton 0.3% and then incubated with the blocking solution (Roche™) for about 45 min at room temperature; the primary antibodies incubation was performed overnight at 4°C with mouse anti-FMRP (Merck Millipore cod. MAB2160), rabbit anti-FXR1P (Abcam; cod. ab129089) and rabbit anti-ADAR2 (Abcam; cod. ab64830) for endogenous PLA experiments in neuronal cultures; rabbit anti-HA (SIGMA; cod. H6908) and mouse c-Myc (Santa Cruz Biotechnology; cod. SC40), recognizing the HA tag for ADAR2 and the Myc tag for FMRP respectively, mouse anti α-synuclein (Santa Cruz cod. sc-12767), for exogenous PLA experiments on transfected HEK293T cells (Supplemental results). On the following day, the samples were washed 3 times in PBS at room temperature and then the cells were incubated 1h at 37°C with the PLA probe containing the secondary antibodies conjugated with the DNA probes. After PLA probe removal, the samples were washed 4 times x 10’ with the Buffer A at 37°C. After a brief wash with Buffer A at 37°C, the samples were incubated with the ligation buffer containing oligonucleotides that hybridize to the PLA probe and the DNA ligase which allows the annealing between probe and oligonucleotides to form a rolling circle DNA strand. This reaction was incubated for 30 min at 37°C. Subsequently the cells were washed with Buffer A at 37°C and then incubated with the amplification-detection solution containing the DNA polymerase for rolling circle amplification (100 min at 37°C). Next, the samples were washed 4 times with Buffer B at room temperature; then the coverslips were incubated for 10 min with mounting buffer containing DAPI and analyzed with a confocal microscope.
Co-immunoprecipitation experiments
FC tissues for WT and fmr1 KO mice at P21 were lysed by sonication in immunoprecipitation buffer (Tris-HCl 50 mM pH 7.4, NaCl 300 mM, 1% Triton X-100, Protease inhibitor Roche® 1x). The extracts were added to 60 µl of Protein G Dynabeads™ (10007D Invitrogen ® by Thermo Fisher Scientific) coupled with 5µg of rabbit anti-FMRP (Abcam ab17722) or rabbit anti-ADAR2 (Abcam ab64830). After 2h of incubation at 4°C on a rotating wheel, 5 washes with immunoprecipitation buffer were performed. The elution step was performed with 50 µl of sample buffer 2x and DTT 10x; then, the samples were denatured at 75°C for 10 min for Western Blot procedure.
This step was performed using rabbit anti FMRP (Abcam ab17722) 1:500 in 5% non-fat dry milk in TBST 0.2%; the Alkaline Phosphatase (AP)- conjugated anti-rabbit secondary antibody was used 1:10000 in TBST 0.2% (Promega cod. S373B); for both the antibodies the incubation was performed 1h at RT. The RNase A treatment was performed by adding to the extracts 340 µg of enzyme in each sample. Then the above mentioned procedure for the co-IP was followed.
Statistical analysis
Statistical analysis of the editing data was performed using unpaired t test with Welch's correction while FACS data were analyzed by one-way ANOVA followed by Bonferroni's post-test. Graph pad software was use to performed the analysis and create the graphs
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by MIUR (PRIN 2012 prot.2012A9T2S9_004) to AB and Fondazione Cariplo, Associazione Italiana Sindrome X Fragile, Telethon GGP15257, Lejeune Foundation to CB. We thanks Prof. Keith Murai for the gift of FXR1P vectors; Dr. Arianna Bellucci for the gift of α-synuclein vector; Prof. Michel Jantsch for the gift of RNAG vector; Prof. Rosa Alba Rana for the support with EM experiments.
References
- 1.Martin JP, Bell J. A pedigree of mental defect showing sex-linkage. J Neurol Psychiatry 1943; 6:154-7; PMID:21611430; https://doi.org/ 10.1136/jnnp.6.3-4.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci 2002; 25:315-38; PMID:12052912; https://doi.org/ 10.1146/annurev.neuro.25.112701.142909 [DOI] [PubMed] [Google Scholar]
- 3.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J Clin Invest 2012; 122:4314-22; PMID:23202739; https://doi.org/ 10.1172/JCI63141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook D, Nuro E, Murai KK. Increasing our understanding of human cognition through the study of Fragile X Syndrome. Dev Neurobiol 2014; 74:147-77; PMID:23723176; https://doi.org/ 10.1002/dneu.22096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurin T, Zongaro S, Bardoni B. Fragile X Syndrome: From molecular pathology to therapy. Neurosci Biobehav Rev 2014; 46:Pt 2:242-55; PMID:24462888; https://doi.org/ 10.1016/j.neubiorev.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 6.Coffee B, Keith K, Albizua I, Malone T, Mowrey J, Sherman SL, Warren ST. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet 2009; 85:503-14; PMID:19804849; https://doi.org/ 10.1016/j.ajhg.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraan CM, Hocking DR, Bradshaw JL, Fielding J, Cohen J, Georgiou-Karistianis N, Cornish KM. Neurobehavioural evidence for the involvement of the FMR1 gene in female carriers of fragile X syndrome. Neurosci Biobehav Rev 2013; 37:522-47; PMID:23352653; https://doi.org/ 10.1016/j.neubiorev.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 8.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 1991; 66:817-22; PMID:1878973; https://doi.org/ 10.1016/0092-8674(91)90125-I [DOI] [PubMed] [Google Scholar]
- 9.Pietrobono R, Tabolacci E, Zalfa F, Zito I, Terracciano A, Moscato U, Bagni C, Oostra B, Chiurazzi P, Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum Mol Genet 2005; 14:267-77; PMID:15563507; https://doi.org/ 10.1093/hmg/ddi024 [DOI] [PubMed] [Google Scholar]
- 10.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet 1992; 1:397-400; PMID:1301913; https://doi.org/ 10.1093/hmg/1.6.397 [DOI] [PubMed] [Google Scholar]
- 11.Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet 1996; 5:1083-91; PMID:8842725; https://doi.org/ 10.1093/hmg/5.8.1083 [DOI] [PubMed] [Google Scholar]
- 12.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008; 60:201-14; PMID:18957214; https://doi.org/ 10.1016/j.neuron.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N. RNA granules. J Cell Biol 2006; 172:803-8; PMID:16520386; https://doi.org/ 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al.. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011; 146:247-61; PMID:21784246; https://doi.org/ 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al.. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001; 107:477-87; PMID:11719188; https://doi.org/ 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- 16.Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 2003; 37:417-31; PMID:12575950; https://doi.org/ 10.1016/S0896-6273(03)00034-5 [DOI] [PubMed] [Google Scholar]
- 17.Zhou LT, Ye SH, Yang HX, Zhou YT, Zhao QH, Sun WW, Gao MM, Yi YH, Long YS. A novel role of fragile X mental retardation protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience 2017; 349:64-75; PMID:28257890; https://doi.org/ 10.1016/j.neuroscience.2017.02.044 [DOI] [PubMed] [Google Scholar]
- 18.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al.. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008; 134:1042-54; PMID:18805096; https://doi.org/ 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- 19.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 2003; 112:317-27; PMID:12581522; https://doi.org/ 10.1016/S0092-8674(03)00079-5 [DOI] [PubMed] [Google Scholar]
- 20.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci 2007; 27:5338-48; PMID:17507556; https://doi.org/ 10.1523/JNEUROSCI.0937-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem 2008; 283:18478-82; PMID:18474609; https://doi.org/ 10.1074/jbc.C800055200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol 2007; 5:e52; PMID:17298186; https://doi.org/ 10.1371/journal.pbio.0050052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacoux C, Di Marino D, Boyl PP, Zalfa F, Yan B, Ciotti MT, Falconi M, Urlaub H, Achsel T, Mougin A, et al.. BC1-FMRP interaction is modulated by 2'-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res 2012; 40:4086-96; PMID:22238374; https://doi.org/ 10.1093/nar/gkr1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centonze D, Rossi S, Mercaldo V, Napoli I, Ciotti MT, De Chiara V, Musella A, Prosperetti C, Calabresi P, Bernardi G, et al.. Abnormal striatal GABA transmission in the mouse model for the fragile X syndrome. Biol Psychiatry 2008; 63:963-73; PMID:18028882; https://doi.org/ 10.1016/j.biopsych.2007.09.008 [DOI] [PubMed] [Google Scholar]
- 25.Gatto CL, Broadie K. Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front Synaptic Neurosci 2010; 2:4; PMID:21423490; https://doi.org/ 10.3389/fnsyn.2010.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhogal B, Jepson JE, Savva YA, Pepper AS, Reenan RA, Jongens TA. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat Neurosci 2011; 14:1517-24; PMID:22037499; https://doi.org/ 10.1038/nn.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol 2012; 13:252; PMID:23273215; https://doi.org/ 10.1186/gb-2012-13-12-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlandi C, Barbon A, Barlati S. Activity regulation of adenosine deaminases acting on RNA (ADARs). Mol Neurobiol 2012; 45:61-75; PMID:22113393; https://doi.org/ 10.1007/s12035-011-8220-2 [DOI] [PubMed] [Google Scholar]
- 29.Rosenthal JJ, Seeburg PH. A-to-I RNA editing: Effects on proteins key to neural excitability. Neuron 2012; 74:432-9; PMID:22578495; https://doi.org/ 10.1016/j.neuron.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eran A, Li JB, Vatalaro K, McCarthy J, Rahimov F, Collins C, Markianos K, Margulies DM, Brown EN, Calvo SE, et al.. Comparative RNA editing in autistic and neurotypical cerebella. Mol Psychiatry 2013; 18:1041-8; PMID:22869036; https://doi.org/ 10.1038/mp.2012.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Urban DJ, Blashka J, McPheeters MT, Kroeze WK, Mieczkowski P, Overholser JC, Jurjus GJ, Dieter L, Mahajan GJ, et al.. Quantitative analysis of focused a-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS One 2012; 7:e43227; PMID:22912834; https://doi.org/ 10.1371/journal.pone.0043227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbon A, Barlati S. Glutamate receptor RNA editing in health and disease. Biochemistry (Mosc) 2011; 76:882-9; PMID:22022961; https://doi.org/ 10.1134/S0006297911080037 [DOI] [PubMed] [Google Scholar]
- 33.Silberberg G, Lundin D, Navon R, Ohman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum Mol Genet 2012; 21:311-21; PMID:21984433; https://doi.org/ 10.1093/hmg/ddr461 [DOI] [PubMed] [Google Scholar]
- 34.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry 2010; 79:321-49; PMID:20192758; https://doi.org/ 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassell GJ. Fragile balance: RNA editing tunes the synapse. Nat Neurosci 2011; 14:1492-4; PMID:22119945; https://doi.org/ 10.1038/nn.2982 [DOI] [PubMed] [Google Scholar]
- 36.Shamay-Ramot A, Khermesh K, Porath HT, Barak M, Pinto Y, Wachtel C, Zilberberg A, Lerer-Goldshtein T, Efroni S, Levanon EY, et al.. Fmrp interacts with Adar and regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet 2015; 11:e1005702; PMID:26637167; https://doi.org/ 10.1371/journal.pgen.1005702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Tan BZ, Shen Y, Tao J, Jiang F, Sung YY, Ng CK, Raida M, Kohr G, Higuchi M, et al.. RNA editing of the IQ domain in Ca(v)1.3 channels modulates their Ca(2)(+)-dependent inactivation. Neuron 2012; 73:304-16; PMID:22284185; https://doi.org/ 10.1016/j.neuron.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA biology 2013; 10:192-204; PMID:23353575; https://doi.org/ 10.4161/rna.23208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tariq A, Garncarz W, Handl C, Balik A, Pusch O, Jantsch MF. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res 2013; 41:2581-93; PMID:23275536; https://doi.org/ 10.1093/nar/gks1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature 1996; 379:460-4; PMID:8559253; https://doi.org/ 10.1038/379460a0 [DOI] [PubMed] [Google Scholar]
- 41.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci 2015; 16:595-605; PMID:26350240; https://doi.org/ 10.1038/nrn4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al.. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods 2006; 3:995-1000; PMID:17072308; https://doi.org/ 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 43.Cook D, Nuro E, Jones EV, Altimimi HF, Farmer WT, Gandin V, Hanna E, Zong R, Barbon A, Nelson DL, et al.. FXR1P limits long-term memory, long-lasting synaptic potentiation, and de novo GluA2 translation. Cell reports 2014; 9:1402-16; PMID:25456134; https://doi.org/ 10.1016/j.celrep.2014.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tariq A, Jantsch MF. Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front Neurosci 2012; 6:99; PMID:22787438; https://doi.org/ 10.3389/fnins.2012.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hood JL, Emeson RB. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol 2012; 353:61-90. PMID:21796513; https://doi.org/ 10.1007/82_2011_157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 1994; 266:1709-13; PMID:7992055; https://doi.org/ 10.1126/science.7992055 [DOI] [PubMed] [Google Scholar]
- 47.Egebjerg J, Heinemann SF. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci U S A 1993; 90:755-9; PMID:7678465; https://doi.org/ 10.1073/pnas.90.2.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speidel D, Varoqueaux F, Enk C, Nojiri M, Grishanin RN, Martin TF, Hofmann K, Brose N, Reim K. A family of Ca2+-dependent activator proteins for secretion: Comparative analysis of structure, expression, localization, and function. J Biol Chem 2003; 278:52802-9; PMID:14530279; https://doi.org/ 10.1074/jbc.M304727200 [DOI] [PubMed] [Google Scholar]
- 49.Miyake K, Ohta T, Nakayama H, Doe N, Terao Y, Oiki E, Nagatomo I, Yamashita Y, Abe T, Nishikura K, et al.. CAPS1 RNA editing promotes dense core vesicle exocytosis. Cell reports 2016; 17:2004-14; PMID:27851964; https://doi.org/ 10.1016/j.celrep.2016.10.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckenstaler R, Lessmann V, Brigadski T. CAPS1 effects on intragranular pH and regulation of BDNF release from secretory granules in hippocampal neurons. Journal of cell science 2016; 129:1378-90; PMID:26869227; https://doi.org/ 10.1242/jcs.178251 [DOI] [PubMed] [Google Scholar]
- 51.Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al.. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: A quantitative examination. Am J Med Genet 2001; 98:161-7; PMID:11223852; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 52.Ascano M Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al.. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 2012; 492:382-6; PMID:23235829; https://doi.org/ 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000; 406:78-81; PMID:10894545; https://doi.org/ 10.1038/35017558 [DOI] [PubMed] [Google Scholar]
- 54.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 2000; 102:437-49; PMID:10966106; https://doi.org/ 10.1016/S0092-8674(00)00049-0 [DOI] [PubMed] [Google Scholar]
- 55.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome medicine 2013; 5:105; PMID:24289319; https://doi.org/ 10.1186/gm508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbon A, Vallini I, La Via L, Marchina E, Barlati S. Glutamate receptor RNA editing: A molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res Mol Brain Res 2003; 117:168-78; PMID:14559151; https://doi.org/ 10.1016/S0169-328X(03)00317-6 [DOI] [PubMed] [Google Scholar]
- 57.Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res 2007; 35:3723-32; PMID:17517775; https://doi.org/ 10.1093/nar/gkm314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Centurione L, Di Baldassarre A, Zingariello M, Bosco D, Gatta V, Rana RA, Langella V, Di Virgilio A, Vannucchi AM, Migliaccio AR. Increased and pathologic emperipolesis of neutrophils within megakaryocytes associated with marrow fibrosis in GATA-1(low) mice. Blood 2004; 104:3573-80; PMID:15292068; https://doi.org/ 10.1182/blood-2004-01-0193 [DOI] [PubMed] [Google Scholar]
- 59.Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol Cell Neurosci 2007; 34:343-54; PMID:17254795; https://doi.org/ 10.1016/j.mcn.2006.11.015 [DOI] [PubMed] [Google Scholar]
- 60.Lesuisse C, Martin LJ. Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. Journal of neurobiology 2002; 51:9-23; PMID:11920724; https://doi.org/ 10.1002/neu.10037 [DOI] [PubMed] [Google Scholar]
- 61.Zaltieri M, Grigoletto J, Longhena F, Navarria L, Favero G, Castrezzati S, Colivicchi MA, Della Corte L, Rezzani R, Pizzi M, et al.. alpha-synuclein and synapsin III cooperatively regulate synaptic function in dopamine neurons. Journal of cell science 2015; 128:2231-43; PMID:25967550; https://doi.org/ 10.1242/jcs.157867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.