Abstract
Isolated left ventricular noncompaction (LVNC) results from excessive trabeculation and impaired myocardial compaction during heart development. The extracellular matrix (ECM) that separates endocardium from myocardium plays a critical but poorly understood role in ventricular trabeculation and compaction. In an attempt to characterize solute carrier family 39 member 8–null (Slc39a8-null) mice, we discovered that homozygous null embryos do not survive embryogenesis and exhibit a cardiac phenotype similar to human LVNC. Slc39a8 encodes a divalent metal cation importer that has been implicated in ECM degradation through the zinc/metal regulatory transcription factor 1 (Zn/MTF1) axis, which promotes the expression of ECM-degrading enzymes, including Adamts metalloproteinases. Here, we have shown that Slc39a8 is expressed by endothelial cells in the developing mouse heart, where it serves to maintain cellular Zn levels. Furthermore, Slc39a8-null hearts exhibited marked ECM accumulation and reduction of several Adamts metalloproteinases. Consistent with the in vivo observations, knockdown of SLC39A8 in HUVECs decreased ADAMTS1 transcription by decreasing cellular Zn uptake and, as a result, MTF1 transcriptional activity. Our study thus identifies a gene underlying ventricular trabeculation and compaction development, and a pathway regulating ECM during myocardial morphogenesis.
Keywords: Cardiology, Development
Keywords: Extracellular matrix, Genetic diseases, Mouse models
Introduction
Cardiac ventricular morphogenesis begins from a tubular structure composed of an outer 1-cell layer of myocardium and an inner 1-cell layer of endocardium separated by extracellular matrix (ECM), or cardiac jelly (1). Trabeculation in mice begins at embryonic day 9 (E9.0), when myocardial cells protrude into cardiac jelly to form projections lined by endocardium: structures named trabeculae. Trabeculae rapidly grow and expand between E9.5 and E13.5. By E14.5, trabeculae are long and thin, with little cardiac jelly between myocardium and endocardium. Concomitant with the development of coronary circulation, trabeculae go through compaction, collapsing into the ventricle wall and becoming part of the compact myocardium. Therefore, the mature ventricle has thick compact myocardium with few trabeculae (2, 3). Endocardium plays unique and critical roles in trabeculation and compaction by secreting signaling molecules that orchestrate cardiomyocyte proliferation and differentiation and enzymes that are involved in ECM production and degradation (1, 4, 5).
Many forms of congenital heart defects result from, or are associated with, abnormal cardiac trabeculation and compaction. For instance, left ventricular noncompaction (LVNC) is a cardiomyopathy caused by arrested compaction. It is characterized by excessive trabeculation with deep intertrabecular recesses and thin compact myocardium (6). It usually affects both ventricles, but is more prominent on the left in humans. In the pediatric population, LVNC is the third most common cardiomyopathy after dilated cardiomyopathy and hypertrophic cardiomyopathy (7). Typical complications of LVNC include heart failure, ventricular arrhythmias, and systemic embolic events (8). Mutations in genes encoding sarcomeric, mitochondrial, cytoskeletal, and nuclear membrane proteins, and Notch signaling pathway components have been identified in LVNC patients (9–13), but the molecular mechanisms remain largely unresolved, reflecting a lack of understanding of the compaction process.
Cardiac jelly serves as the media for the signal exchanges between endocardium and myocardium (14). Perturbation of cardiac jelly production or degradation has been shown to cause abnormal trabeculation and compaction. Two major components of cardiac jelly are hyaluronan, a mucopolysaccharide, and versican, a chondroitin sulfate proteoglycan. Mice lacking hyaluronan synthase-2 or versican do not develop trabeculae (15–17). On the other hand, a substantial amount of cardiac jelly needs to be degraded prior to ventricular compaction. Cardiac jelly degradation is mediated by metalloproteinases including Adamts family members. Adamts1-knockout mice develop hypertrabeculation and Adamts9-haploinsufficient mice develop ventricular noncompaction accompanied by cardiac jelly accumulation (5, 18). Conversely, mice overexpressing Adamts1 or Adamts5 exhibit sparse trabeculation due to excessive cardiac jelly degradation (5, 19). However, the upstream factors regulating Adamts expression, and the mechanism by which cardiac jelly regulates trabeculation and compaction are not well understood.
Zinc (Zn) is required for the structure and function of a variety of enzymes and transcription factors (20). Zn deficiency has been shown to result in developmental defects including multiple types of cardiac abnormalities (21, 22). Zn homeostasis is primarily regulated by 10 Zn exporters and 14 Zn importers (23). Solute carrier family 39 member 8 (Slc39a8, which encodes a protein historically termed ZIP8) is a cellular Zn importer (24) and has been shown to play a causal role in the pathogenesis of osteoarthritis by promoting cellular Zn uptake, resulting in increased metal-regulatory transcription factor 1 (Mtf1) activity, increased Adamts expression, and ECM degradation (25). Here, we found that Slc39a8-null mice exhibit typical ventricular noncompaction phenotypes associated with reduced Adamts transcription and reduced cardiac jelly degradation. Our study demonstrates that ZIP8 is crucial for ventricular trabeculation and compaction, reveals a potentially novel regulator of ventricular myocardial development, and provides a valuable model to study ventricular noncompaction.
Results
Slc39a8 is expressed in the developing heart and regulates Zn levels.
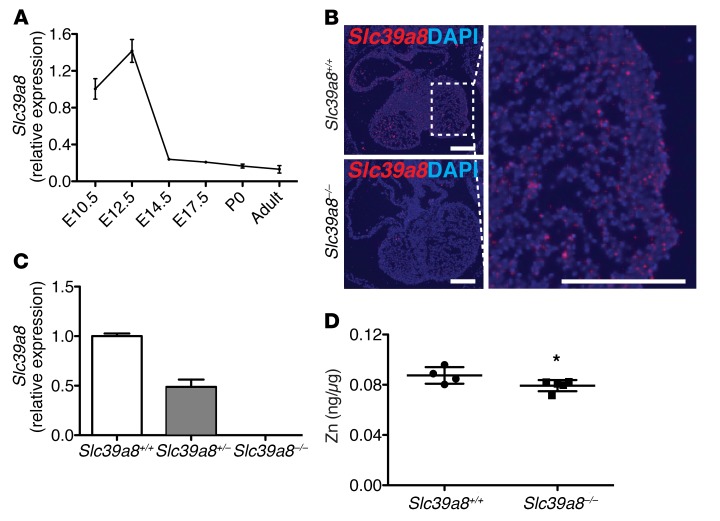
By quantitative reverse transcription PCR (qRT-PCR) analysis on whole-heart lysates, we found that Slc39a8 was expressed in the developing heart. The expression peaked at E12.5 and then gradually declined to low levels in adult heart (Figure 1A). RNA in situ hybridization further verified the expression of Slc39a8 in E12.5 heart (Figure 1B). Further qRT-PCR analysis on single-cell populations isolated by fluorescence-activated cell sorting (FACS) demonstrated that Slc39a8 was expressed in cardiac endothelial cells of E12.5 hearts (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI96993DS1). To study the role of Slc39a8 in cardiac ventricular morphogenesis, we generated Slc39a8–/– mice by removing the third exon of the Slc39a8 gene (26). Slc39a8 mRNA was efficiently deleted in the Slc39a8–/– hearts (Figure 1C). Consistent with the function of ZIP8 to promote cellular Zn uptake (24), Zn levels were lower in E14.5 Slc39a8–/– hearts compared with Slc398+/+ hearts (P = 0.06) (Figure 1D).
Figure 1. Slc39a8 is expressed in the developing hearts and regulates Zn levels.
(A) qRT-PCR analysis of Slc39a8 in the whole heart at different developmental stages. Gapdh was used as cDNA loading control. n = 3 for each time point. (B) RNA in situ hybridization showed that Slc39a8 is expressed in the trabecular region of E12.5 hearts. Scale bars: 250 μm. (C) qRT-PCR analysis showed that Slc39a8 was efficiently deleted in E12.5 Slc39a8–/– hearts. n = 3 for each genotype. (D) ICP-MS analysis showed that Zn was reduced in E14.5 Slc39a8–/– hearts as compared with Slc39a8+/+ hearts. Results were normalized to protein content. *P = 0.06 by Student’s t test.
Slc39a8 deletion results in hypertrabeculation and noncompaction.
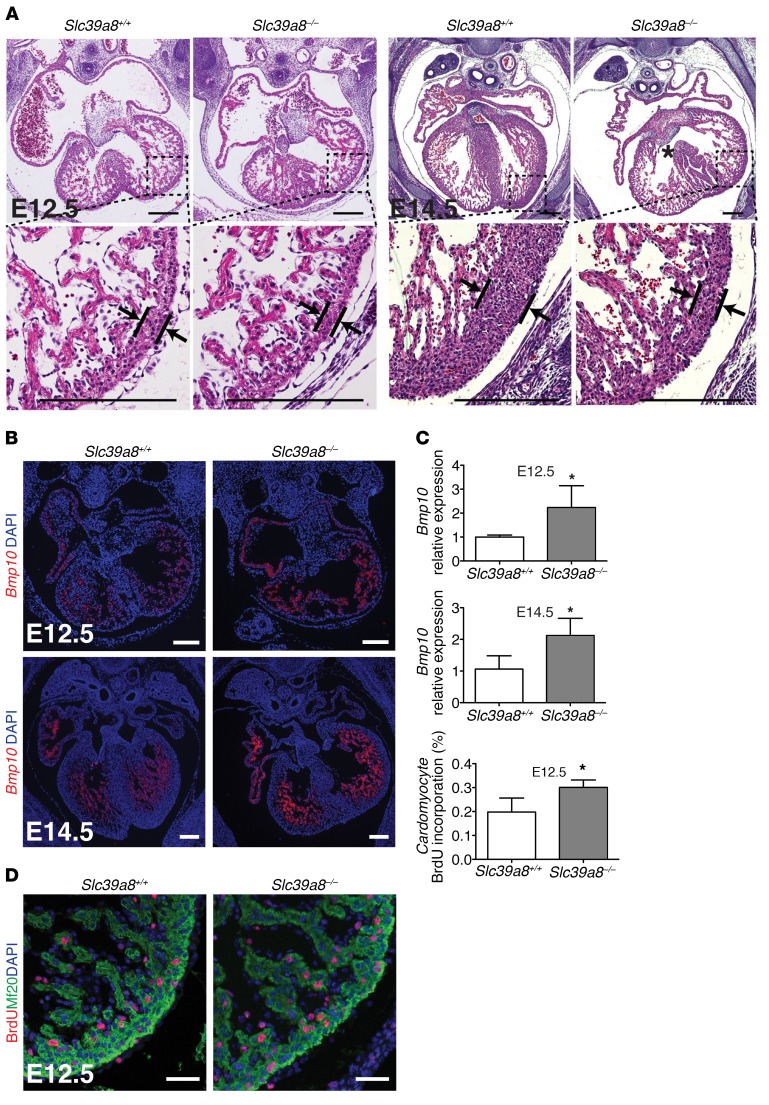
Slc39a8+/– mice appeared phenotypically indistinguishable from Slc39a8+/+ mice. Slc39a8+/– mice were fertile and had normal life spans. Genotyping of Slc39a8+/– breeding progenies at different embryonic time points and at weaning revealed that Slc39a8–/– mice were embryonic lethal by E16.5 (Supplemental Table 1). Histological analysis revealed that compared with Slc39a8+/+ hearts, Slc39a8–/– hearts exhibited hypertrabeculation and noncompaction phenotypes including excessive trabeculae (+24.9%, P < 0.01 for E12.5 and +44.2%, P < 0.05 for E14.5) and thin compact myocardium (–53.9%, P < 0.001 for E12.5 and –72.7%, P < 0.001 for E14.5), which are the hallmarks of LVNC (Figure 2A and Supplemental Figure 2). These phenotypes were evident at E12.5 and prominent at E14.5, and Slc39a8–/– embryos that survived until E16.5 had an even stronger phenotype (Supplemental Figure 3). We also observed ventricular septal defects (VSDs) in Slc39a8–/– hearts (Figure 2A). Some E14.5 Slc39a8–/– embryos exhibited body edema, suggesting that cardiac muscle function was compromised. Further analysis by both in situ hybridization and qRT-PCR demonstrated that Slc39a8–/– hearts had elevated expression of Bmp10, a cytokine that is an established trabecular marker, and which is known to promote cell proliferation during trabeculation (Figure 2, B and C, and ref. 4). In agreement with this, BrdU immunostaining demonstrated that cardiomyocyte proliferation was significantly increased in E12.5 Slc39a8–/– hearts compared with that of Slc39a8+/+ hearts (Figure 2D), while cell death was similar between groups at the same stage (Supplemental Figure 4).
Figure 2. Slc39a8 deletion results in LVNC.
(A) H&E staining showed that Slc39a8–/– hearts exhibited hypertrabeculation and noncompaction at E12.5 and E14.5. Arrows indicate compact myocardium. The asterisk indicates a ventricular septal defect (VSD). (B) RNA in situ hybridization demonstrated that Bmp10 transcriptional signal was increased in Slc39a8–/– hearts as compared with Slc39a8+/+ hearts at E12.5 and E14.5. (C) qRT-PCR analysis showed that Bmp10 was significantly increased in E12.5 and E14.5 Slc39a8–/– hearts. n = 4 for each genotype. (D) IF staining showed that BrdU+ cardiomyocyte number was significantly increased in Slc39a8–/– hearts as compared with Slc39a8+/+ hearts at E12.5. Embryos were harvested 3 hours after the pregnant mice were intraperitoneally injected with BrdU (100 mg/kg body weight). Mf20 marks cardiomyocytes. Ratio of BrdU+ cardiomyocytes to total cardiomyocytes per section at similar anatomical levels was calculated. n = 4 for each genotype. *P < 0.05 by Student’s t test. Scale bars: 250 μm (A and B) and 50 μm (D).
Slc39a8 deletion leads to decreased expression of Adamts metalloproteinases and impaired cardiac ECM degradation.
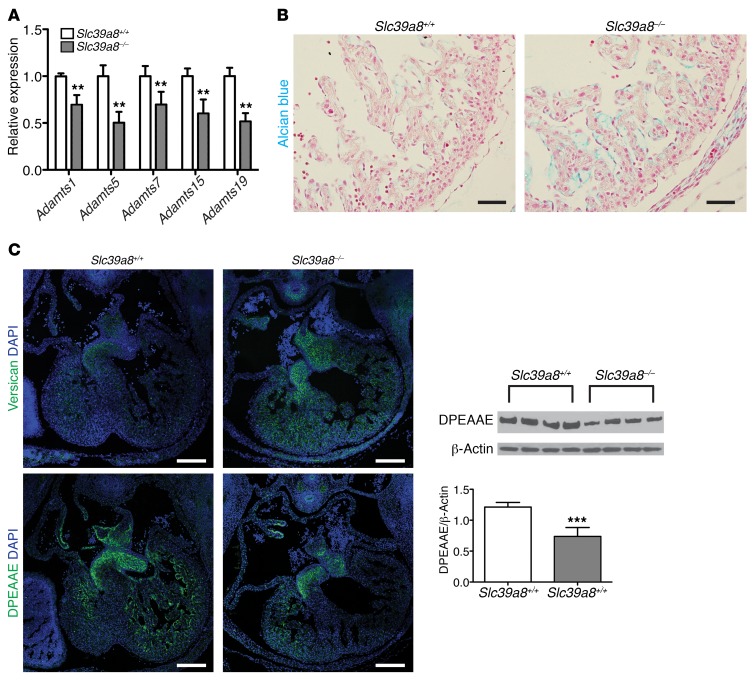
To understand the potential molecular mechanisms by which Slc39a8 regulates ventricular trabeculation and compaction, we performed a microarray analysis with E12.5 Slc39a8+/+ and Slc39a8–/– hearts. Interestingly, multiple Adamts metalloproteinases, including Adamts5, -7, -15, and -19, were found to be downregulated in Slc39a8–/– hearts (Supplemental Figure 5A). These findings were further verified by qRT-PCR analysis, which showed a 31% decrease of Adamts1 (P = 0.001), 50% decrease of Adamts5 (P < 0.001), 30% decrease of Adamts7 (P = 0.013), 40% decrease of Adamts15 (P = 0.003), and 48% decrease of Adamts19 (P < 0.001) in Slc39a8–/– hearts compared with Slc39a8+/+ hearts (Figure 3A). Adamts metalloproteinases are key proteinases that mediate cardiac ECM degradation (5, 19). Consistent with this, the ECM pathway was the top downregulated pathway in our GO analysis of the microarray results (Supplemental Figure 5B). We therefore examined ECM by Alcian blue staining, which detects major cardiac ECM components including hyaluronan and versican (18, 19). Consistent with decreased expression of Adamts metalloproteinases, the intensity of Alcian blue staining was substantially increased in E12.5 Slc39a8–/– hearts compared with Slc39a8+/+ hearts (Figure 3B). Furthermore, immunofluorescence (IF) staining demonstrated that versican was markedly increased, while DPEAAE, a cleavage product of versican by Adamts, was markedly decreased in E12.5 Slc39a8–/– hearts compared with Slc39a8+/+ hearts (Figure 3C). The decrease in DPEAAE was further verified and quantified by Western blot analysis (–39.1%, P = 0.001) (Figure 3C). These observations provided strong evidence that cardiac jelly degradation was impaired in Slc39a8–/– hearts. Aberrant cardiac ECM accumulation can perturb signal exchanges between endocardium and myocardium (5). In agreement with this, IF staining and Western blot analysis demonstrated that NICD1, the intracellular domain of Notch receptor 1 and an indicator of activated Notch signaling, was significantly increased in E12.5 Slc39a8–/– hearts (Supplemental Figure 6). Taken together, our results demonstrated that Slc39a8 deletion resulted in aberrant cardiac ECM accumulation due to impaired degradation as a result of decreased expression of Adamts metalloproteinases.
Figure 3. Slc39a8 deletion leads to decreased expression of Adamts metalloproteinases and impaired cardiac ECM degradation.
(A) qRT-PCR analysis showed decreased expression of Adamts metalloproteinases in Slc39a8–/– hearts at E12.5. n = 3 for each genotype. **P < 0.01. (B) Alcian blue staining showed that cardiac ECM was substantially increased in Slc39a8–/– hearts as compared with Slc39a8+/+ hearts at E12.5. Scale bars: 50 μm. (C) IF staining demonstrated that the immunoreactive signal of versican was greatly increased in Slc39a8–/– hearts, whereas DPEAAE was markedly reduced in Slc39a8–/– hearts as compared with Slc39a8+/+ hearts at E12.5. Scale bars: 200 μm. Western blot of DPEAAE on E12.5 Slc39a8+/+ and Slc39a8–/– hearts is shown on the right. n = 4 for each genotype. β-Actin was used as protein loading control. The densitometry analysis is shown at the right bottom panel. ***P < 0.001 by Student’s t test.
SLC39A8 knockdown in HUVECs results in decreased ADAMTS1 expression and MTF1 transcriptional activity.
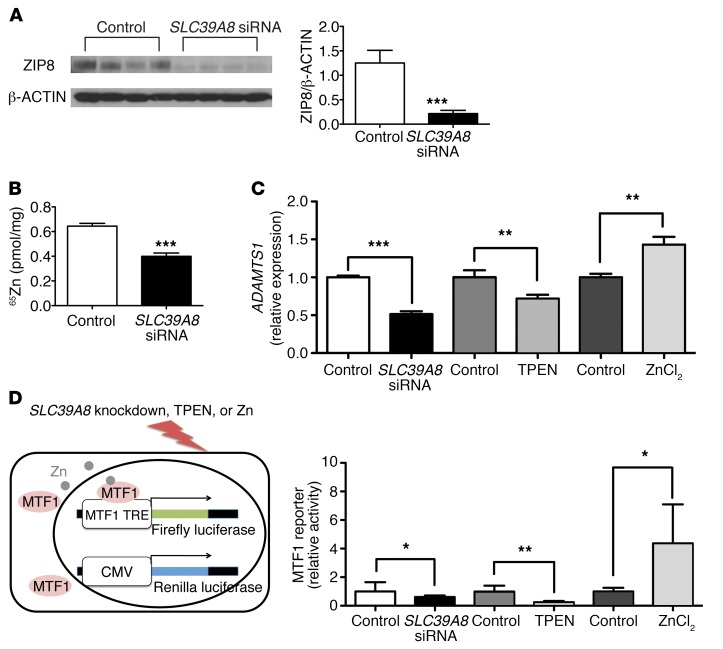
Adamts metalloproteinases are primarily expressed in endocardial cells in the developing heart, where ZIP8 is also expressed. To further explore the potential molecular mechanism by which ZIP8 regulates Adamts expression, we knocked down SLC39A8 in human umbilical vein endothelial cells (HUVECs) using siRNA. Western blot analysis confirmed effective knockdown of SLC39A8 (Figure 4A). Consistent with decreased Adamts in embryonic Slc39a8–/– hearts, SLC39A8 knockdown resulted in decreased ADAMTS1 expression in HUVECs (–49%, P < 0.001) (Figure 4C). It has been reported that ZIP8 regulates Adamts expression by modulating cellular Zn uptake and subsequently Mtf1 transcriptional activity during osteoarthritis (25). Consistent with this model and the in vivo observation of reduced Zn in Slc39a8–/– embryonic hearts, SLC39A8 knockdown led to reduced cellular Zn uptake (–38%, P = 0.0002) (Figure 4B).
Figure 4. SLC39A8 knockdown in HUVECs results in decreased ADAMTS1 expression and MTF1 transcriptional activity.
(A) Western blot and densitometry analysis showed that SLC39A8 siRNA treatment resulted in efficient SLC39A8 knockdown in HUVECs. HUVECs were treated with SLC39A8 siRNAs or control scramble siRNAs for 24 hours. n = 4 for each condition. ***P < 0.001 by Student’s t test. (B) 65Zn uptake was significantly reduced in SLC39A8 siRNA–treated HUVECs as compared with that in control siRNA–treated cells. n = 4 for each group. ***P < 0.001 by Student’s t test. (C) qRT-PCR analysis of ADAMTS1 expression in HUVECs treated with SLC39A8 siRNA, TPEN, or ZnCl2. ADAMTS1 expression was significantly decreased in SLC39A8 siRNA–treated or TPEN-treated HUVECs, but increased in ZnCl2-treated HUVECs. n = 4 for each condition. **P < 0.01; ***P < 0.001 by Student’s t test. (D) A schematic representation of the MTF1 reporter assay is shown on the left. TRE, tandem response element. MTF1 transcriptional activities were significantly decreased in HUVECs treated with SLC39A8 siRNA or TPEN but significantly increased in HUVECs treated with ZnCl2 as compared with the corresponding controls (n = 8, n = 6, and n = 4 respectively). Firefly luciferase activities were normalized to that of Renilla. MTF1 reporter activities in each treatment condition were normalized to the average of the corresponding control groups. *P < 0.05, **P < 0.01 by Student’s t test.
To test whether reduced Zn influx contributes to the reduction of ADAMTS1 expression by SLC39A8 knockdown, we treated HUVECs with TPEN, a Zn-specific metal ion chelator (27). Interestingly, TPEN treatment also resulted in reduced ADAMTS1 expression (–28%, P = 0.009). Conversely, ZnCl2 treatment increased ADAMTS1 expression (+43%, P = 0.003) (Figure 4C). Lastly, we sought to understand whether SLC39A8 regulates the expression of ADAMTS1 through modulating MTF1 transcriptional activity by Zn influx. We examined MTF1 transcriptional activity using a reporter construct in which firefly luciferase reporter gene expression is under the control of an MTF1 transcriptional response element (ref. 27 and Figure 4D). Both SLC39A8 knockdown and TPEN treatment resulted in significantly decreased MTF1 transcriptional activity (–39%, P = 0.05 and –74%, P = 0.001), while ZnCl2 treatment greatly enhanced MTF1 transcriptional activity (4.37-fold, P = 0.05) (Figure 4E). Taken together, these results strongly suggest that SLC39A8 regulates ADAMTS1 expression by modulating cellular Zn uptake and consequently MTF1 transcriptional activity.
Discussion
Ventricular noncompaction is a cardiomyopathy that can develop severe complications over time. Excessive cardiac jelly degradation leads to hypotrabeculation, while impaired cardiac jelly degradation results in hypertrabeculation and noncompaction. While the amount of cardiac jelly is clearly both important and under dynamic temporal control, it is not well understood how its degradation is regulated as myocardial morphogenesis progresses. In this study, we demonstrated that Slc39a8–/– mice developed ventricular hypertrabeculation and noncompaction and impaired cardiac jelly degradation as a result of decreased Adamts metalloproteinase expression. Consistently, knocking down SLC39A8 in HUVECs decreased ADAMTS1 expression by reducing cellular Zn uptake and MTF1 transcriptional activity.
Slc39a8 hypomorphic mice have been reported to exhibit diminished Zn uptake, multiple-organ hypoplasia, anemia, and perinatal death, although it was not reported whether there were cardiac defects in those mice (28). There were quite a few similarities in phenotypes between our Slc39a8–/– mice and the Slc39a8 hypomorphic mice, such as early embryonic/neonatal lethality and reduced cellular Zn uptake. However, it does appear that the phenotypes of Slc39a8–/– mice are more severe than those of the Slc39a8 hypomorphic mice, which is likely a reflection of the degree of the loss of gene function in these 2 different lines of mice.
One key finding of our study is that deletion of Slc39a8, a Zn transporter, in mouse leads to ventricular noncompaction. LVNC is classified by the American Heart Association as a primary genetic cardiomyopathy (29). Familial occurrence is frequent, and autosomal dominant, autosomal recessive, X-linked, and maternal transmissions have been observed (30). Multiple genetic mutations have been identified in LVNC patients, but the molecular mechanism underlying LVNC remains elusive (30). Our results support the hypothesis that LVNC is a distinct genetic cardiomyopathy with a developmental basis. It establishes the causal relationship between Slc39a8 loss of function and LVNC in mouse and provides insight into the mechanism underlying the disease. Our study suggests that SLC39A8 might be considered as a candidate gene when screening LVNC patients for gene mutations. The clinical features of LVNC are often heterogeneous: with or without association with other heart defects or cardiomyopathy (31). Based on our current Slc39a8 genetic deletion findings, we expect that patients carrying SLC39A8 loss-of-function mutations might present typical ventricular noncompaction features, accompanied by other heart defects such as VSD and cardiomyopathy, though the severity and spectrum of the clinical presentation is likely to be affected by the nature of the SLC39A8 mutations and epigenetic factors. Our study also suggests that Zn deficiency (regardless of its cause) may be involved in the pathogenesis of LVNC. This is in agreement with previous studies showing that maternal Zn deficiency resulted in multiple forms of cardiac abnormalities including thin ventricular wall and irregular trabeculae (21, 22). Genetic testing for SLC39A8 mutations in cases of LVNC may be worth consideration.
Another key finding of our study is that the Zn/ZIP8/MTF1 axis regulates the genes encoding multiple Adamts metalloproteinases, and consequently impacts cardiac jelly degradation during trabeculation and compaction. It is known that Adamts metalloproteinases are repressed at the initial stage of trabeculation when cardiac jelly is required (5, 19). However, it is unknown how they are de-repressed at the end of the trabeculation stage to degrade cardiac jelly and to terminate trabeculation. In chondrocytes, the Zn/ZIP8/MTF1 axis has been reported to upregulate matrix-degrading enzymes including Adamts5 during osteoarthritis (25). We demonstrated that this regulation is conserved in endothelial cells and that it plays a potentially novel and important role in the regulation of Adamts, cardiac jelly degradation, and myocardial morphogenesis. An interesting observation is that Slc39a8 expression peaked at E12.5, which matched very well with the initiation of Adamts1 de-repression, cardiac jelly degradation, and trabeculation termination (5).
Various mouse models have established a critical role of cardiac jelly in trabeculation and compaction. Although the mechanism is not completely understood, one hypothesis is that cardiac jelly defects can disrupt signaling transduction between endocardium and myocardium. Our observation of elevated Notch signaling and Bmp10 in Slc39a8–/– heart provide support for this hypothesis. Further studies are required to elucidate the mechanism underlying the interaction between cardiac jelly and signaling transduction and the impact it has on myocardial morphogenesis.
Collectively, our study revealed a potentially novel gene underlying ventricular noncompaction and a regulatory pathway mediating ECM degradation during myocardial morphogenesis. It suggests that SLC39A8 may be added to the list of genes to be screened in patients with ventricular noncompaction or other diseases involving dysregulation of ECM degradation.
Methods
Mice.
Mice harboring an Slc39a8 constitutive knockout allele, C57BL/6-Slc39a8tm1.2 mrl, were generated and provided by Merck. Details of the design of the Slc39a8–/– mice can be found at http://www.taconic.com/mouse-model/slc39a8-ko The constitutive knockout allele was detected using genotyping primers 2476_27 (5′-CAGGGTTTCTCTGTGTAACAGG-3′) and 2476_32 (5′-CCAATATGGCCATAACAGATAGG-3′). The R26REYFP mice, which have a loxP-flanked STOP sequence followed by the enhanced yellow fluorescent protein (EYFP) gene inserted into the Gt(ROSA)26Sor locus, were purchased from the Jackson Laboratory (stock 006148) and crossed with Tie2-Cre transgenic mice (stock 004128), also obtained from the Jackson Laboratory.
Cell culture.
HUVECs (Lonza) were cultured in VascuLife EnGS medium (Lifeline, catalog LL-0002). To knock down SLC39A8, cells were plated at a density of 2.5 × 104 per well in a 12-well plate. The next day (at ~70% confluence), cells were transfected either with 10 nM Ambion Silencer Select control siRNA (Life Technologies, catalog 4390843) or SLC39A8 siRNA (Life Technologies, catalog 4392420) using RNAiMax (Life Technologies, catalog 13778075). Cells were used for experiment 8 hours after siRNA treatment. To chelate Zn, cells were treated with 1 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) (Sigma-Aldrich, catalog P4413) for 24 hours. To load cells with Zn, cells were treated with ZnCl2 (25 μM) for 4 hours.
For the MTF1 reporter assay (Qiagen, catalog CCS-5033L) (25), cells were transfected with a mixture of a MTF1-responsive firefly luciferase construct and a constitutive Renilla luciferase construct (40:1) by reverse transfection using Lipofectamine 3000 (Thermo Fisher Scientific, catalog L3000015). Cells (1 × 104), 120 ng reporter construct mixture, 0.12 μl P3000, and 0.2 μl Lipofectamine 3000 were used per well of a 96-well plate. To study the effect of SLC39A8 knockdown on MTF1 reporter activity, cells were transfected with MTF1 reporter constructs for 24 hours and then treated with 20 nM control siRNA or SLC39A8 siRNA using RNAiMax for 8 hours before being harvested for the MTF1 reporter assay. To study the effect of Zn chelation, cells were treated with TPEN (1 μM) 24 hours after transfection of the reporter construct and the MTF1 reporter assay was performed 24 hours later. To study the effect of Zn overload, cells were treated with ZnCl2 (25 μM) 24 hours after transfection of the reporter construct and the MTF1 reporter assay was performed 4 hours later. The MTF1 reporter assay was performed using Dual-Luciferase Reporter Assay System (Promega, catalog E1910).
Histology, IF staining, RNA in situ hybridization, and Alcian blue staining.
Histology, IF staining, and Alcian blue staining were performed as was previously described (19). Embryos were fixed in 2% paraformaldehyde overnight, dehydrated using graded ethanol, and embedded in paraffin. Eight-micrometer-thick sections were collected. The antibodies used for IF staining were rat anti–PECAM-1 (1:500, BD PharMingen, catalog 553370), rabbit anti-versican (1:200, Millipore, catalog AB1033), rabbit anti-DPEAAE (1:200, Pierce, catalog PA1-1748A), rat anti-BrdU (1:20, Accurate Chemical & Scientific Corp, catalog OBT0030S), and rabbit anti-NICD1 (1:50, Cell Signaling Technology, catalog 4147). Bmp10 in situ hybridization probe was obtained from Weinian Shou (Indiana University, Indianapolis, Indiana, USA). Primers used to generate Slc39a8 in situ hybridization probe are 5′-TGTGACTTGCTATGCCAACC-3′ and 5′-TCTCCTGCGTACAAGGTGAT-3′. IF images were acquired on a Zeiss LSM 710 confocal microscope.
FACS.
E12.5 embryos were isolated and kept in ice-cold PBS. EYFP-positive embryos were identified by their EYFP epifluorescence. Hearts were dissected and digested with collagenase I (0.5 mg/ml in DMEM basal medium) for 30 minutes on a shaker at 37°C and dissociated by pipetting every 5 to 10 minutes during the digestion. Dissociated cells were added into 10 ml DMEM, filtered through a 40-μm cell strainer, centrifuged, and resuspended in sorting buffer (Ca2+/Mg2+–free PBS, 1.5% BSA, and 25 mM HEPES). Cells from EYFP-negative embryos were used to set the EYFP-negative gate, and EYFP-positive and EYFP-negative cells from EYFP-positive embryonic hearts were sorted and collected in RLT buffer of the RNeasy Mini kit (Qiagen, catalog 74104).
Zn measurements.
To measure Zn, mouse embryonic hearts were homogenized in 100 μl PBS (Ca2+/Mg2+ free) and Zn was measured by inductively coupled plasma mass spectrometry (ICP-MS) using an Elan 6100 (PerkinElmer) at the PADLS New Bolton Center Toxicology Laboratory, School of Veterinary Medicine of the University of Pennsylvania, as was previously described (32). Briefly, after the weight and volume of E14.5 hearts were measured, the sample was digested overnight with twice the amount (weight/volume) of 70% nitric acid at 70°C and 0.15 ml of the digested sample was diluted with deionized water to a final volume of 5 ml for analysis. The concentration was measured using a calibration curve of aqueous standards prepared at 4 different concentrations of Zn. Results are reported in ppm on a weight basis. Total amount of Zn in a heart was calculated and normalized to total amount of protein in the heart measured by BCA.
To study cellular Zn uptake, ZnCl2 and 65ZnCl2 (PerkinElmer, catalog NEZ111) were added into HBSS buffer (Ca2+/Mg2+ free) to make the uptake buffer containing 0.1 μM of ZnCl2 and 0.04 μCi/ml of radioactivity. Cells were washed with HBSS buffer (Ca2+/Mg2+ free) twice and incubated with the uptake buffer at 37°C. Five minutes later, the uptake buffer was removed and cells were washed 3 times with HBSS buffer (Ca2+/Mg2+ free). Five hundred microliters of 0.1N NaOH was added into each well to digest the cells. Two hours later, 400 μl of cell lysate was used for liquid scintillation counting and 50 μl of cell lysate was used for BCA protein assay.
Microarray and qRT-PCR analysis.
Total RNA was isolated using an RNeasy Mini kit. Microarray analysis was performed using Affymetrix Mouse Gene 2.0 ST chips, as previously described (33). Functional annotation was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (34). The data set can be accessed via the NCBI’s gene expression omnibus (GEO GSE103673). cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, catalog 4368814). qPCR was performed using Fast SYBR Green Mastermix (Life Technologies, catalog 4385614). A list of SYBR primers used can be found in Supplemental Table 2.
Western blot.
Embryonic mouse hearts were homogenized in 100 μl PBS with protease and phosphatase inhibitors. Protein (15 μg) was separated via a NuPage SDS Page system (Invitrogen, catalog NP0322BOX). Primary antibodies used were anti-NICD1 (1:1,000, Cell Signaling Technology, catalog 4147S), anti-DPEAAE (1:2,500, Thermo Fisher Scientific, catalog PA1-1748A), and anti–β-actin (1:1,000, Santa Cruz Biotechnology, catalog sc-81178). HRP-conjugated anti-rabbit and anti-mouse secondary antibodies (1:2,500, GE Healthcare Life Sciences, catalog NA934V and NA931V) and SuperSignal West Femto Maximum Sensitivity Substrate (Life Technologies, catalog 34094) were used to visualize the signal.
Statistics.
Two-tailed Student’s t tests were used to detect differences between 2 groups in all experiments unless otherwise noted. The χ2 test was performed to compare the observed genotype distribution of Slc39a8+/– breeding progenies with expected Mendelian ratios. All data represent the mean ± SD unless otherwise noted. P less than 0.05 was considered to be significant.
Study approval.
All mouse experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania following guidelines described in the NIH Guide for the Care and Use of Laboratory Animals.
Author contributions
WL, DL, JAE, and DJR designed experiments, analyzed the data, and wrote the manuscript. WL and DL performed the experiments. NJH supervised experiments, analyzed data, and edited the manuscript. LC, LL, and FL provided technical help.
Supplementary Material
Acknowledgments
We thank Ying Tian and Zinan Zhou for helpful discussions. We thank Christi Kern for providing important reagents. We thank the Penn CVI Histology Core and the Flow Cytometry and Cell Sorting Resource Laboratory for technical assistance and the Department of Cell and Developmental Biology Microscopy Core for confocal imaging. We thank the Penn New Bolton Center Toxicology Lab for performing the ICP-MS experiment. This work was supported by the NIH (grants RC2HL101864 and R01HL089309 to DJR and U01 HL100405 to JAE), the Cotswold Foundation, and the WW Smith Endowed Chair to JAE.
Version 1. 01/16/2018
Electronic publication
Version 2. 02/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(2):826–833.https://doi.org/10.1172/JCI96993.
Contributor Information
Wen Lin, Email: linwen@mail.med.upenn.edu.
Deqiang Li, Email: deqli@mail.med.upenn.edu.
Lan Cheng, Email: lancheng@mail.med.upenn.edu.
Li Li, Email: li10@mail.med.upenn.edu.
Feiyan Liu, Email: feiyan@mail.med.upenn.edu.
Jonathan A. Epstein, Email: epsteinj@mail.med.upenn.edu.
Daniel J. Rader, Email: rader@mail.med.upenn.edu.
References
- 1.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C(3):144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258(4):319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Oechslin E, Jenni R. Left ventricular non-compaction revisited: a distinct phenotype with genetic heterogeneity? Eur Heart J. 2011;32(12):1446–1456. doi: 10.1093/eurheartj/ehq508. [DOI] [PubMed] [Google Scholar]
- 4.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.[No authors listed] Retraction notice to “Activation and silencing of matrix metalloproteinases” [Semin. Cell Dev. Biol. 19 (2008) 2–13] Semin Cell Dev Biol. 2009;20(3):375. doi: 10.1016/j.semcdb.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Towbin JA. Left ventricular noncompaction: a new form of heart failure. Heart Fail Clin. 2010;6(4):453–69, viii. doi: 10.1016/j.hfc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Nugent AW, et al. The epidemiology of childhood cardiomyopathy in Australia. N Engl J Med. 2003;348(17):1639–1646. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 8.Oechslin EN, Attenhofer Jost CH, Rojas JR, Kaufmann PA, Jenni R. Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis. J Am Coll Cardiol. 2000;36(2):493–500. doi: 10.1016/S0735-1097(00)00755-5. [DOI] [PubMed] [Google Scholar]
- 9.Klaassen S, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117(22):2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, et al. Mutation analysis of the G4.5 gene in patients with isolated left ventricular noncompaction. Mol Genet Metab. 2002;77(4):319–325. doi: 10.1016/S1096-7192(02)00195-6. [DOI] [PubMed] [Google Scholar]
- 11.Ichida F, et al. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103(9):1256–1263. doi: 10.1161/01.CIR.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 12.Hermida-Prieto M, et al. Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am J Cardiol. 2004;94(1):50–54. doi: 10.1016/j.amjcard.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 13.Schlegelberger B. In memoriam: Prof. Dr. rer. nat. Dr. med. h.c. Lore Zech; 24.9.1923 - 13.3.2013: Honorary member of the European Society of Human Genetics, Honorary member of the German Society of Human Genetics, Doctor laureate, the University of Kiel, Germany. Mol Cytogenet. 2013;6(1):20. doi: 10.1186/1755-8166-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart M, Wirrig E, Phelps A, Wessels A. Extracellular matrix and heart development. Birth Defects Res Part A Clin Mol Teratol. 2011;91(6):535–550. doi: 10.1002/bdra.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camenisch TD, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106(3):349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooley MA, et al. Fibulin-1 is required during cardiac ventricular morphogenesis for versican cleavage, suppression of ErbB2 and Erk1/2 activation, and to attenuate trabecular cardiomyocyte proliferation. Dev Dyn. 2012;241(2):303–314. doi: 10.1002/dvdy.23716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatano S, et al. Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology. 2012;22(9):1268–1277. doi: 10.1093/glycob/cws095. [DOI] [PubMed] [Google Scholar]
- 18.Kern CB, et al. Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 2010;29(4):304–316. doi: 10.1016/j.matbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell. 2015;32(2):168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc-proteins encoded in the human genome. J Proteome Res. 2006;5(1):196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 21.Duffy JY, Overmann GJ, Keen CL, Clegg MS, Daston GP. Cardiac abnormalities induced by zinc deficiency are associated with alterations in the expression of genes regulated by the zinc-finger transcription factor GATA-4. Birth Defects Res B Dev Reprod Toxicol. 2004;71(2):102–109. doi: 10.1002/bdrb.20004. [DOI] [PubMed] [Google Scholar]
- 22.Lopez V, Keen CL, Lanoue L. Prenatal zinc deficiency: influence on heart morphology and distribution of key heart proteins in a rat model. Biol Trace Elem Res. 2008;122(3):238–255. doi: 10.1007/s12011-007-8079-2. [DOI] [PubMed] [Google Scholar]
- 23.Sekler I, Sensi SL, Hershfinkel M, Silverman WF. Mechanism and regulation of cellular zinc transport. Mol Med. 2007;13(7–8):337–343. doi: 10.2119/2007-00037.Sekler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begum NA, Kobayashi M, Moriwaki Y, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80(6):630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, et al. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell. 2014;156(4):730–743. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Lin W, et al. Hepatic metal ion transporter ZIP8 regulates manganese homeostasis and manganese-dependent enzyme activity. J Clin Invest. 2017;127(6):2407–2417. doi: 10.1172/JCI90896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radford RJ, Lippard SJ. Chelators for investigating zinc metalloneurochemistry. Curr Opin Chem Biol. 2013;17(2):129–136. doi: 10.1016/j.cbpa.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CY, et al. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J Biol Chem. 2012;287(41):34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maron BJ, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113(14):1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 30.Finsterer J, Stöllberger C, Towbin JA. Left ventricular noncompaction cardiomyopathy: cardiac, neuromuscular, and genetic factors. Nat Rev Cardiol. 2017;14(4):224–237. doi: 10.1038/nrcardio.2016.207. [DOI] [PubMed] [Google Scholar]
- 31.Captur G, Nihoyannopoulos P. Left ventricular non-compaction: genetic heterogeneity, diagnosis and clinical course. Int J Cardiol. 2010;140(2):145–153. doi: 10.1016/j.ijcard.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Stout JD, Brinker DF, Driscoll CP, Davison S, Murphy LA. Serum biochemistry values, plasma mineral levels, and whole blood heavy metal measurements in wild northern goshawks (Accipiter gentilis) J Zoo Wildl Med. 2010;41(4):649–655. doi: 10.1638/2009-0258.1. [DOI] [PubMed] [Google Scholar]
- 33.Barbarulo A, et al. Hedgehog signalling in the embryonic mouse thymus. J Dev Biol. 2016;4(3):22. doi: 10.3390/jdb4030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis G, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.