ABSTRACT
Dual-specificity phosphatase 11 (DUSP11) is a conserved protein tyrosine phosphatase (PTP) in metazoans. The cellular substrates and physiologic activities of DUSP11 remain largely unknown. In nematodes, DUSP11 is required for normal development and RNA interference against endogenous RNAs (endo-RNAi) via molecular mechanisms that are not well understood. However, mammals lack analogous endo-RNAi pathways and consequently, a role for DUSP11 in mammalian RNA silencing was unanticipated. Recent work from our laboratory demonstrated that DUSP11 activity alters the silencing potential of noncanonical viral miRNAs in mammalian cells. Our studies further uncovered direct cellular substrates of DUSP11 and suggest that DUSP11 is part of regulatory pathway that controls the abundance of select triphosphorylated noncoding RNAs. Here, we highlight recent findings and present new data that advance understanding of mammalian DUSP11 during gene silencing and discuss the emerging biological activities of DUSP11 in mammalian cells.
KEYWORDS: Argonaute, BLV, dicer, DUSP11, miRNA, miRNA biogenesis, PIR-1, RNAi, triphosphate, VA RNA
Introduction
Specifically processed non-protein-coding RNAs (ncRNAs) play important roles in protein biosynthesis and regulation of gene expression. For example, the abundant 5S rRNA forms a key structural component of the ribosome1 and microRNAs (miRNAs) generated from larger hairpin precursors dock to specific mRNAs and repress translation.2,3 In addition, less specifically processed ncRNAs with particular structural or biochemical features are sensed by cells to detect the presence of pathogens. Double-stranded RNAs (dsRNAs), RNAs with features of transposons, and 5′ tri-phosphorylated products of viral replicative intermediates all can trigger inflammatory responses.4-10 Consequently, sensitive detection and proper control of these ncRNAs are important to avoid infectious or autoimmune diseases. The mounting interest in ncRNA biology emphasizes the importance of understanding the proteins that modify and regulate ncRNAs. Recently, our laboratory characterized the role of the DUSP11 RNA tri-phosphatase activity during noncanonical miRNA biogenesis pathways and in control of select non-protein-coding RNA substrates.11 Here we highlight these findings, provide additional data advancing understanding of the basic properties of DUSP11, and discuss the activities of DUSP11 relevant to diverse areas of ncRNA biology.
DUSP11
DUSP11 belongs to the DUSP subgroup of type-I cysteine-based protein tyrosine phosphatases (PTPs). All DUSPs share a highly conserved catalytic domain containing a consensus phosphatase sequence (HCXXXXXR), which gives them the ability to dephosphorylate both tyrosine and serine/threonine residues.12 Based on sequence and/or function, DUSPs are categorized into 6 subgroups including: the slingshot phosphatases, phosphatase of regenerating liver (PRL), Cdc14 phosphatases, PTEN (phosphatase and tensin homolog deleted on chromosome 10)-like and myotubularin phosphatases, the mitogen-activated protein kinase phosphatases (MKPs), and the atypical DUSPs (For a review, see ref. 13). DUSP11 is a member of the atypical DUSP subgroup, which is composed of 16 members. The “atypical” DUSPs are so called because their members do not share a common proximal ancestor and they lack conserved/characterized domains (e.g., such as the N-terminal CH2 domain of MPKs) outside the DUSP catalytic domain.13 Consequently, the substrates, cellular functions, and physiologic roles of atypical DUSPs remain largely unknown and are likely diverse.
The first insight into the substrate preference of DUSP11 was the discovery of its high affinity association with RNA in vitro.14 This accounts for its alternative (common) gene name, “PIR-1” which stands for phosphatase that interacts with RNA–ribonucleoprotein complex 1. Loss of nuclear localization of ectopically expressed DUSP11 with RNaseA treatment provided evidence for its association with RNA and/or RNP complexes in mammalian cells.14 Subsequently, DUSP11 was shown to remove the γ- and β- phosphates from 5′-triphosphorylated RNAs in vitro.15 Of note, the phosphatase activity of DUSP11 on tri- and di- phosphorylated RNA was several orders of magnitude greater than that with phospho-protein substrates, implying that dephosphorylation of RNA substrates is physiologically relevant. Consistent with this, DUSP11 shares extensive sequence homology to a well-studied RNA phosphatase, Baculovirus phosphatase (BVP).14 Both DUSP11 and BVP contain the consensus RNA tri-phosphatase domain (HCTHGXNRT) and share more structural similarity with the metazoan mRNA capping enzymes than other PTPs.16-18 Nevertheless, whether DUSP11 functions as an RNA tri-phosphatase in mammalian cells had remained undetermined until recently.
DUSP11 and RNAi
RNA silencing is a conserved eukaryotic mechanism of post-transcriptional gene expression regulation (For review, see ref. 19). During RNA silencing, small RNAs (∼22–30 nts) guide the RNA-induced silencing complex (RISC), whose main component is an Argonaute protein that binds to miRNAs and connects to mRNA transcripts via base pairing. Upon mRNA association, RISC generally represses gene expression of the mRNA by direct cleavage of the mRNA20 or by repression of translation followed by increased turnover of the mRNA.21,22 Small interfering RNAs (siRNAs), microRNAs (miRNAs), and piwi-interacting RNAs (pi-RNAs), which differ in their biogenesis and/or silencing mechanisms, are effectors of RNA silencing. siRNAs mediate RNA interference (RNAi), typically to defend against transposons and viruses. While it is clear that RNAi is a major antiviral response in invertebrates, the relevant contribution of RNAi to antiviral defense in vertebrates remains debated.23,24
In C. elegans, DUSP11 suppresses Orsay virus replication, presumably by an RNAi-based mechanism.18 DUSP11 has been shown to interact with Dicer and ERI-1 complexes to promote RNAi.18,25 Initially, knockout of DUSP11 was shown to abolish siRNA production triggered by exogenous RNAs.25 However, subsequent studies showed that knockout of DUSP11 more specifically reduced endo-RNAi pathways in germline cells, while RNAi stimulated by exogenous RNAs remained largely intact.18 During endo-RNAi, DUSP11 is required for generation and/or function of 26G-RNAs, which are primary siRNAs that initiate 22G-RNA amplification. 26G-RNAs are predominately 5′-monophosphorylated but are thought to be directly synthesized by RNA dependent RNA polymerases (RdRPs) and initially contain a 5′-triphosphate.26,27 Therefore, DUSP11 RNA tri-phosphatase activity has been postulated to function in 26G-RNA maturation26 by two non-mutually exclusive mechanisms: 1) Dephosphorylation of an RdRP-generated precursor is required for 26G-RNA biogenesis. 2) Dephosphorylation of the mature 26G-RNA permits stable association with primary Argonaute proteins.18 A similar role for DUSP11 in mammalian cells was unanticipated since mammals do not encode an RdRp and lack analogous siRNA biogenesis pathways.28
DUSP11 and noncanonical microRNAs
miRNAs are a class of small RNAs conserved in metazoans that regulate diverse cellular processes vital for development and homeostasis, such as cell differentiation and metabolism.2 Unlike siRNAs, miRNAs are encoded in defined genomic loci and mature through a conserved step-wise miRNA biogenesis pathway (Fig. 1).2,3 Initially, miRNAs are embedded within stem-loop structures in RNA polymerase II-transcribed primary miRNA (pri-miRNA) transcripts.29 Multiple structural features of pri-miRNAs are common and necessary for efficient miRNA maturation, including a basal single-stranded RNA (ssRNA) region, a double-stranded RNA (dsRNA) stem (∼3 helices), a terminal loop, and primary sequences within the basal ssRNA and terminal loop.30-34 Through cooperation between the flanking ssRNA regions and internal structures,31,32,35,36 the nuclear Microprocessor complex – which includes the core RNase III endonuclease Drosha and its RNA binding partner DGCR8 – cleaves the dsRNA stem ∼11 nts from the basal ssRNA and ∼22 nts from the terminal loop, liberating a precursor miRNA (pre-miRNA) hairpin.37-40 In the cytosol, Dicer processes the pre-miRNA41-43 into duplexed (5p:3p) miRNAs,37 each containing a 5′ monophosphate and 3′ hydroxyl termini.42,44 Through a process termed strand selection, one strand (guide strand) is efficiently loaded into an Argonaute protein, a core component of RISC, while the other strand (passenger strand) is rapidly turned over.45,46 Once loaded into Argonaute, miRNAs form stable RISCs, termed miRISC.
Figure 1.
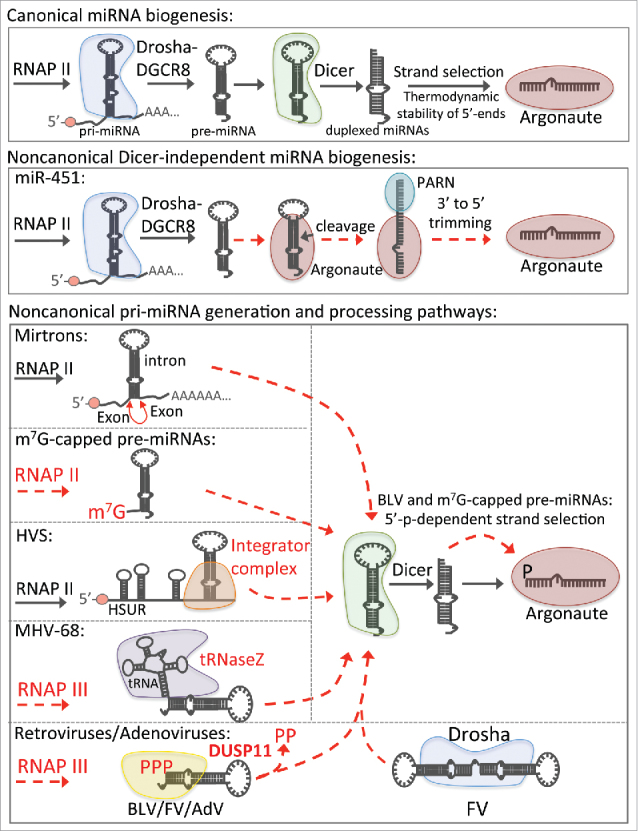
Canonical and noncanonical host and viral miRNA biogenesis pathways. Schematic diagrams of the canonical and noncanonical host and viral miRNA biogenesis in animals. Dashed red lines indicate noncanonical processing mechanisms. Enzymes in red indicate noncanonical miRNA biogenesis enzymes or mechanisms used by those enzymes to generate miRNA RNA species.
Although most miRNAs mature through the canonical miRNA biogenesis pathway, there are several conserved cellular miRNAs generated via unconventional mechanisms (Fig. 1; for review see ref. 47). Pre-miR-451 is directly cleaved by Argonaute 2 and trimmed by PARN to generate miR-451, bypassing the Dicer processing step. 48,49 Mirtrons are a class of miRNAs that are encoded in short hairpin introns that generate pre-miRNAs upon splicing.50 The miR-320 and miR-484 pre-miRNAs are directly transcribed by RNAP II and contain a 5′-m7G-cap, which only permits loading of the 3p arm into Argonaute.47,51
Diverse viruses also encode miRNAs (For review, see refs. 52 and 53). Like host miRNAs, most viral miRNA mature through the canonical miRNA biogenesis pathway. However, some viruses utilize noncanonical mechanisms to generate miRNAs. Murid gammaherpesvirus 68 (MHV-68) expresses RNAP III-transcribed transcripts with a tRNA structure upstream of a pre-miRNA hairpin,54-57 which is liberated via tRNaseZ-mediated cleavage of the tRNA structure.58 Herpesvirus saimiri (HVS) expresses RNAP II-transcribed Sm class U RNAs (HSURs) that are processed by the integrator complex to generate pre-miRNAs.59 Retroviruses, including members of the foamy virus (FV) family and bovine leukemia virus (BLV), express subgenomic pri-miRNA transcripts via RNAP III-mediated transcription from the proviral genome.60-63 Some FV pri-miRNA transcripts resemble a “dumbbell” structure composed of 2 juxtaposed stem-loops that are processed by Drosha to generate pre-miRNAs. Other FV pre-miRNAs and all of the BLV pre-miRNAs are directly transcribed by RNAP III and processed by Dicer, analogous to typical RNAP III-transcribed short hairpin RNAs (shRNAs).62 Similarly, Adenoviruses (AdV) express viral-associated RNAs (VA RNAs) via RNAP III-mediated transcription that are processed by Dicer to generate miRNAs.64-67 The retroviral pre-miRNAs and AdV VA RNAs initially contain a 5′ triphosphate, typical of RNAP III primary transcripts.62,68 However, the BLV and AdV 5p miRNAs, which share the same 5′-end as their 5′-triphosphorylated precursors, are predominantly 5′-monophosphorylated.62,69 This implicated a tri-phosphatase step during their maturation. We recently characterized this phenomenon and identified DUSP11 as the RNA tri-phosphatase responsible for the dephosphorylation of the BLV and AdV 5p miRNAs.11
In DUSP11-null human cell lines, we observed a substantial reduction in the steady-state levels, RISC activity, and AGO-association of the BLV and AdV 5p miRNAs but not the corresponding 3p miRNAs. This indicates that DUSP11 is not required for Dicer-mediated processing of the precursors, but rather alters incorporation of the 5p miRNAs into RISC. Several lines of evidence support that DUSP11-mediated dephosphorylation of the BLV/AdV 5p miRNAs promotes their association with Argonaute proteins: 1) 5p miRNAs that mature through the canonical biogenesis pathway were unaffected by DUSP11, indicating that DUSP11 is specific for 5′ triphosphorylated pre-miRNAs. 2) BLV/AdV 5p miRNA biogenesis was restored in DUSP11-null cells by ectopically expressing wild type DUSP11, but not a catalytic mutant (C152S). This demonstrates that the RNA tri-phosphatase activity of DUSP11 is required for efficient BLV/AdV5p miRNA biogenesis. 3) Using pre-miRNA mimics, we demonstrated that 5p miRNAs derived from 5′-ppp-pre-miRNAs, but not 5′-p-pre-miRNAs, are inefficiently loaded into Argonaute proteins. These results support a model whereby DUSP11-mediated dephosphorylation of 5′-ppp miRNA precursors promotes AGO association with the 5p miRNAs. This occurs via a 5´-monophosphate-dependent-strand selection mechanism that is coupled to or enhanced by Dicer processing (Fig. 2). Combined, these findings raise new questions, including the relevant subcellular location and the subdomains/motifs of DUSP11 that are involved in DUSP11-dependent RNA silencing.
Figure 2.
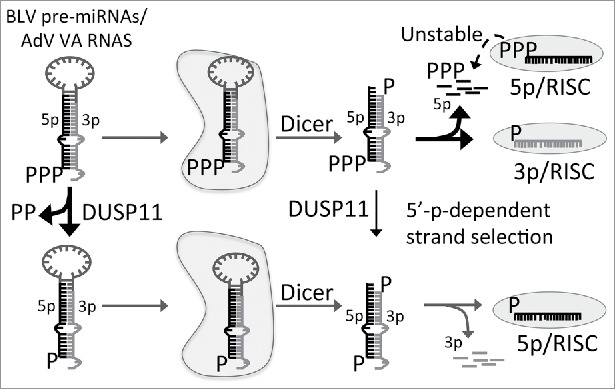
Model of DUSP11-dependent Argonaute association of BLV and AdV 5p miRNAs. The BLV and Adenovirus miRNA precursors are transcribed by RNAP III and initially contain a 5′ triphosphate. In the absence of DUSP11-mediated dephosphorylation, the 5′ triphosphorylated precursors are processed by Dicer to yield mature 5p:3p duplex miRNAs. Through a 5′-monophosphate-dependent strand selection mechanism that is coupled to Dicer processing, the triphosphorylated 5p miRNAs are excluded from being stably loaded into Argonaute proteins, resulting in preferential loading of the 3p strand. The dashed line indicates that 5′ triphosphorylated 5p miRNAs may have a reduced stable association with Argonaute proteins, resulting in their increased turnover. However, miRNA precursors that are dephosphorylated by DUSP11 produce 5′ monophosphorylated 5p miRNAs that are more efficiently loaded and/or more stably associate with Argonaute proteins.
Characterization of DUSP11 regions important for localization and RNA tri-phosphatase activity in cells
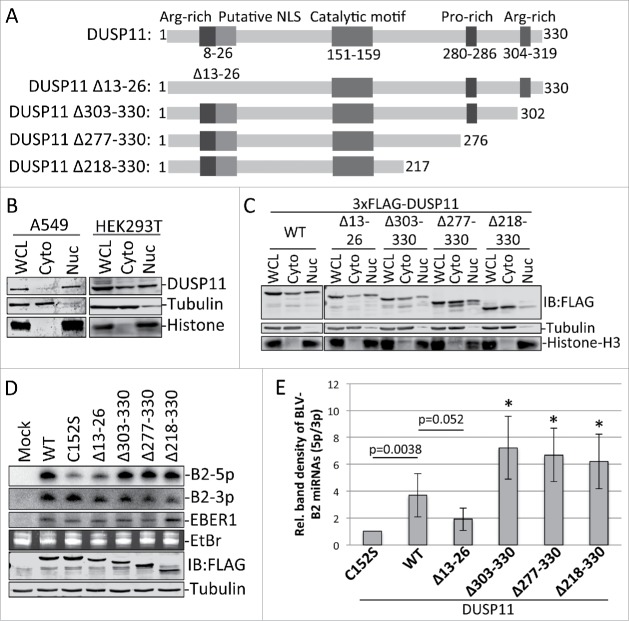
DUSP11 contains 2 arginine-rich sequences [one near the N-terminus (residues 8–20) and the other near the C-terminus (residues 304–319)] that are characteristic of RNA-binding domains (Fig. 3A). The N-terminal arginine-rich region also overlaps with a putative nuclear localization signal (NLS) at residues 13–26.17,18 A proline-rich region, with similarities to motifs known to interact with Src homology 3 (SH3) domains, is located near the c-terminus (residues 280–286). To gain insight into the function of these regions, we generated several deletion mutants of DUSP11. We engineered a mutant that removed an internal stretch of amino acids in N-terminal NLS/arginine-rich region (Δ13–26). We also made a series of C-terminal truncation mutants that deleted the C-terminal arginine-rich region (Δ303–330), the arginine-rich region and the proline-rich region (Δ277–330), or the majority of the C-terminal region after the catalytic domain (Δ218–330; Fig. 3A). We then analyzed the subcellular localization and ability of these mutants to rescue the reduced 5p:3p miRNA ratios in DUSP11-null cells.
Figure 3.
DUSP11 motifs/domains involved in localization and RNA phosphatase activity in cells. (A) Schematic diagram of DUSP11 (not to scale) illustrating the relative location of the arginine-rich motifs, putative NLS, catalytic domain, and proline-rich motif. Below are the DUSP11 mutants that we generated, which are described in the text. We cloned the constructs into pcDNA3.1+ and tagged the N-terminus via PCR-based engineering with a 3xFLAG epitope. (B) Nuclear/cytosolic fractionation of A549 and HEK293T cells to determine localization of DUSP11. Cells from one well of 12-well format plate were trypsinized, washed with phosphate-buffered solution (PBS), resuspended in 50-ul of CSKT buffer [10mM PIPES (pH 6.8), 100mM NaCl, 300mM sucrose, 3mM MgCl2, 1mM EDTA, 1mM DTT, 0.5% (vol/vol) tritonX-100, protease inhibitors (Roche)], and placed on ice for 10 minutes. Nuclei were then pelleted by centrifugation at 5000xg for 5 minutes. The supernatant [cytosolic fraction (Cyto)] was removed (50-ul) and added to 50-ul of SDS lysis buffer (1% SDS; 2% 2-mercaptoethanol). The nuclei were then washed in 50-ul of CSKT buffer and centrifuged at 5000xg for 5 minutes. The nuclei were then resuspended in 50-ul of water followed by addition of 50-ul of SDS buffer to make the nuclear fraction (Nuc). A whole cell lysate (WCL) was collected in parallel by trypsinization and washing of the cells, which were then resuspended in 50-ul of water, followed by addition of 50-ul of SDS lysis buffer. Samples were boiled for 10 minutes followed by vortexing for 30 seconds. Equal volumes (10-ul) of fractions were then fractionated via 12.5% SDS-PAGE. Protein was then transferred to nitrocellulose membrane (Bio-Rad). Anti-DUSP11 polyclonal rabbit antibody (1:2000 dilution; Proteintech, catalog no. 10204–2-AP), anti-α-Tubulin monoclonal mouse antibody (1:10,000 dilution; Sigma-Aldrich, catalog no. T6199), and anti-Histone H3 (D1H2) rabbit antibody (Cell signaling; mAb #4499) in phosphate-buffered saline (PBS) with 0.1% Tween20 (PBST) and 5% BSA were used to blot for DUSP11, α-Tubulin (cytosolic localized control), and Histone H3 (nuclear localized control). After washing with PBST, membranes were blotted with IRDye 800CW and IRDye 680LT secondary antibodies (1:10,000 dilution; LI-COR) in PBST with 5% BSA. Blots were washed 4 times with PBST and then scanned on an Odyssey CLx infrared imaging system (LI-COR). (C) Nuclear/cytosolic fractionation as described in (B) of HEK293T cells (12-well format) transfected with 200-ng/well of the 3xFLAG-tagged DUSP11 constructs from (A). The anti-FLAG M2 mouse monoclonal antibody (Sigma-Aldrich) was used to stain for the 3xFLAG-tagged DUSP11 proteins. (D) Analysis of the BLV-B2 5p:3p miRNA ratios with co-expression of the DUSP11 constructs from (A). DUSP11-null HEK293T cells (24-well format; 70% confluent) were co-transfected with 5-ng/well of the EBER1 transfection control expression vector (pEBV RIJ),83 400-ng/well of pBLV-B2, and 100-ng/well of pDUSP11, pDUSP11-C152, or the DUSP11 construct shown in (A). Northern blot analysis was then performed as described in refs. 11 and 84. The membrane was probed with the BLV-miR-B2–5p probe, stripped and re-probed with the BLV-miR-B2–3p probe, and stripped and re-probed with the EBER1 probe as a loading/transfection control. Below is immunoblot analysis that was performed on duplicate lysates to confirm and quantitate DUSP11 expression from each construct using the anti-FLAG M2 mouse monoclonal antibody (Sigma-Aldrich). (E) Bar graph of the relative band density from 4 independent RNA gel blot analyses, as shown in D (except for DUSP11-Δ13–26, in which 3 replicates were performed). Bars represent the average band density ratio (5p/3p) +/− SD of the BLV-B2 miRNAs. The ratio with co-expression of each DUSP11 construct was normalized to the C152S catalytic mutant. Asterisks represent statistical significance (p<0.05) in comparison to wild type DUSP11 (WT). P-values were determined using student's t-test.
Crude subcellular fractionation followed by immunoblot analysis revealed that endogenous DUSP11 is primarily localized to the nucleus (∼90%) in A549 cells, while ∼10% is localized to the cytoplasm (Fig. 3B). This is consistent with previous findings of DUSP11 being predominantly nuclear.14 Substantially more endogenous DUSP11 localized to the cytoplasm (∼40%) in HEK293T cells, although the majority (∼60%) still localized to the nucleus (Fig. 3B). We observed a similar localization pattern of endogenous un-tagged and exogenous 3xFLAG-tagged full length DUSP11 in HEK293T cells, indicating that the 3xFLAG did not grossly alter localization of DUSP11 (Fig. 3C). Deletion of the residues 13–26, which contains a putative nuclear localization signal,17,18 did not affect localization of DUSP11. In contrast, the C-terminal truncation mutants showed reduced nuclear localization. Deleting residues 303–330, which contains the C-terminal arginine-rich region, reduced localization of DUSP11 in the nucleus 2-fold, with ∼30% localizing to the nucleus. A similar reduction in nuclear localization was observed with deletion of residues 277–330. However, we observed a further reduction in nuclear localization with deletion of residues 218–330, which showed ∼75% (or greater) localization in the cytoplasm (Fig. 3C). These data indicate that typical nuclear localization of DUSP11 requires regions in the C-terminus of portion of DUSP11.
To determine whether these deleted DUSP11 regions are required for the RNA tri-phosphatase activity of DUSP11 in cells, we analyzed the 5p to 3p ratio of miRNAs derived from BLV-pre-miR-B2. We expressed 3xFLAG-tagged wild type DUSP11, each DUSP11 deletion mutant, or the C152S catalytic mutant in DUSP11 knock out HEK293T cells and performed northern blot analysis. Wild type DUSP11 and all the C-terminal truncation mutants increased the 5p to 3p ratio of BLV-pre-miR-B2 as compared with the negative control DUSP11-C152S catalytic mutant (Fig. 3D). In contrast, a smaller increase in the 5p to 3p ratio was observed with DUSP11Δ13-26 in comparison to wild type DUSP11 (Fig. 3D). These data indicate that residues 13–26, which contain an arginine-rich motif, are required for efficient DUSP11-mediated dephosphorylation of the BLV 5′-ppp substrate RNAs in cells, whereas the C-terminal portion of DUSP11 is not.
Our data confirm that DUSP11 is predominately localized to the nucleus, albeit the magnitude of this varies by cell type. Future analysis in additional cell types will be informative to the extent and the mechanism of this phenomenon. Unexpectedly, deletion of the C-terminus, and not the putative N-terminal NLS-like motif, reduced nuclear localization. However, the C-terminus is dispensable for RNA tri-phosphatase activity in cells, at least during BLV miRNA biogenesis. In fact, truncation of the C-terminus reproducibly increased (∼1.5-fold) DUSP11 RNA tri-phosphatase activity (Fig. 3E). This could be due to deletion of residues in the C-terminus that negatively regulate DUSP11 activity and/or due to the increased cytosolic levels of DUSP11 being better positioned to dephosphorylate the BLV pre-miRNAs/miRNAs. Interestingly, truncation of the C-terminus results in 2-fold higher RNA tri-phosphatase activity in vitro,15 supporting that the C-terminus can alter catalytic activity. While deletion of residues 13–26 did not alter localization of the DUSP11, we did observe a trend in which deletion of these residues slightly reduced DUSP11 RNA tri-phosphatase activity on the BLV miRNAs (Fig. 3D,E). Barring that this deletion did not grossly alter the structure of the DUSP11, these results are consistent with residues 13–26 performing an important function in cells. Given the positive charge of these residues, one speculation is that these residues comprise part of a direct RNA binding function. Alternatively, these residues may be important for protein-protein interactions. Further analysis is warranted to better understand the mechanisms of DUSP11 domains/motifs.
Mammalian substrates of DUSP11
Small RNA sequencing demonstrated that DUSP11 expression does not alter endogenous miRNAs,11 likely since they harbor 5′-monophosphates as a result of endonucleolytic cleavage by Drosha and Dicer. Our analysis of RNAP III-transcribed RNAs, which at least initially contain a 5′ triphosphate, did reveal significant differences of some transcripts. Using an RNA-sequencing platform able to quantify highly structured RNAs and differences in triphosphate status (TGIRT-seq),11,70-73 we observed increased overall levels of vault RNAs and Alu RNAs in DUSP11 knockout cells. We also observed a higher triphosphorylated portion of these ncRNAs in DUSP11 knockout cells. Northern blot analysis confirmed increased vault RNA steady-state levels in DUSP11 knockout cells and demonstrated that reconstitution of DUSP11 restored vault RNAs levels to wild-type levels. These observations support a model whereby DUSP11-mediated conversion of select 5′-triphorylated RNAs to monophosphates increases their susceptibility to exonuclease attack or otherwise alters their stability (Fig. 4).
Figure 4.
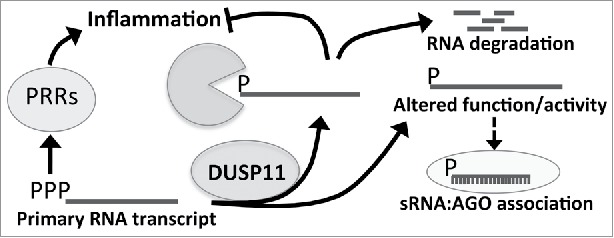
Model for putative roles of DUSP11 in mammalian cells. Illustration of the model for DUSP11s role in mammalian cells. DUSP11 dephosphorylates exogenous or endogenous 5′-triphosphorylated RNAs. This activity results in 5′ monophosphorylated RNAs, which can alter the function/activity of the RNA (for example loading into RISC) or permit degradation of the RNA by nucleases. This process may also reduce activation of PRRs, which recognize 5′ triphosphorylated RNAs and trigger the innate immune response.
Conclusions and perspectives
Recent studies have convincingly demonstrated that DUSP11 has RNA tri-phosphatase activity in the context of nematodes and mammalian cells.11,18 In mammalian cells, DUSP11-mediated conversion of 5′ triphosphorylated small RNAs and/or their precursors to a 5′ monophosphate increases their association with AGO proteins. This occurs through a Dicer-coupled loading mechanism and/or through stabilization of the miRNA:AGO complex.11 Although performed in mammalian cells, this model may also apply to 26G RNAs in nematodes. However, the extent of DUSP11-mediated RNA silencing in mammalian cells remains unclear since DUSP11-dependent RISC activity has only been shown for a small subset of noncanonical viral miRNAs. Some RNAP III-transcribed ncRNAs – such as the 5S rRNA, vault RNAs, Alu RNAs, and Y-RNAs – generate abundant small RNA derivatives that can associate at low levels with RISC complexes in certain contexts.11,74-76 Since DUSP11 modulates the steady-state levels and 5′ tri-phosphorylation status of these RNAs and/or their small RNA derivatives,11 it is conceivable that DUSP11 may be required for the biogenesis or stable AGO association of these sRNAs. As these less abundant RNAs could regulate gene expression via RNA silencing mechanisms that require only low levels of loaded AGOs – for example, such as in chromatin regulation77 – further investigation into DUSP11s role in RNA silencing in various mammalian contexts is warranted.
Our analysis of the mRNA profiles in DUSP11-null cells did not reveal consistent changes in mRNAs levels shared between the two different parental cell backgrounds that we analyzed, nor did we observe any differences in patterns of mRNA splicing. This indicates that DUSP11 does not grossly alter mRNA splicing under the contexts we examined. Nevertheless, the association between DUSP11 and multiple splicing factors14,78 suggests that DUSP11 could alter splicing in other still-to-be-determined contexts, such as stress responses. Although we did not observe differences in mRNA processing in DUSP11-null cells, we did observe an increase in the levels and 5′ triphosphorylation status of select RNAP III-transcribed RNAs. Consistent with these results, a previous study demonstrated that siRNA-mediated knockdown of DUSP11 increased levels of the RNAP I-transcribed 47S primary transcript, indicating that DUSP11 is involved in 47S rRNA biogenesis.79 Furthermore, a significant increase in a 47S RNA 5′-fragment [+1 to cleavage site 01 of the 5′ external transcribed spacer (ETS)] was observed with DUSP11 knock down, suggesting DUSP11 is required for turning over this by-product RNA. Since primary RNA transcripts, such as the 47S RNA, vault RNAs and Alu RNAs, initially contain a triphosphate, these observations are consistent with a model in which DUSP11-mediatied dephosphorylation promotes processing and/or turnover of diverse 5′-triphosphoylated RNAs (Fig. 4).
The possible association of DUSP11 with disease remains unknown. Increases in SINE transcript levels, such as Alu transcripts, are associated with increasing DNA damage, inducing inflammation, and the age-associated autoimmune disease, macular degeneration.8-10,80,81 Therefore, the turnover of 5′ triphosphorylated RNAs by DUSP11 may be an important mechanism against excessive inflammation and transposon activity. Additionally, as 5′ triphosphorylated transcripts are produced as RNA virus replication intermediates,5-7 DUSP11 also may play a role in regulating the response to pathogens. DUSP11 has been reported to be elevated in an in vitro model of transformation,82 but it is also a weakly inducible p53-responsive gene that has been reported to reduce cell proliferation.78 Thus, whether and in what contexts DUSP11 activity relates to tumor biology remain unresolved. Combined, these observations merit further analysis of the role of DUSP11 during development and disease.
Numerous aspects of DUSP11 biology remain incompletely understood. These include the complexes that DUSP11 associates with, its substrates, and its potential contribution to development, homeostasis, aging, and the immune response. Although still early days, the future study of DUSP11 shows promise for better understanding diverse aspects of RNA biology.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Joan Steitz for the pEBV RIJ plasmid, Nicole Welch for plasmid reagents, and Joon Choi and Adam Hilterbrand for comments regarding this manuscript.
Funding
The work described in this article was supported by a Burroughs Wellcome Investigators in Pathogenesis Award and a grant from the Cancer Prevention and Research Institute of Texas [RP140842].
References
- 1.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S ribosomal RNA database. Nucleic Acids Res 2002; 30(1):176-78; PMID:11752286; https://doi.org/ 10.1093/nar/30.1.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116(2):281-97; PMID:14744438; https://doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6(5):376-85; PMID:15852042; https://doi.org/ 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- 4.Julkunen I, Sareneva T, Pirhonen J, Ronni T, Melen K, Matikainen S. Molecular pathogenesis of influenza a virus infection and virus-induce regulation of cytokine gene expression. Cytokine Growth Factor Rev 2001; 12(2-3):171-80; PMID:11325600; https://doi.org/ 10.1016/S1359-6101(00)00026-5 [DOI] [PubMed] [Google Scholar]
- 5.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Martin S, et al.. Triphosphate RNA is the ligand for RIG-I. Science 2006; 314(5801):994-7; PMID:17038590; https://doi.org/ 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′ phosphates. Science 2006; 314(5801):997-1001; PMID:17038589; https://doi.org/ 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 7.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 2007; 318(5855):1455-8; PMID:18048689; https://doi.org/ 10.1126/science.1147347 [DOI] [PubMed] [Google Scholar]
- 8.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al.. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011; 471(7338):325-30; PMID:21297615; https://doi.org/ 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al.. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012; 149(4):847-59; PMID:22541070; https://doi.org/ 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerur N, Hirano Y, Tarallo V, Fowler BJ, Bastos-Carvalho A, Yasuma T, Yasuma R, Kim Y, Hinton DR, Kirschning CJ, et al.. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest Ophthalmol Vis Sci 2013; 54(12):7395-401; PMID:24114535; https://doi.org/ 10.1167/iovs.13-12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke JM, Kincaid RP, Nottingham RM, Lambowitz AM, Sullivan CS. DUSP11 activity on triphosphorylated transcripts promotes argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev 2016; 30(18):2076-92; PMID:27798849; https://doi.org/ 10.1101/gad.282616.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denu JM, Dixon JE. A catalytic mechanism for the dual-specific phosphatases. Proc Natl Acad Sci U S A 1995; 92(13):5910-4; PMID:7597052; https://doi.org/ 10.1073/pnas.92.13.5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson KI1, Brummer T, O'Brien PM, Daly RJ. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem J 2009; 418(3):475-89; PMID:19228121; https://doi.org/ 10.1042/BJ20082234 [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Li DM, Sun H. PIR1, a novel phosphatase that exhibits high affinity to RNA ribonucleoprotein complexes. J Biol Chem 1998; 273(32):20347-53; PMID:9685386; https://doi.org/ 10.1074/jbc.273.32.20347 [DOI] [PubMed] [Google Scholar]
- 15.Deshpande T, Takagi T, Hao L, Buratowski S, Charbonneau H. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J Biol Chem 1999; 274(23):16590-4; PMID:10347225; https://doi.org/ 10.1074/jbc.274.23.16590 [DOI] [PubMed] [Google Scholar]
- 16.Changela A, Martins A, Shuman S, Mondragón A. Crystal structure of baculovirus RNA triphosphatase complexed with phosphate. J Biol Chem 2005; 280(18):17848-56; PMID:15713658; https://doi.org/ 10.1074/jbc.M500885200 [DOI] [PubMed] [Google Scholar]
- 17.Sankhala RS, Lokareddy RK, Cingolani G. Structure of human PIR1, an atypical dual-specificity phosphatase. Biochemistry 2014; 53(5):862-71; PMID:24447265; https://doi.org/ 10.1021/bi401240x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaves D. The RNA 5′ phosphatase PIR-1 cooperates with dicer to produce endogenous small RNAs and suppress viral replication in C. elegans [Doctoral dissertation] University of Lisbon; 2015. [Google Scholar]
- 19.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136(4):642-55; PMID:19239886; https://doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000; 101(1):25-33; PMID:10778853; https://doi.org/ 10.1016/S0092-8674(00)80620-0 [DOI] [PubMed] [Google Scholar]
- 21.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336(6078):233-7; PMID:22422859; https://doi.org/ 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012; 336(6078):237-40; PMID:22499947; https://doi.org/ 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullen BR, Cherry S, tenOever BR. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 2013; 14(4):374-8; PMID:24139396; https://doi.org/ 10.1016/j.chom.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 24.Pare JM, Sullivan CS. Distinct antiviral responses in pluripotent versus differentiated cells. PLOS Pathog 2014; 10(2):e1003865; PMID:24516379; https://doi.org/ 10.1371/journal.ppat.1003865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al.. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 2006; 124(2):343-54; PMID:16439208; https://doi.org/ 10.1016/j.cell.2005.11.036 [DOI] [PubMed] [Google Scholar]
- 26.Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 2010; 37(5):679-89; PMID:20116306; https://doi.org/ 10.1016/j.molcel.2010.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A 2010; 107(8):3582-7; PMID:20133583; https://doi.org/ 10.1073/pnas.0911908107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the drosophila and human RNAi pathways. Mol Cell 2002; 10(3):537-48; PMID:12408822; https://doi.org/ 10.1016/S1097-2765(02)00651-2 [DOI] [PubMed] [Google Scholar]
- 29.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 2002; 21(17):4663-70; PMID:12198168; https://doi.org/ 10.1093/emboj/cdf476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by drosha requires flanking nonstructured RNA sequences. J Biol Chem 2005; 280(30):27595-603; PMID:15932881; https://doi.org/ 10.1074/jbc.M504714200 [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme drosha. EMBO J 2005; 24(1):138-48; PMID:15565168; https://doi.org/ 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell 2006; 125(5):887-901; PMID:16751099; https://doi.org/ 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Zeng Y. The terminal loop region controls microRNA processing by drosha and dicer. Nucleic Acids Res 2010; 38(21):7689-97; PMID:20660014; https://doi.org/ 10.1093/nar/gkq645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013; 152(4):844-58; PMID:23415231; https://doi.org/ 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma H, Wu Y, Jang-Gi C, Wu H. Lower and upper stem–single-stranded RNA junctions together determine the drosha cleavage site. Proc Natl Acad Sci U S A 2013; 110(51):20687-92; PMID:24297910; https://doi.org/ 10.1073/pnas.1311639110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke JM, Kelenis DP, Kincaid RP, Sullivan CS. A central role for the primary microRNA stem in guiding the position and efficiency of drosha processing of a viral pri-miRNA. RNA 2014; 20(7):1068-77; PMID:24854622; https://doi.org/ 10.1261/rna.044537.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III drosha initiates microRNA processing. Nature 2003; 425(6956):415-9; PMID:14508493; https://doi.org/ 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 38.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004; 432(7014):231-5; PMID:15531879; https://doi.org/ 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 39.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432(7014):235-40; PMID:15531877; https://doi.org/ 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- 40.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004; 18(24):3016-27; PMID:15574589; https://doi.org/ 10.1101/gad.1262504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001; 106(1):23-34; PMID:11461699; https://doi.org/ 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- 42.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293(5531):834-8; PMID:11452083; https://doi.org/ 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- 43.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001; 15(20):2654-9; PMID:11641272; https://doi.org/ 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basyuk E, Suavet F, Doglio A, Bordonne R, Bertrand E. Human let-7 stem–loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res 2003; 31(22):6593-7; PMID:14602919; https://doi.org/ 10.1093/nar/gkg855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115(2):209-16; PMID:14567918; https://doi.org/ 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- 46.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003; 115(2):199-208; PMID:14567917; https://doi.org/ 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- 47.Xie M, Steitz JA. Versatile microRNA biogenesis in animals and their viruses. RNA Biol 2014; 11(6):673-81; PMID:24823351; https://doi.org/ 10.4161/rna.28985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature 2010; 465:584-9; PMID:20424607; https://doi.org/ 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. PARN mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep 2013; 5(3):715-26; PMID:24209750; https://doi.org/ 10.1016/j.celrep.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell 2007; 28(2):328-36; PMID:17964270; https://doi.org/ 10.1016/j.molcel.2007.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Šestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 2013; 155(7):1568-80; PMID:24360278; https://doi.org/ 10.1016/j.cell.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology 2011; 411(2):325-43; PMID:21277611; https://doi.org/ 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog 2012; 8(12):e1003018; PMID:23308061; https://doi.org/ 10.1371/journal.ppat.1003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al.. Identification of microRNAs of the herpesvirus family. Nat Methods 2005; 2:269-76; PMID:15782219; https://doi.org/ 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- 55.Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 2010; 16(1):170-85; PMID:19948768; https://doi.org/ 10.1261/rna.1873910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diebel KW, Claypool DJ, van Dyk LF. A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene 2014; 544(1):8-18; PMID:24747015; https://doi.org/ 10.1016/j.gene.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feldman ER, Kara M, Coleman CB, Grau KR, Oko LM, Krueger BJ, Renne R, van Dyk LF, Tibbetts SA. Virus-encoded MicroRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. mBio 2014; 5(3):e00981-14; PMID:24865551; https://doi.org/ 10.1128/mBio.00981-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses non-canonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 2010; 37(1):135-42; PMID:20129062; https://doi.org/ 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cazalla D, Xie M, Steitz JA. A primate herpesvirus uses the integrator complex to generate viral MicroRNAs. Mol Cell 2011; 43(6):982-92; PMID:21925386; https://doi.org/ 10.1016/j.molcel.2011.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A 2012; 109(8):3077-82; PMID:22308400; https://doi.org/ 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS. Non-canonical miRNA biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio 2014; 5(2):e00074-14; PMID:24713319; https://doi.org/ 10.1128/mBio.00074-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burke JM, Bass CR, Kincaid RP, Sullivan CS. Identification of tri-phosphatase activity in the biogenesis of retroviral microRNAs and RNAP III-generated shRNAs. Nucleic Acids Res 2014; 42(22):13949-62; PMID:25428356; https://doi.org/ 10.1093/nar/gku1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Löchelt M, Cullen BR. Identification of novel, highly expressed retroviral MicroRNAs in cells infected by bovine foamy virus. J Virol 2014; 88(9):4679-86; PMID:24522910; https://doi.org/ 10.1128/JVI.03587-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 2004; 78(23):12868-76; PMID:15542639; https://doi.org/ 10.1128/JVI.78.23.12868-12876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 2006; 80(3):1376-84; PMID:16415015; https://doi.org/ 10.1128/JVI.80.3.1376-1384.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu N, Segerman B, Zhou X, Akusjärvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol 2007; 81(19):10540-9; PMID:17652395; https://doi.org/ 10.1128/JVI.00885-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furuse Y, Ornelles DA, Cullen BR. Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA. Virology 2013; 447(1-2):140-5; PMID:24210108; https://doi.org/ 10.1016/j.virol.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vennström B, Pettersson U, Philipson L. Two initiation sites for adenovirus 5.5S RNA. Nucleic Acids Res 1978; 5(1):195-204; PMID:643608; https://doi.org/ 10.1093/nar/5.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu N, Gkountela S, Saeed K, Akusjärvi G. The 5′-end heterogeneity of adenovirus virus-associated RNAI contributes to the asymmetric guide strand incorporation into the RNA-induced silencing complex. Nucleic Acids Res 2009; 37(20):6950-9; PMID:19755500; https://doi.org/ 10.1093/nar/gkp764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al.. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 2013; 19(7):958-70; PMID:23697550; https://doi.org/ 10.1261/rna.039743.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katibah GE, Qin Y, Sidote DJ, Yao J, Lambowitz AM, Collins K. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci U S A 2014; 111(33):12025-30; PMID:25092312; https://doi.org/ 10.1073/pnas.1412842111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, Lambowitz AM. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 2016; 22(4):597-613; PMID:26826130; https://doi.org/ 10.1261/rna.055558.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, Lambowitz AM. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 2016; 22(2):111-28; PMID:26554030; https://doi.org/ 10.1261/rna.054809.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol 2009; 11(10):1268-71; PMID:19749744; https://doi.org/ 10.1038/ncb1972 [DOI] [PubMed] [Google Scholar]
- 75.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141(1):129-41; PMID:20371350; https://doi.org/ 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs (riRNAs) regulates human stem cell proliferation. Nat Struct Mol Biol 2012; 19(11):1168-75; PMID:23064648; https://doi.org/ 10.1038/nsmb.2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woolnough JL, Atwood BL, Giles KE. Argonaute 2 binds directly to tRNA genes and promotes gene repression in cis. Mol Cell Biol 2015; 35(13):2278-94; PMID:25918241; https://doi.org/ 10.1128/MCB.00076-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caprara G, Zamponi R, Melixetian M, Helin K. Isolation and characterization of DUSP11, a novel p53 target gene. J Cell Mol Med 2009; 13(8B):2158-70; PMID:19120688; https://doi.org/ 10.1111/j.1582-4934.2008.00616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 2013; 51(4):539-51; PMID:23973377; https://doi.org/ 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al.. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011; 10(17):3016-30; PMID:21862875; https://doi.org/ 10.4161/cc.10.17.17543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karijolich J, Abernathy E, Glaunsinger BA. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-κB pathway. PLoS Pathog 2015; 11(11):e1005260; PMID:26584434; https://doi.org/ 10.1371/journal.ppat.1005260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dardousis K, Voolstra C, Roengvoraphoj M, Sekandarzad A, Mesghenna S, Winkler J, Ko Y, Hescheler J, Sachinidis A. Identification of differentially expressed genes involved in the formation of multicellular tumor spheroids by HT-29 colon carcinoma cells. Mol Ther 2007; 15(1):94-102; PMID:17164780; https://doi.org/ 10.1038/sj.mt.6300003 [DOI] [PubMed] [Google Scholar]
- 83.Tycowski KT1, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 1998; 2(5):629-38; PMID:9844635; https://doi.org/ 10.1016/S1097-2765(00)80161-6 [DOI] [PubMed] [Google Scholar]
- 84.Burke JM, Kuny CV, Kincaid RP, Sullivan CS. Identification, validation, and characterization of noncanonical miRNAs. Methods 2015; 91:57-68; PMID:26210399; https://doi.org/ 10.1016/j.ymeth.2015.07.013 [DOI] [PubMed] [Google Scholar]