Abstract
The nonerythrocytic α-spectrin-1 (SPTAN1) gene encodes the cytoskeletal protein αII spectrin. Mutations in SPTAN1 cause early infantile epileptic encephalopathy type 5 (EIEE5); however, the role of αII spectrin in neurodevelopment and EIEE5 pathogenesis is unknown. Prior work suggests that αII spectrin is absent in the axon initial segment (AIS) and contributes to a diffusion barrier in the distal axon. Here, we have shown that αII spectrin is expressed ubiquitously in rodent and human somatodendritic and axonal domains. CRISPR-mediated deletion of Sptan1 in embryonic rat forebrain by in utero electroporation caused altered dendritic and axonal development, loss of the AIS, and decreased inhibitory innervation. Overexpression of human EIEE5 mutant SPTAN1 in embryonic rat forebrain and mouse hippocampal neurons led to similar developmental defects that were also observed in EIEE5 patient-derived neurons. Additionally, patient-derived neurons displayed aggregation of spectrin complexes. Taken together, these findings implicate αII spectrin in critical aspects of dendritic and axonal development and synaptogenesis, and support a dominant-negative mechanism of SPTAN1 mutations in EIEE5.
Keywords: Development, Neuroscience
Keywords: Epilepsy, Genetic diseases
Introduction
Early infantile epileptic encephalopathies (EIEEs) are a devastating group of epilepsies characterized by refractory seizures and cognitive arrest or regression, and they typically carry a poor prognosis (1). Recent progress in genetic analysis reveals that a significant proportion of previously cryptogenic EIEEs are single-gene disorders, with de novo mutations in numerous genes recently identified (2). In 2010, de novo in-frame microinsertions/microdeletions (indels) in SPTAN1, encoding the nonerythrocyte αII spectrin, were identified in 2 patients [c.6619_6621del/p.(Glu2207del) and c.6923_6928dup/p.(Arg2308_Met2309dup)] exhibiting a distinctive form of early-onset West syndrome with infantile spasms, severe developmental delay, spastic quadriplegia, poor visual attention, and distinctive MRI features that was termed EIEE5 (MIM 613477) (3, 4). The most recent genetic study identified 20 patients with pathogenic or likely pathogenic SPTAN1 variants that exhibited a broad spectrum of neurodevelopmental phenotypes, comprising 6 patients with mild to moderate intellectual disability, with or without epilepsy and behavioral disorders, and 14 patients with EIEE, of whom 13 had severe neurodevelopmental impairment and 4 died in early childhood (5). The majority of pathological mutations discovered reside in the last 2 spectrin repeats in the C-terminal region critical for the formation of α/β spectrin heterotetramers, and some lead to aggregation of spectrin complexes in vitro (3, 4). Although these observations suggest dominant-negative effects, little is known about the role of αII spectrin in brain development and the pathogenesis of EIEE5-producing SPTAN1 mutations.
SPTAN1 consists of 57 exons and encodes αII spectrin, the primary α spectrin subunit in the nervous system (6). αII Spectrin reportedly is absent in the axon initial segment (AIS) and localized specifically to the distal axon, where ankyrin-B, αII spectrin, and βII spectrin define a boundary limiting ankyrin-G to the proximal axon (7). Functional spectrin heterotetramers consist of end-to-end heterodimers between β subunits (II–IV) and αII spectrin in neurons (6), and βIV is predominantly localized to the AIS (8). Thus, it is counterintuitive that αII spectrin is absent in the AIS domain. Previous studies have also shown that αII spectrin knockout does not impair axon development in Drosophila (9), and RNAi knockdown in mouse neocortex alters only AIS length and not its formation (7). In contrast, brain MRIs from patients with SPTAN1 mutations consistently show agenesis of the corpus callosum and hypomyelination, implicating a role for αII spectrin in axonal growth and myelination. Recent stochastic optical reconstruction microscopy (STORM) studies reveal that axons are supported by submembranous actin organized in a periodic ring structure linked by βII spectrins, the obligatory binding partner of αII spectrin, further suggesting that αII/βII spectrin complexes have indispensable structural roles in axon development (10). Because Sptan1–/– mice die before embryonic day 16 (E16) with cardiac and neural tube malformations, while Sptan1+/– mice have no detectable phenotypic abnormalities, studying αII spectrin function in vivo in rodents has been challenging (11).
In this study, we used a mouse monoclonal antibody that specifically recognizes the αII spectrin SRC homology 3 (SH3) domain to show that αII spectrin is ubiquitously expressed in all neuronal domains, including the AIS, in rodent and human neurons. We then focally deleted Sptan1 in vivo using CRISPR and Cas9 endonuclease gene editing, overexpressed pathogenic SPTAN1 in vivo, and generated neurons derived from EIEE5 patient induced pluripotent stem cells to show that αII spectrin is required for neuronal process development and inhibitory synapse formation. These findings suggest a multifaceted and critical role for αII spectrin in neural development, and they suggest that dominant-negative mutations in EIEE5 disrupt αII spectrin, leading to brain dysgenesis and seizures.
Results
Sptan1 is expressed in both axonal and somatodendritic domains.
Spectrin complexes typically represent α/β heterotetramers consisting of end-to-end heterodimers with two α and two β subunits. β Spectrins are encoded by 5 different genes (SPTB, SPTBN1, SPTBN2, SPTBN4, and SPTBN5) (6). βII Spectrin (encoded by SPTBN1) and βIII spectrin (encoded by SPTBN2) localize primarily to distal axonal and somatodendritic domains, respectively (7, 12). βIV Spectrin (encoded by SPTBN4) is more specifically localized to the AIS in mature neurons (8). α Spectrins, the obligatory binding partner for β spectrins, are encoded by 2 genes in mammals (SPTA1 and SPTAN1). αI Spectrin (encoded by SPTA1) is preferentially expressed in erythroid cells, and the nonerythroid αII spectrin (encoded by SPTAN1) is expressed in most other tissues (6). Immunostaining data have suggested a lack of αII spectrin in the AIS, leading to the diffusion barrier model: ankyrin-B and αII and βII spectrins define a boundary limiting ankyrin-G to the proximal axon (7). Because βIV spectrin has to bind to αII spectrin in order to form functional complexes, we hypothesized that the lack of immunostaining of αII spectrin in the AIS is due to specific antigen masking in this highly protein-enriched subcellular domain. Here, we identified a monoclonal antibody (D8B7) that recognizes the unique SH3 domain in αII spectrin, and detects both intact and cleaved forms. Although the sequences of αI and αII spectrin are similar in the SH3 domain, this antibody does not detect human ghost red blood cells or a GST–αI spectrin SH3 fusion protein (13). In addition, the αII–SH3 domain fusion protein (SPTAN1_SH3) used for antibody production contains 2 sequence insertions that are not present in αI spectrin (SPTA1_SH3) (13–16).
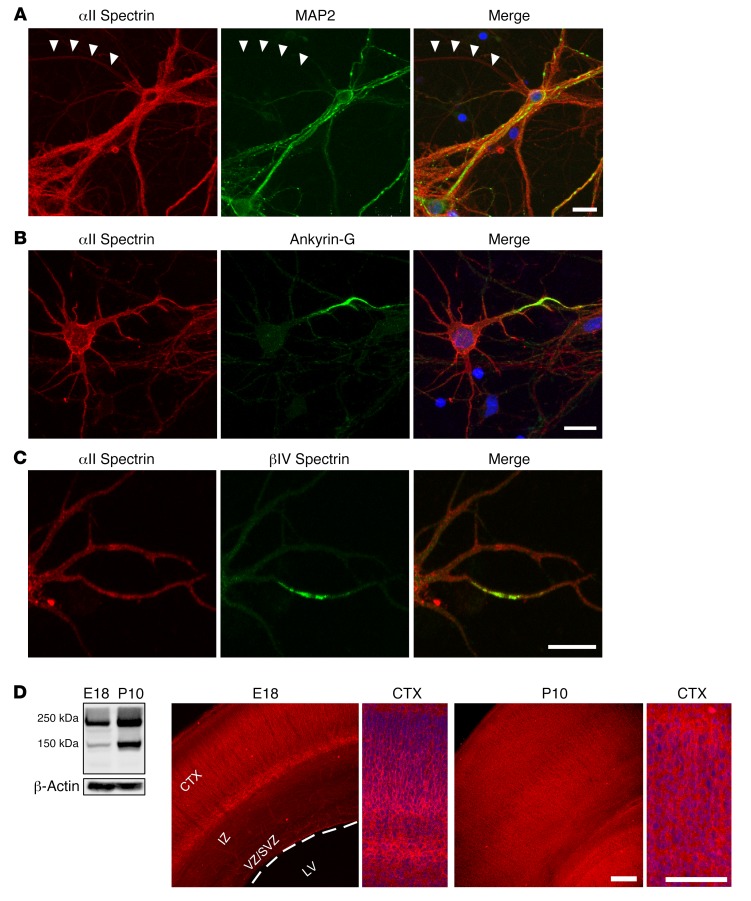
In cultured rat cortical neurons, αII spectrin immunoreactivity was seen ubiquitously in both axonal and somatodendritic domains, but expression was more intense in dendrites than axons (Figure 1A). Immunoreactivity for αII spectrin was observed at the AIS, colabeling with ankyrin-G and βIV spectrin (Figure 1, B and C, and Supplemental Figure 1, C and E; supplemental material available online with this article; https://doi.org/10.1172/JCI95743DS1). Consistent with previous studies (11), abundant αII spectrin expression was detected in the embryonic cerebral cortex (Figure 1D and Supplemental Figure 1). The expression was enriched in the cortical plate but low in the intermediate zone and ventricular zone/subventricular zone. The expression pattern also overlapped with that of MAP2, ankyrin-G, and βIV spectrin (Supplemental Figure 1). Levels of αII spectrin increased in the early postnatal cerebral cortex, with more diffuse labeling in both the cerebral cortex and white matter (Figure 1D). These expression data suggested that αII spectrin has a much broader set of functions than previously thought.
Figure 1. αII Spectrin expression in vitro and in vivo.
(A) αII Spectrin (red) is ubiquitously expressed in all neuronal domains of cultured rat cortical neurons, including the axon, dendrite, and soma. Immunoreactivity is intense in dendrites marked by MAP2 (green), as well as in the axon that is negative for MAP2 staining (arrowheads). (B and C) Ankyrin-G (B) and βIV spectrin (C) (green) specifically localize to the AIS and overlap with αII spectrin. Lower-magnification images are shown in Supplemental Figure 1. (D) Western blot shows that 240-kDa αII spectrin and its 150-kDa cleavage form are expressed in both the embryonic and the postnatal rat brain, with higher expression of both forms at P10 than at E18. See complete unedited blots in the supplemental material. In the embryonic brain, αII spectrin is highly expressed in the cortical plate (CTX), with low expression in the intermediate zone (IZ) and ventricular zone (VZ). LV, lateral ventricle. At P10, αII spectrin expression is more ubiquitous. Cortex is also shown with higher magnification. Scale bars: 10 μm in A–C, 100 μm in D.
CRISPR-mediated Sptan1 deletion in embryonic forebrain.
Studying in vivo function of SPTAN1 has been difficult because Sptan1–/– mice die before E16 with cardiac and neural tube malformations, and Sptan1+/– mice are reportedly normal (11). Moreover, spectrins are membrane proteins that provide a membrane-cytoskeleton interface and mediate cell-cell interaction, adhesion, and mechanical stress adaptation, functions not easily recapitulated by in vitro cell culture (6). Therefore, we used in utero electroporation (IUE) of CRISPR/Cas9 constructs to delete Sptan1 in a subset of rat dorsal cerebral cortex neural progenitors (17, 18). We designed 2 guide RNAs (gRNAs) to target rat Sptan1 at either exon 2 or exon 8 (Supplemental Figure 2A), regions that are present in all Sptan1 variants. Based on an online algorithm (crispr.mit.edu), the on-target faithfulness score is 96 for gRNA A and 90 for gRNA B with 100 as the highest and 60 as a minimally acceptable score. Although Sptan1 and Spta1 share certain homology, the sequences of gRNAs A and B share minimal similarity with SPTA1 and are therefore unlikely to cause off-target effects on SPTA1.
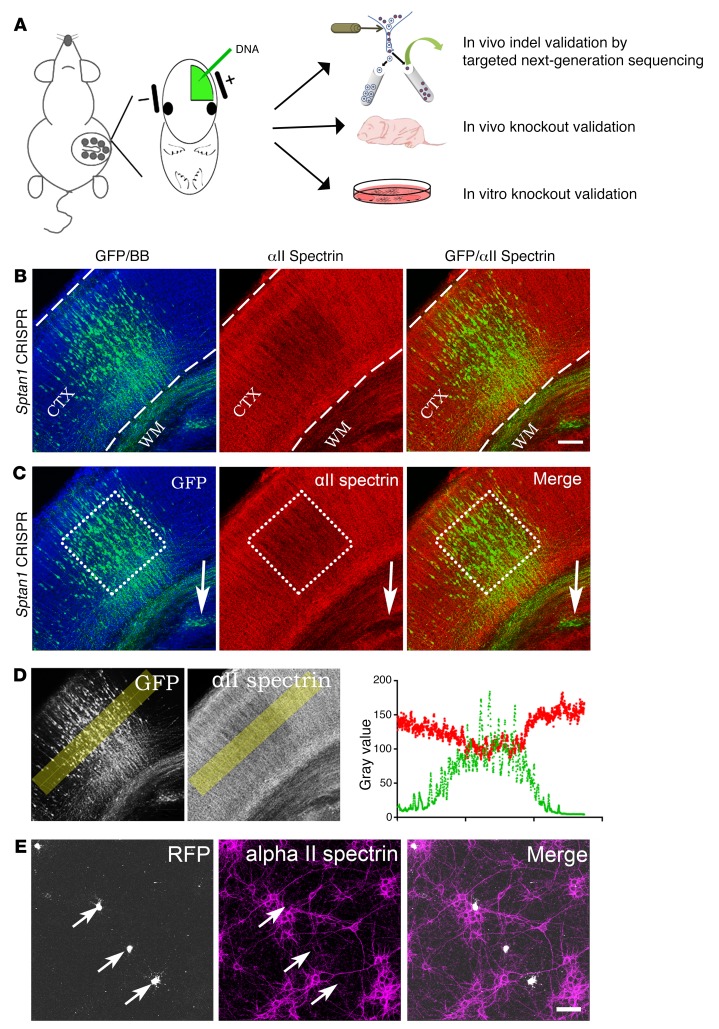
To test whether the gRNAs successfully introduced out-of-frame indels, we first used IUE to deliver Sptan1 gRNAs and Cas9, along with a plasmid encoding GFP, into E13–14 dorsal neocortical neural progenitors (Figure 2, A and B) and harvested the brains at postnatal days 3–5 (P3–5). GFP-labeled cells were isolated by FACS following cortical neuron dissociation. Genomic DNA was extracted, and an approximately 200-bp amplicon spanning the CRISPR cut site was sent for next-generation sequencing at the Center for Computational and Integrative Biology DNA Core of Massachusetts General Hospital. Among about 70,000 reads, 59.6% of CRISPR A–transfected and 47.8% of CRISPR B–transfected cells showed out-of-frame indels (Supplemental Figure 2B). Next, we harvested P1 brains after E13–14 Sptan1 IUE CRISPR gene editing and performed αII spectrin immunohistochemistry on brain sections. In these brains, heavily transfected regions marked by GFP labeling showed substantially reduced αII spectrin immunoreactivity qualitatively (Figure 2, B and C) and by semiquantitative line-graph analysis in ImageJ (NIH) (Figure 2D).
Figure 2. Sptan1 CRISPR in utero electroporation.
(A) Schematic representation of IUE and the CRISPR validation methods. FACS-sorted GFP-labeled cells are processed for next-generation sequencing to identify specific mutations. Brain sections are immunostained to visualize the in vivo qualitative change in αII spectrin immunostaining. Immunostaining on dissociated cortical neurons is performed to quantify the efficiency of Sptan1 CRISPR knockout. (B) Representative confocal image from a P1 brain section after prior transfection with Sptan1 CRISPR A at E13–14. Sptan1 CRISPR–transfected cells are marked by GFP (green) in the cerebral cortex (CTX, between the dashed lines). These cells also project axons to the contralateral hemisphere through white matter (WM). Red, αII spectrin; blue, bisbenzimide (BB) nuclear stain. (C) The area outlined by the dotted square (same image as in B) with a high density of GFP labeling has a marked reduction in αII spectrin immunoreactivity. Another area with a cluster of GFP+ cells in the WM (arrow) also demonstrates a clear lack of αII spectrin staining compared with the surrounding nontransfected (GFP–) area. Note that these regions do not have homogeneous loss of αII spectrin owing to the mosaic mutation introduced by CRISPR IUE. (D) Quantification of fluorescence intensity across the transfected regions (yellow box) shows that the intensities of αII spectrin and GFP signals are inversely correlated. (E) Representative confocal image of αII spectrin (magenta) and RFP expression (white) in rat cortical neurons dissociated from brains 4 days after IUE with Sptan1 CRISPR gRNA and RFP (to label the cells) at E14–15, then cultured for 10 days. Three RFP+ cells lack αII spectrin immunoreactivity, whereas other RFP– cells are αII spectrin positive. Scale bars: 100 μm in B (for B–D), 50 μm in E.
To more quantitatively calculate Sptan1 gRNA knockout efficiency, we dissociated labeled cortical regions 4–5 days after transfection, cultured the cortical neurons for 10 days, and then immunostained them for αII spectrin (Figure 2E and Supplemental Figure 3, A and B). The percentage of transfected cells (GFP+) that were also immunopositive for αII spectrin was determined by counting of about 100 GFP+ cells (3 embryos per transfection condition). With empty PX330 control plasmid, nearly all GFP+ cells (96.6%) were αII spectrin positive. In contrast, a small fraction of GFP+ cells (16.5%) coexpressed αII spectrin when transfected with Sptan1 gRNAs and Cas9 (Supplemental Figure 3B). It is important to point out that, as in previous studies (18), the percentages of cells with indels after FACS purification of IUE gene-edited neurons substantially underestimate the degree of CRISPR deletion because of technical limitations in isolating transfected cells. Thus, quantification of immunostaining for the protein gave us a more accurate measure of about 80% deletion in vivo (Supplemental Figure 3B). To test whether in vivo Sptan1 deletion led to compensatory changes of other spectrins, we immunostained Sptan1 CRISPR–transfected neurons with αI and βI–IV spectrin antibodies. As expected, αI and βI spectrins were not present in neurons (Supplemental Figure 3C and Supplemental Figure 4A). In contrast, the expression of βII and βIII spectrins was high and ubiquitous in nontransfected cultured cortical neurons, while βIV spectrin was enriched at the AIS (Supplemental Figure 4); however, all of these β spectrins were absent in Sptan1 CRISPR–transfected neurons (Supplemental Figure 4). We also examined the efficacy of Sptan1 CRISPR deletion in vitro by transfecting cultured mouse primary hippocampal neurons with GFP and either empty PX330 control or Sptan1 CRISPR A plasmids. Immunostaining showed that approximately 75% of Sptan1-transfected cells were negative for αII spectrin (Supplemental Figure 5, A and B). Taken together, these results indicate that Sptan1 CRISPR gene editing efficiently introduces out-of-frame mutations in vivo and in vitro that result in loss of αII spectrin expression.
Impaired neuronal morphogenesis after Sptan1 deletion.
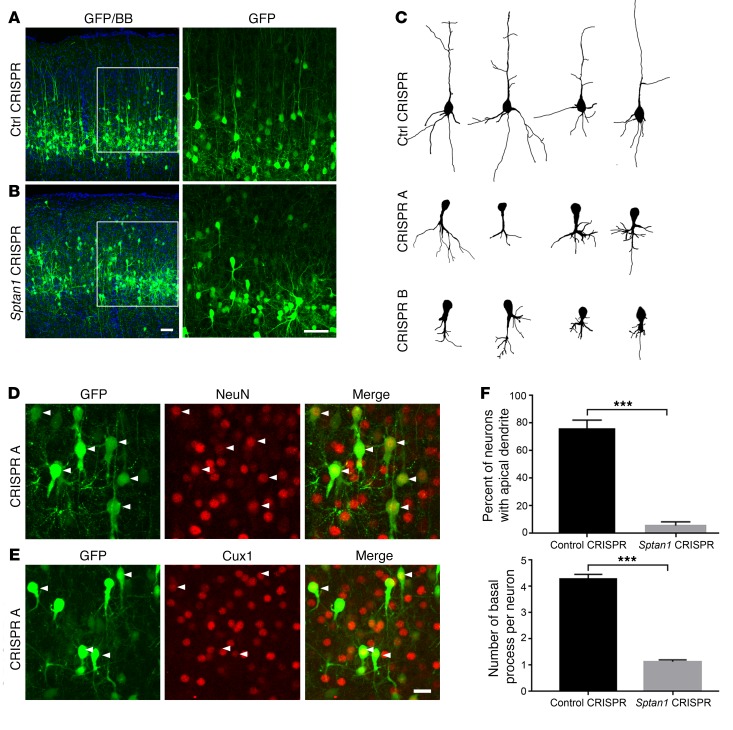
We next assessed the phenotypes of neurons generated from E14–15 neural progenitors transfected with Sptan1 CRISPR A, CRISPR B, or control plasmids (empty PX330 or unrelated gRNAs, n ≥ 3 for each group). In all conditions, transfected cells migrated normally to their correct laminar positions by P21 and were positive for the neuronal marker NeuN and the superficial cortical layer marker Cux1, suggesting that they acquired a birthdate-appropriate neuronal identity (Figure 3, A, B, D, and E). However, the morphologies were strikingly different between GFP-labeled control and CRISPR Sptan1-deleted cells (Figure 3, A–C and F). In control neurons, the apical dendrites extended to the pial surface with apical tufts, and basal dendrites developed into a classical tree-like structure that often consisted of 4–5 primary dendrites (all controls were similar, and representative examples from the empty PX330 vector are shown). In a subset of control neurons (23.0%), the apical dendrite was not well visualized, likely because of weak GFP signal or cell orientation relative to tissue sectioning. After Sptan1 deletion, in contrast, apical dendrites were absent from nearly all cells (~94.4%), and basal dendritic architecture often consisted of a single shortened and hypertrophic basal process that gave rise to supernumerary small branches. Our in vitro experiments also showed that dendritic number and length were significantly decreased in Sptan1 CRISPR A–transfected hippocampal neurons as compared with control neurons (Supplemental Figure 5, C–E). Loss of αII spectrin also led to defects in axon development. In control brains, GFP-labeled axons ran ventrally toward the white matter with extensive arborization in layer 5, and many of them then projected toward the midline and fasciculated to form the corpus callosum (Supplemental Figure 6, A and B). In Sptan1 CRISPR–transfected brains, both callosal axons and arborization in the contralateral hemisphere were decreased and disorganized (Supplemental Figure 6C), resembling agenesis or thinning of the corpus callosum that has been observed in patients with SPTAN1 mutation–associated EIEE5.
Figure 3. Altered neuronal morphology after Sptan1 deletion.
(A) Confocal images of P21 brain sections after control CRISPR transfection at E14–15 show normal-appearing GFP-labeled cells in cortical layer II/III (left). A higher-magnification image of the boxed region (right) reveals typical pyramidal neuron morphology with a single apical dendrite and multiple basal dendrites. (B) Neurons transfected with Sptan1 CRISPR A reach the appropriate cortical layers but appear disorganized, lack apical dendrites, and have abnormal basal dendritic architecture that typically consists of a single short, thickened but exuberant process. (C) Reconstructed neurons transfected with control CRISPR or Sptan1 CRISPR A or B. Note that neurons transfected in vivo with Sptan1 CRISPR A or B are morphologically similar, indicating the specificity of the gene editing. (D and E) Although Sptan1 deletion severely alters neuronal morphology, the GFP+ cells coexpress the neuronal marker NeuN (red in D) and the superficial cortical layer marker Cux1 (red in E), suggesting preserved neuronal differentiation. Scale bars: 25 μm in B (for A and B), 10 μm in E (for D and E). (F) Quantification of percentages of GFP-labeled cells with apical dendrites and numbers of basal dendrites per GFP+ cell in neurons transfected with control plasmid or Sptan1 CRISPR A. A total of 39 neurons from 3 brains with Sptan1 CRISPR A transfection and 43 neurons from 3 brains from the CRISPR control group were quantified. Percentage data are transformed to arcsine value for statistical analysis. One-tailed t test: ***P < 0.001.
Loss of αII spectrin impairs AIS formation and disrupts neuronal polarity.
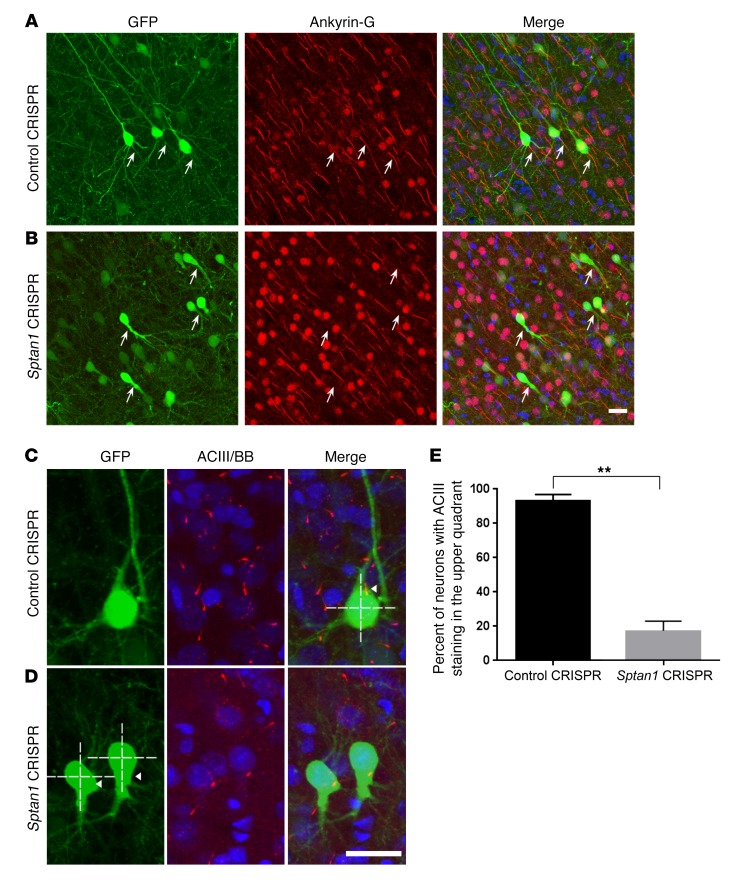
The severely altered neuronal morphology, particularly the absence of an apical dendrite, suggests that deletion of αII spectrin may alter the polarization of developing neurons (19). We first asked whether clustering of ankyrin-G, a master regulator at the AIS that recruits AIS-specific proteins and maintains neuronal polarity, is intact in the absence of αII spectrin. Because ankyrin-G binds to βIV spectrin through its ankyrin-G binding domain (20), we hypothesized that proper αII/βIV spectrin complex formation is required for maintenance of ankyrin-G anchoring at the AIS. In line with previous work showing undetectable or very faint ankyrin-G staining in βIV spectrin–null mice (8), we found that ankyrin-G staining was absent in αII spectrin–deleted neurons (Figure 4, A and B). A similar loss of ankyrin-G was seen after CRISPR-mediated αII spectrin deletion in mouse hippocampal neurons in vitro (Supplemental Figure 6, D and E). These data suggest that αII/βIV spectrin complex formation is one of the earliest events during AIS assembly, and is required for ankyrin-G clustering.
Figure 4. Sptan1 deletion disrupts AIS formation and neuronal polarity in vivo.
(A) Confocal images from a P21 control after prior transfection at E14–15 showing GFP+ cortical neurons (green) with typical pyramidal morphology and ankyrin-G immunolabeling (red) of their proximal axon (arrows). (B) In contrast, Sptan1 CRISPR–transfected cells lack ankyrin-G staining. (C and D) Adenylyl cyclase III (ACIII, red), a marker for primary neuronal cilia, is seen in the typical apical location of a control GFP-labeled neuron (C, arrowhead) but is mislocalized (D, arrowheads) to the basal pole in GFP+ neurons after αII spectrin deletion, indicating disrupted neuronal polarity. Blue is bisbenzimide (BB) nuclear stain. Scale bars: 10 μm in B (for A and B), 10 μm in D (for C and D). (E) Quantification of the percentages of cells with ACIII staining in the upper quadrants of the soma in control (92.9%) and Sptan1 CRISPR groups (22.0%). n = 42 neurons from 4 brains with Sptan1 CRIPSR A transfection, n = 39 from 3 brains with control CRISPR transfection. The percentage data are transformed to arcsine value for statistical analysis with a 1-tailed t test; **P < 0.01.
Neuronal primary cilia are recognized as key integrators of extracellular ligand-based signaling required for appropriate cellular polarity development, and in cortical neurons the primary cilium invariably extends from the base of the apical dendrite (21). Therefore, we used adenylyl cyclase III (ACIII), a well-established marker of neuronal cilia, to ask whether this highly polarized organelle is also disrupted (21). We noted that in contrast to the disappearance of ankyrin-G and βIV spectrin clustering, ACIII immunoreactivity was well preserved, but its distribution was mislocalized to the basal aspect of cortical neurons after Sptan1 deletion (Figure 4, C–E). Together, these results indicate that αII spectrin is critical for the formation of the AIS and appropriate polarity of cortical pyramidal neurons.
Loss of αII spectrin impairs inhibitory synaptogenesis.
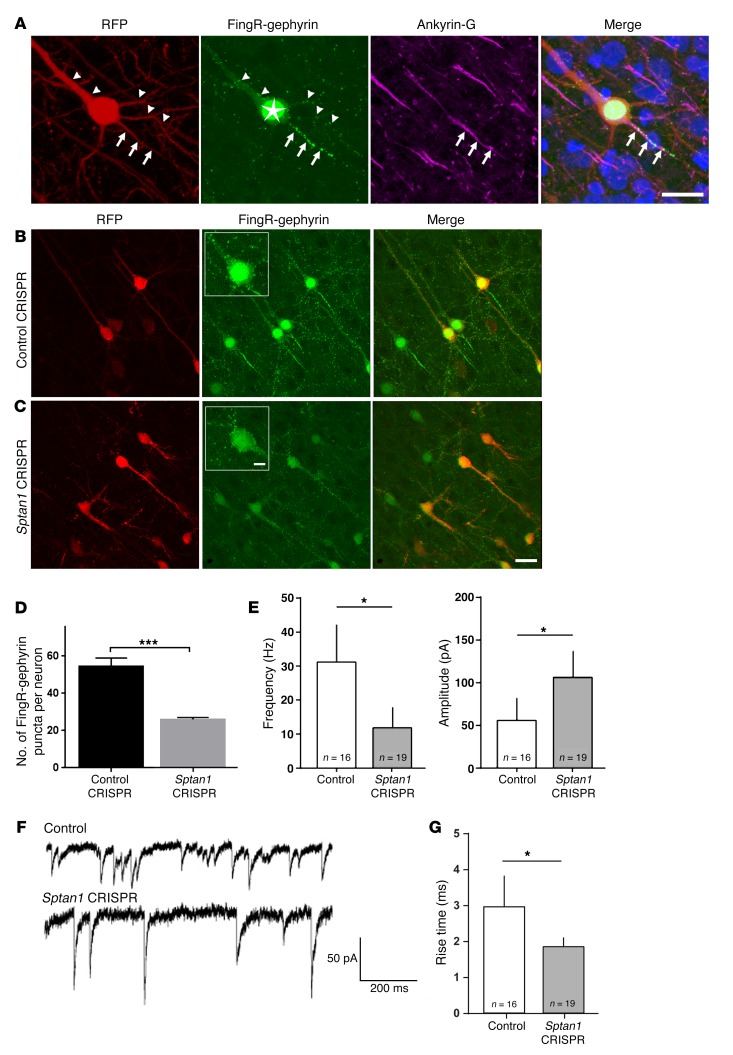
A potential epilepsy mechanism is disinhibition of cortical pyramidal neurons leading to hyperexcitability. To test whether Sptan1-deleted cortical pyramidal cells exhibit a defect in inhibitory input, we first used the FingR (fibronectin intrabodies generated with mRNA display) system to label endogenous postsynaptic gephyrin, a GABA type A receptor (GABAAR) anchoring protein, as a surrogate for GABAergic synapses (22). The gephyrin-FingR-GFP construct incorporates a transcriptional regulation system that ties expression to the level of the target, allowing inhibitory synapses to be visualized without altering localization, morphology, and function, in either fixed or living neurons (22). To test the construct, we cotransfected gephyrin-FingR-GFP and RFP plasmids (the latter to fill cells) via IUE at E13–14 and examined brain sections at P21, a time when the majority of synaptogenesis has occurred. The AIS is known to have a high density of GABAergic innervation with GABAARs and gephyrin colocalized on the postsynaptic densities (23). As expected, we found gephyrin-FingR-GFP clustering at the AIS, which was identified by colabeling with ankyrin-G antibody (Figure 5A, arrows). GFP-labeled puncta were also seen on the soma and dendrites (Figure 5A, arrowheads), concordant with the previously described distribution of endogenous gephyrin (22).
Figure 5. Loss of αII spectrin alters inhibitory innervation.
(A) Confocal image of a neuron in P21 rat cortex that displays typical pyramidal cell morphology after co-electroporation of gephyrin-FingR-GFP (green) and RFP (red) by IUE at E13–14. Arrows indicate the AIS where GFP+ puncta (green) and ankyrin-G (purple) cluster and colocalize; arrowheads indicate punctate GFP labeling along the dendrites. The star indicates excess nuclear gephyrin-FingR-GFP, caused by saturation of endogenous gephyrin binding that leads to a feedback mechanism within the FingR system. Bisbenzimide nuclear stain in blue. (B) Neurons cotransfected with control CRISPR, RFP, and gephyrin-FingR-GFP show abundant GFP+ puncta along axonal and somatodendritic domains. (C) Sptan1 CRISPR/Cas9–transfected neurons, in contrast, show dramatically decreased GFP+ puncta. Insets in middle panels of B and C are higher-magnification images. Scale bars: 10 μm in A, 10 μm in C (for B and C), 5 μm in C inset for insets in B and C. (D) Quantification of GFP+ puncta density on proximal somatodendritic domains and axons (AIS excluded) shows a 45.8% reduction in Sptan1-deleted neurons. One-tailed t test. (E) Frequency (left) and amplitude (right) of miniature inhibitory postsynaptic currents (mIPSCs) are altered in the Sptan1-deleted neurons. Two-tailed t test. (F) Examples of voltage-clamp mIPSC recordings from control (upper trace) and Sptan1-deleted (lower trace) neurons. (G) mIPSC rise times for GFP+ cells from control and Sptan1 CRISPR–transfected cells. The mean rise time is significantly shorter for Sptan1 CRISPR cells. Two-tailed t test performed in G. One-tailed t test: *P < 0.05; ***P < 0.001. In FingR-gephyrin experiments, n = 21 neurons (3 brains) with Sptan1 CRISPR A transfection and n = 19 neurons (3 brains) with control CRISPR transfection were analyzed. In patch-clamp experiments, n = 19 Sptan1-deleted and n = 16 control neurons (from ≥3 brains and 3 neurons per brain per condition) were analyzed.
To examine whether Sptan1 deletion alters the distribution of gephyrin-FingR-GFP puncta, we cotransfected brains by IUE with Sptan1 CRISPR gRNAs and Cas9, RFP to fill the cells, and gephyrin-FingR-GFP at E13–14, and then examined brain sections at P21. Compared with CRISPR controls, the tightly clustered gephyrin puncta at the proximal axon disappeared in Sptan1 CRISPR–transfected neurons, consistent with loss of the AIS, and somatodendritic punctate GFP labeling also markedly decreased (Figure 5, B and C). We quantified the density of GFP+ puncta on the soma, proximal dendrites, and proximal axon (excluding the AIS cluster), and observed a 45.8% reduction after Sptan1 deletion (Figure 5D). To confirm a functional decrease in synaptic inhibition onto these cells, we performed whole-cell patch-clamp recordings of miniature inhibitory postsynaptic currents (mIPSCs). We targeted cortical layer II/III gephyrin-FingR-GFP/RFP–labeled Sptan1 CRISPR or control pyramidal neurons for recordings. The frequency of mIPSCs was significantly reduced in Sptan1-deleted cells (Figure 5, E and F), consistent with decreased inhibitory innervation. Interestingly, the mIPSC amplitude was increased in these cells (Figure 5, E and F), likely reflecting loss of the apical dendritic branches (Figure 3) leading to the capture of events during voltage clamp recordings predominantly from perisomatic regions. To provide evidence supporting this idea, we measured the mIPSC rise times and found significantly shorter rise times in the Sptan1-knockout group (Figure 5G). This result suggests that most mIPSCs recorded in this group originated closer to the recording electrode in the soma rather than more distally in dendrites, which would lead to increased mIPSC amplitudes. Together with our morphological studies, these data demonstrate defective GABAergic innervation of cortical pyramidal neurons lacking αII spectrin. Our findings are also supported by the observation that ankyrin-G promotes stability of somatodendritic GABAergic synapses in vitro and in vivo through opposing endocytosis of GABAARs via interaction with GABAAR-associated protein (GABARAP) (24).
Dominant-negative effects of pathogenic SPTAN1 mutations.
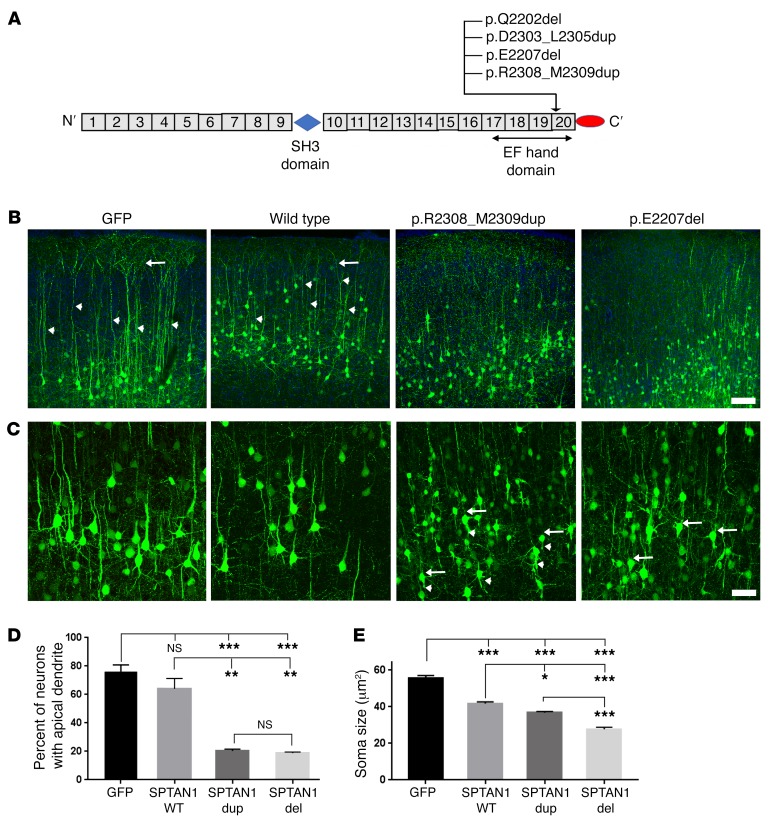
All identified pathogenic SPTAN1 mutations causing EIEE5 reside in the last 2 spectrin repeats critical for α/β spectrin heterodimerization (Figure 6A) (4). In vitro data suggest that these mutations impair α/β spectrin complex formation (4). However, the effects of EIEE5 mutations in vivo in animal models or in cultured human neurons are unknown. We hypothesized that αII spectrin mutations would give rise to comparable, or more severe, in vivo phenotypes compared with those observed in Sptan1 deletion experiments. To test this idea, we first overexpressed human WT or mutant SPTAN1 in rat cerebral cortex by IUE at E14–15 and examined the brains at P10. We chose 2 mutations, p.R2308_M2309dup and p.E2207, for this experiment because they were the first 2 identified in SPTAN1 epileptic encephalopathy and the patients were well described clinically. Other mutations, such as p.D2303_L2305dup and p.Q2202del, are in close proximity and would be expected to have the same cellular pathogenesis. Pyramidal neuron morphology was similar in cells transfected with GFP either with or without WT SPTAN1, except for reduced soma size with WT overexpression (Figure 6, B–E). Neurons cotransfected with GFP and EIEE5 mutant SPTAN1 containing either a microduplication or a microdeletion, in contrast, displayed lack of apical dendrites and severe disorganization of basal dendrites, as well as a more profound decrease in soma size (Figure 6, B–E). In vitro overexpression confirmed the phenotypes observed in vivo (Supplemental Figure 7).
Figure 6. Pathogenic SPTAN1 mutations produce phenotypes similar to Sptan1 deletion.
(A) All reported αII spectrin mutations reside at the C-terminal, a critical region required for spectrin heterodimer formation. (B) Neurons cotransfected with WT SPTAN1 and GFP show normal morphology compared with GFP alone except for decreased soma size. Neurons transfected with p.R2308_M2309dup or p.E2207del are smaller and have shortened or absent apical dendrites and abnormal basal dendrites. Arrowheads indicate well-formed apical dendrites in GFP and WT SPTAN1 groups, and arrows indicate the apical tufts in layer I cortex (seen best in the GFP group). Apical dendrites and tufts in the mutant groups are minimal. (C) Higher-magnification images reveal the severely altered neuronal morphology in the mutant overexpression groups. Arrows indicate the lack of apical dendrites, and arrowheads denote impaired basal dendritic trees. Scale bars: 100 μm in B, 50 μm in C. (D) Most neurons transfected with GFP (76.2%) or WT SPTAN1 (63.9%) display intact apical dendrites. Neurons transfected with mutant SPTAN1 constructs typically lack apical dendrites (20.3% in p.R2308_M2309dup; 19.0% in p.E2207del overexpression group). Percentage data are transformed to arcsine values for statistical comparisons. (E) The soma size of neurons transfected with WT or mutant SPTAN1 is significantly smaller than that of GFP controls, and both mutants show significantly reduced soma size compared with WT SPTAN1. For apical dendrite analysis, n = 32 neurons from 3 brains for GFP control; n = 20 neurons from 3 brains with WT SPTAN1; n = 43 neurons from 4 brains with SPTAN1 p.R2308_M2309dup; n = 37 neurons from 3 brains with SPTAN1 deletion were used. For soma size analysis, 25 neurons from 3 GFP brains, 53 neurons from 3 brains with WT SPTAN1, 47 neurons from 3 brains with SPTAN1 p.R2308_M2309dup, and 50 neurons from 4 brains with SPTAN1 p.E2207del were used. ANOVA test: *P < 0.05; **P < 0.01; ***P < 0.001.
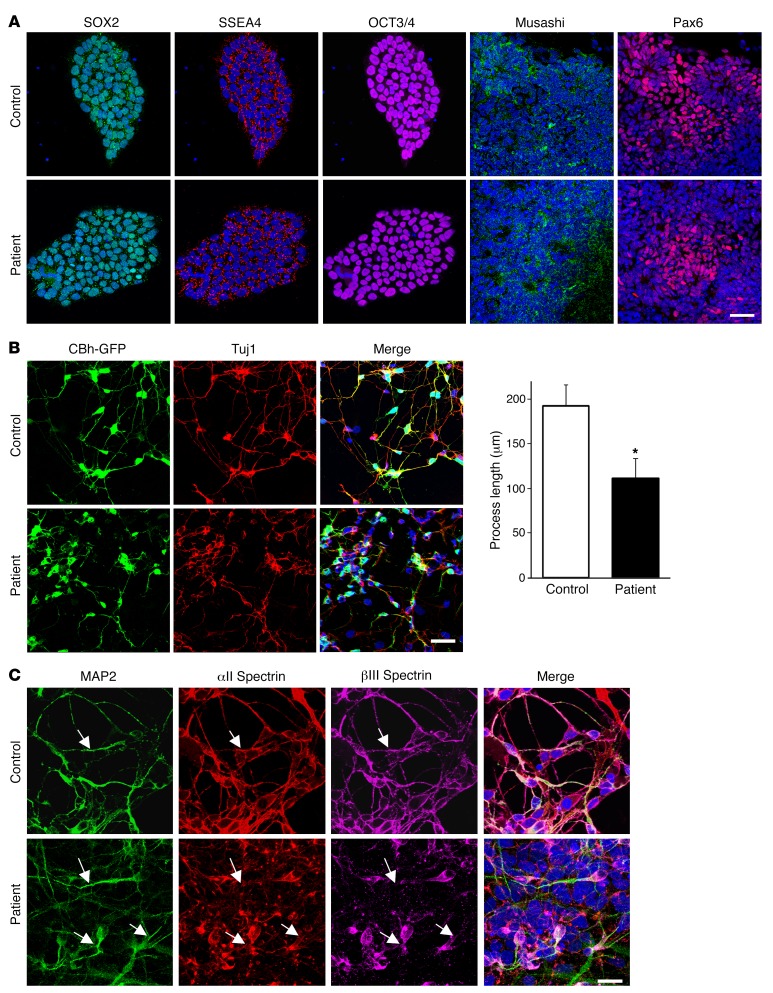
To determine whether the developmental defects observed in rat neurons expressing EIEE5 SPTAN1 mutations are also present in human neurons, we generated induced pluripotent stem cells (iPSCs) from fibroblasts obtained from 2 control individuals and an EIEE5 subject carrying a pathogenic SPTAN1 mutation (p.D2303_L2305dup). This mutation, a 9-bp in-frame duplication at the C-terminal domain, was previously reported in patients presenting with West syndrome, a severe infantile epileptic encephalopathy (4). The subject was a boy (1 year of age at the time of skin biopsy) with EIEE5 manifest with severe developmental delay and daily seizures who died at age 2 years. We reprogrammed his fibroblasts into iPSCs and differentiated them into excitatory cortical-like neurons, along with 2 control human iPSC lines. No differences were apparent between mutant and control iPSCs or neural progenitors (Figure 7A). Immunostaining of iPSC-derived neurons showed that the majority were glutamatergic and expressed the excitatory cortical deep layer projection neuron marker CTIP2 (Supplemental Figure 8). After transduction with a GFP-expressing lentivirus, the control neurons displayed a normal morphology with long and complex processes, along with a ubiquitous pattern of αII and βIII spectrin immunoreactivity (Figure 7, B and C) similar to that observed in rat cortical or mouse hippocampal neurons. In contrast, cortical neurons derived from the EIEE5 patient exhibited shortened and less complex neuronal processes, as well as aggregation of αII and βIII spectrins that were absent from GFP+ processes (Figure 7, B and C). These findings suggest dominant-negative effects of the SPTAN1 mutation in human cortical-like neurons leading to a severe morphological phenotype.
Figure 7. Patient iPSC-derived neurons show aberrant neurite development and spectrin aggregation.
(A) Patient and control iPSCs express the pluripotency markers SOX2, SSEA4, and OCT3/4. After differentiation into neural progenitors, both control and EIEE5 patient cells form similarly appearing neural rosettes that express Musashi and Pax6. (B) Cells were transduced with GFP-expressing lentivirus (CBh-GFP, green), and neurons were identified by Tuj1 immunoreactivity (red). Total process length per cell was measured in GFP+ cells and was significantly shorter in patient neurons. Two-tailed t test: *P < 0.05. n = 44 neurons per group from 4 biological replicates per group. (C) Confocal images showing coexpression of the neuronal marker MAP2 (green), αII spectrin (red), and βIII spectrin (magenta) in control (top row) and patient (bottom row) iPSC-derived neurons. Note the overlap of GFP and spectrins in control processes (arrows in top panels), but aggregation of spectrins in patient cells, with lack of spectrin immunoreactivity in GFP+ processes (arrows in bottom panels). Two different iPSC lines derived from patient and control fibroblasts were used. Scale bars: 20 μm.
Discussion
Here we demonstrate a unique platform that combines in vivo rodent genome editing and human iPSC models to study the function of an epileptic encephalopathy gene during brain development. This approach is particularly useful in diseases in which traditional genetically modified mice exhibit prenatal death, as is the case with many EIEE genes. Our data indicate that αII spectrin is a critical structural protein in neurons. In vivo deletion of αII spectrin impairs dendritic and axonal development, cell polarity, and inhibitory innervation of developing cortical pyramidal neurons. In vivo and in vitro overexpression of human mutant SPTAN1 in rodent cortical and mouse hippocampal neurons, respectively, produces similar abnormalities that are also exhibited by EIEE5 patient–derived neurons. Our findings further suggest that dominant-negative effects of SPTAN1 mutations contribute to the pathogenesis of EIEE5, and implicate disinhibition of cortical pyramidal neurons as a potential seizure mechanism.
Our CRISPR constructs delivered by IUE achieved a very high efficiency of introducing out-of-frame indels, consistent with previous reports using the same technique (18, 25–27). Although off-target effects are a concern, CRISPR/Cas9 is much less prone to off-target effects compared with RNAi or transcription activator–like effector nuclease (TALEN) methods. When in vitro Cas9-digested whole-genome sequencing (Digenome-seq) was used to profile CRISPR/Cas9 off-target editing in human cells, off-target mutations were induced at only a few rather than thousands of sites in the genome (28). Because of the insensitivity of the Surveyor/T7 assay and difficulty predicting off-target sites by sequence similarity (29–31), we chose instead to validate the specificity of our CRISPR/Cas9 gene editing with multiple independent experiments. First, we showed that 2 different CRISPR gRNAs targeting different Sptan1 exons generated similar phenotypes. Second, IUE of Cas9 alone or with other CRISPR gRNAs targeting different genes (data not shown) failed to generate the phenotypes. Finally, we found that overexpression of SPTAN1 pathogenic mutations in rodent neurons and generation of EIEE5 patient iPSC-derived neurons produced similar results.
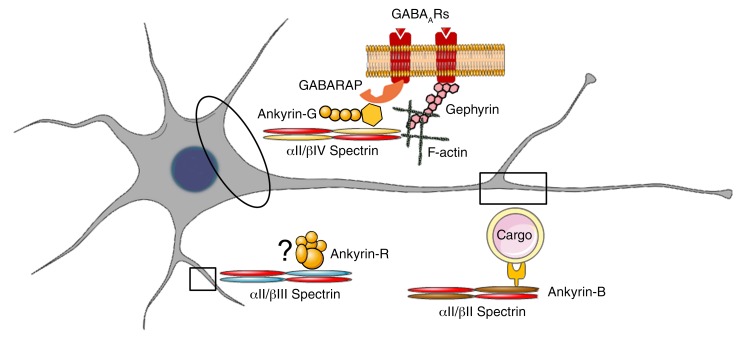
Based on a lack of immunostaining evidence supporting its presence at the AIS, αII spectrin was proposed to be part of a diffusion barrier in the distal axon (7). However, its absence in the AIS is counterintuitive because βIV spectrin, an obligatory binding partner for αII spectrin, is specifically localized to the AIS (8). To our knowledge, we show for the first time ubiquitous neuronal αII spectrin expression in both axonal and somatodendritic domains, including the AIS. This finding is concordant with the established distribution of different β spectrin subunits in different neuronal domains (βII in the distal axon, βIII in dendrites, and βIV in the AIS). αII Spectrin probably provides a universal structural support in all of these neuronal domains (Figure 8).
Figure 8. αII Spectrin is a universal neuronal structural protein.
In neurons, αII spectrin is ubiquitously expressed and interacts with different β spectrins in different neuronal microdomains. In the AIS, αII/βIV spectrin complexes are required to stabilize ankyrin-G that also interacts with GABARAP to stabilize GABAARs in somatodendritic domains. The αII/βIV spectrin complex likely also stabilizes gephyrin in the AIS through F-actin. In the more distal axon, αII/βII spectrin complexes interact with ankyrin-B to modulate organelle transport, and particularly retrograde transport. In dendrites, αII/βIII spectrin complexes function in dendritic development, likely through interactions with ankyrin-R.
To serve as the master AIS organizer for almost all other AIS proteins, ankyrin-G needs to bind to spectrin repeats 14 and 15 of βIV spectrin. Therefore, either loss of αII spectrin or the disruption of αII/βIV spectrin complex formation will disrupt ankyrin-G binding. Our data show that both Sptan1 CRISPR deletion and disruption of spectrin complexes by dominant-negative SPTAN1 overexpression lead to disrupted ankyrin-G clustering. The more distal axonal defects in Sptan1-knockout cells in vivo may result from an early failure to initiate axon outgrowth, impairment of axon elongation, or degeneration following initial extension, all of which depend on stable axonal submembrane and plasma cytoskeletal structures where βII/αII spectrin complexes predominate (10). Indeed, βII spectrin has binding sites for ankyrin-B (32), and in skeletal muscle localizes ankyrin-B to costameres where ankyrin-B in turn localizes dynactin-4 and dystrophin (33). This interaction is required for proper organization and function of dystrophin and dystroglycan and for costamere-associated microtubules. In neurons, ankyrin-B is required for achieving normal axon length, and loss of ankyrin-B impairs organelle transport, particularly retrograde transport (34). Therefore, disrupted ankyrin-B function probably also contributes to the observed axonal defects in Sptan1-deleted animals. Interestingly, we did not observe changes in ankyrin-B localization or expression (data not shown). This result suggests that unlike βII–IV spectrin and ankyrin-G, the expression and localization of ankyrin-B are not dependent on intact αII spectrin or α/β spectrin complex formation. Also, βIII spectrin is reportedly present throughout the dendritic tree of cerebellar Purkinje cells, and is essential for the recruitment and maintenance of ankyrin-R at the plasma membrane. The loss of function of βIII spectrin results in thinner dendrites and excessive dendritic protrusions with loss of planarity, and leads to spinocerebellar ataxia type 5 (35, 36). Taken together, the perturbation in ankyrin and β spectrin interactions in distinct neuronal domains underpins the pathogenesis of loss of function of αII spectrin, their universal binding partner (Figure 8).
Previous work suggested that αII spectrin RNAi knockdown in neurons altered ankyrin-G expression, leading to a diffuse, fragmented pattern that lacked clustering at the AIS (7). Our αII spectrin CRISPR deletion experiments showed a more severe phenotype. The difference may reflect a partial knockdown with RNAi compared with a more complete loss of αII spectrin with CRISPR gene editing. In addition, the lack of a phenotype in heterozygous Sptan1-knockout mice argues that complete loss of αII spectrin is probably essential to study its function (11). Interestingly, αII spectrin RNAi experiments apparently showed no axonal defects (7). Such a finding is unexpected given the proposed model based on STORM imaging studies that show that axons are supported by submembranous αII/βII spectrin complexes linking periodic actin rings (10), and that brain MRIs of patients with SPTAN1 mutations often display agenesis of the corpus callosum (4). Our findings of axonal deficits after Sptan1 deletion are consistent with an essential role of αII spectrin in supporting axonal development or maintaining axonal structure.
Gephyrin, the main structural scaffold protein of inhibitory postsynaptic densities, is localized at the subsynaptic compartment in contact with the cytoskeleton (e.g., spectrin complexes). Gephyrin is required for the organization, stability, and clustering of GABAARs (37, 38). Genetic mutations in both GABAARs and gephyrin have been identified in different epilepsy syndromes, including severe epileptic encephalopathies (39–44). Our data suggest that defective gephyrin clustering and GABAergic innervation of cortical pyramidal neurons contribute to the pathogenesis of EIEE5, providing further evidence that inhibitory transmission plays critical roles in EIEEs. Previous studies also identified a mutation (R290H) in collybistin, a regulator of gephyrin localization, that causes epilepsy and intellectual disability (45, 46). In addition, the 480-kDa ankyrin-G protein itself promotes stability of somatodendritic GABAergic synapses in vitro and in vivo through opposing endocytosis of GABAA via interaction with GABARAP (24). These observations fit well with our data showing that loss of αII spectrin disrupts inhibitory innervation by perturbing gephyrin and ankyrin-G.
Several important questions remain unanswered. First, how are different β spectrin subunits recruited to different neuronal domains to bind to αII spectrin? One possibility is that different αII spectrin variants have specific domain localizations and affinity preferences for different β spectrins. The αII spectrin antibody we used, based on the unique SH3 domain, probably recognizes all αII spectrin variants and therefore shows a ubiquitous expression pattern. Second, our in vivo and in vitro experiments did not focus on the functional or morphological phenotypes of interneurons and glial cells that may play important roles in disease pathogenesis. Third, missense mutations at the SPTAN1 C-terminus have been identified through whole exome sequencing in EIEE5 patients, but their significance remains undetermined. It will be interesting to test whether these missense mutations cause similar phenotypes, and if so, the prevalence of EIEE5 is likely much higher than is currently appreciated.
Methods
Plasmids and IUE.
Sptan1 gRNA sequences for CRISPR gene editing were designed using an online program (http://crispr.mit.edu) as previously described (47). Guide sequences were subcloned into the pX330 vector (Addgene, plasmid 42230). The 2 Sptan1 guide sequences used were Sptan1 A (targeting exon 2): 5′-GGATCGGTACCACCGCTTCA-3′; and Sptan1 B (targeting exon 8): 5′-GGCGGTCAGCCTCGGCACAC-3′. We obtained C-terminal FLAG-tag WT SPTAN1 and mutant SPTAN1 including c.6619_6621del (p.E2207del) and c.6923_6928dup (p.R2308_M2309dup) expression plasmids with a pCIG vector backbone and expressing nuclear-localized EGFP from Hirotomo Saitsu (Yokohama City University, Yokohama, Japan). Site-directed mutagenesis was performed to generate SPTAN1 mutant p.D2303_L2305dup from WT SPTAN1. pCAG-GFP was obtained from Joseph LoTurco (University of Connecticut, Mansfield, Connecticut, USA). FingR-gephyrin plasmid was obtained from Addgene (plasmid 46296). IUE was performed on rats (Sprague-Dawley; Charles River Laboratories) as previously described (17, 48). The final concentrations used for transfections were 0.5 μg/μl for EGFP, 1.5 μg/μl for Sptan1 CRISPRs, 2 μg/μl for WT or mutant SPTAN, and 1 μg/μl for gephyrin-FingR-GFP.
FACS and sequencing.
Reporter-labeled cortical regions from 4 animals transfected with pCAG-EGFP, Cas9, and Sptan1 gRNA at E13–14 via IUE were dissected at P3 under an epifluorescence dissection microscope and enzymatically dissociated into a single-cell suspension. FACS was performed on a Beckman Coulter MoFlo Astrios Flow Cytometer. Sorted cells were pelleted by centrifugation, and genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen). Targeted genomic areas flanking gRNA binding sites were PCR amplified using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, 11304-011). PCR products were sent for deep sequencing at the Center for Computational and Integrative Biology, Massachusetts General Hospital, Harvard. Primers used for targeted sequencing were as follows: CRISPR A forward: 5′-ATCCAAGTGGGGTCAAAGTGC-3′; CRISPR A reverse: 5′-CTGCAAGTTGGTGGGGTCTT-3′; CRISPR B forward: 5′-CGATTAGCAAGGGCCGGAG-3′; CRISPR B reverse: 5′-AGCACCACCACATCATTTGC-3′.
Generation of iPSCs and patient-specific neurons.
Patient fibroblasts with a pathogenic SPTAN1 mutation (p.D2303_L2305dup) were derived from a skin biopsy obtained under IRB approval of McMaster University, Hamilton, Ontario, Canada. Neonatal human foreskin fibroblasts P9-NuFF-1 (GlobalStem) were used as healthy control. To generate iPSCs from human fibroblasts, the following episomal vectors were electroporated into 3 × 105 cells using a Neon transfection device (Invitrogen): pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, and pCXLE-hUL (all from Addgene). Electroporation conditions used were 1,600 V, 10 mm width, 3 pulses. The cells were maintained in fibroblast medium (DMEM supplemented with 10% FBS) for 3 days after electroporation and then changed to 50% fibroblast medium and 50% TeSR-E7 medium (STEMCELL Technologies). At day 7, the medium was changed to 100% TeSR-E7 for a week, then switched to TeSR-E8 medium (STEMCELL Technologies). Seven days later, iPSC-like colonies were selected for further propagation and evaluation. Cortical excitatory neurons were generated using a published protocol with minor modifications (49). Briefly, for neural induction, iPSCs were passaged at high density to ensure the cells reached 100% confluence 1 day after plating. Then the culture medium was switched to neural induction medium (3N medium supplemented with 1 μM dorsomorphin and 10 μM SB431542). Between days 8 and 12 after plating, a uniform neuroepithelial sheet formed, which was dissociated into cell aggregates and replated in 3N medium including 20 ng/ml FGF2. After 4 days, neural rosettes were manually picked and expanded in 3N medium. Three weeks later, 10 ng/ml BDNF and 10 ng/ml GDNF were added to maintain differentiated neurons.
Neuronal culture and transfection.
Briefly, P0 mouse dissociated hippocampal neurons were plated onto poly-d-lysine– and laminin-coated MatTek dishes in Neurobasal-A medium containing 10% (vol/vol) FBS, B27 supplement, 2 mM glutamine, and penicillin/streptomycin. The next day, the medium was replaced with fresh Neurobasal-A medium containing B27, glutamine, penicillin/streptomycin, and 1% FBS. A mixture of Sptan1 CRISPR A and GFP plasmids at a ratio of 3:1 was added to 100 μl of Neurobasal-A, along with Lipofectamine 2000.
Immunocytochemistry.
Brains were removed and fixed in 4% paraformaldehyde in PBS after transcardial perfusion, sectioned at 50–80 μm on a vibratome (Leica VT1000S), and processed for immunocytochemistry as free-floating sections. For acutely dissociated cell preparations, hemispheres were dissected and cells were dissociated and processed as previously described (48). Primary antibodies included mouse anti–αI spectrin (1:1,000; Abcam ab11751/17C7), mouse anti–αII spectrin (1:2,000; Abcam ab11755/D8B7), mouse anti–βI spectrin (1:1,000; Abcam ab2808/4C3), mouse anti–βII spectrin (1:1,000; Santa Cruz Biotechnology sc136074/42), goat anti–βIII spectrin (1:1,000; Santa Cruz Biotechnology sc9660/N19), rabbit anti–βIV spectrin (1:1,500; provided by Paul Jenkins) (50), chicken anti-GFP (1:1,000; Aves GFP-1020), rabbit anti–ankyrin-G (1:500; provided by Paul Jenkins [50] or 1:200; Santa Cruz Biotechnology sc28561/H215), rabbit anti-SOX2 (1:5,000; Chemicon AB5603), mouse anti-SSEA4 (1:200; Developmental Studies Hybridoma Bank MC-813-70), goat anti-OCT3/4 (1:1,000; Santa Cruz Biotechnology sc8628/N19), rabbit anti-Pax6 (1:500; Covence PRB-278P), mouse anti-Tuj1 (1:400; Covence MMS-435p), mouse anti-MAP2 (1:500; Sigma-Aldrich M2320/AP-20), rabbit anti-GABA (1:2,000; Sigma-Aldrich A2052), rat anti-CTIP2 (1:1,000; Abcam ab18465/26B6), and guinea pig anti-VGLUT1 (1:200; Synaptic Systems 135304). Fluorescently conjugated secondary antibodies (Alexa Fluor 488, 594, or 647) were obtained from Molecular Probes, and nuclei were labeled with bisbenzimide (Molecular Probes H1398).
Brain slice preparation, electrophysiological recordings, and data analysis.
Male and female rats (P21–28) that previously underwent IUE with control or Sptan1 CRISPR/Cas9 plus gephyrin-FingR-GFP plasmids at E13.5–14.5 were used for all electrophysiological recordings. Slices were prepared similarly to a previous description (51).
Electrophysiological recordings were made in a recording chamber mounted on the stage of an upright microscope (customized Scientifica Optical Platform). Slices were continuously perfused (MINIPULS 3, Gilson Inc.) with artificial cerebrospinal fluid containing 1 μM tetrodotoxin, 20 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), and 20 μM d–2-amino-5-phosphonovaleric acid (AP-5) to block action potential firing and ionotropic glutamate currents at a rate of 5–6 ml/min at 31°C–32°C (in-line heater TC-324B, Warner Instrument Co.). GFP-expressing cells in cortical layers II and III were targeted for recordings. Recordings were made with micropipettes (3–5 MΩ resistence) filled with an internal solution containing 100 mM Cs-gluconate, 0.2 mM EGTA, 5 mM MgCl2, 40 mM HEPES, 2 mM Mg-ATP, 0.3 mM Na-GTP, 1 mM QX-314, pH 7.2. Data were acquired at 20 kHz and filtered at 5 kHz. mIPSCs were then recorded for 5 minutes in whole-cell voltage-clamp mode at –60 mV. Holding potential was corrected for the liquid junctional potential. Only cells with stable passive properties were further analyzed. We collected data from a maximum of 3 neurons per animal to avoid bias.
Image acquisition and statistics.
Multichannel imaging was performed using a Leica SP5 confocal microscope. All the images were further processed in Adobe Photoshop CS3 software. Statistical analysis was performed using Microsoft Excel and GraphPad Prism (GraphPad Software Inc.). Percentage data were first transformed to arcsine values that were subsequently used for statistical analysis. At least 3 biological replicates were used for all studies. For mIPSC analysis, miniAnalysis software (Synaptosoft Inc.) was used to detect mIPSCs. Automatic analysis followed by manual screening was used to determine mIPSC amplitude, frequency, and rise time for each 5-minute recording. Statistical analysis was performed in GraphPad Prism. Average value for each of the variables was calculated per cell and further averaged for each experimental group. Parametric and nonparametric statistical analysis was performed as dictated by data distribution. Figures were generated in Adobe Illustrator (Adobe Systems Inc.). A confidence interval of 95% (P < 0.05) was required for values to be considered statistically significant. All data are presented as mean ± SEM.
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Michigan.
Author contributions
YW and JMP designed the experiments, analyzed the data, and wrote the manuscript with contributions from all authors. YW performed all in vivo experiments. TJ performed iPSC experiments. ADN and PMJ performed in vitro hippocampal neuron culture experiments. KG and GGM performed patch-clamp experiments.
Supplementary Material
Acknowledgments
We thank Joseph LoTurco and Hirotomo Saitsu for providing plasmids. We thank Mark Tarnopololsky for providing patients’ fibroblasts. This research was supported by NIH grant 1K08NS099379-01 (to YW), a grant from the Pediatric Epilepsy Research Foundation (to JMP and YW), the Michigan Predoctoral Training in Genetics (T32GM007544 to ADN), the Heinz C. Prechter Bipolar Research program, and the Richard Tam Foundation, University of Michigan Depression Center (PMJ).
Version 1. 01/16/2018
Electronic publication
Version 2. 02/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(2):760–773.https://doi.org/10.1172/JCI95743.
Contributor Information
Yu Wang, Email: eegwang@med.umich.edu.
Tuo Ji, Email: tuoji@med.umich.edu.
References
- 1.Epi4K Consortium. et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvill GL, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saitsu H, et al. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet. 2010;86(6):881–891. doi: 10.1016/j.ajhg.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tohyama J, et al. SPTAN1 encephalopathy: distinct phenotypes and genotypes. J Hum Genet. 2015;60(4):167–173. doi: 10.1038/jhg.2015.5. [DOI] [PubMed] [Google Scholar]
- 5.Syrbe S, et al. Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain. 2017;140(9):2322–2336. doi: 10.1093/brain/awx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81(3):1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 7.Galiano MR, et al. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell. 2012;149(5):1125–1139. doi: 10.1016/j.cell.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komada M, Soriano P. βIV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J Cell Biol. 2002;156(2):337–348. doi: 10.1083/jcb.200110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hülsmeier J, Pielage J, Rickert C, Technau GM, Klämbt C, Stork T. Distinct functions of α-Spectrin and β-Spectrin during axonal pathfinding. Development. 2007;134(4):713–722. doi: 10.1242/dev.02758. [DOI] [PubMed] [Google Scholar]
- 10.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339(6118):452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stankewich MC, Cianci CD, Stabach PR, Ji L, Nath A, Morrow JS. Cell organization, growth, and neural and cardiac development require αII-spectrin. J Cell Sci. 2011;124(pt 23):3956–3966. doi: 10.1242/jcs.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, et al. β-III spectrin is critical for development of purkinje cell dendritic tree and spine morphogenesis. J Neurosci. 2011;31(46):16581–16590. doi: 10.1523/JNEUROSCI.3332-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Ziemnicka D, Scalia J, Kotula L. Monoclonal antibodies to alphaI spectrin Src homology 3 domain associate with macropinocytic vesicles in nonerythroid cells. Brain Res. 2001;898(1):171–177. doi: 10.1016/S0006-8993(01)02156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahr KE, et al. The complete cDNA and polypeptide sequences of human erythroid alpha-spectrin. J Biol Chem. 1990;265(8):4434–4443. [PubMed] [Google Scholar]
- 15.Moon RT, McMahon AP. Generation of diversity in nonerythroid spectrins. Multiple polypeptides are predicted by sequence analysis of cDNAs encompassing the coding region of human nonerythroid alpha-spectrin. J Biol Chem. 1990;265(8):4427–4433. [PubMed] [Google Scholar]
- 16.Ziemnicka-Kotula D, et al. Identification of a candidate human spectrin Src homology 3 domain-binding protein suggests a general mechanism of association of tyrosine kinases with the spectrin-based membrane skeleton. J Biol Chem. 1998;273(22):13681–13692. doi: 10.1074/jbc.273.22.13681. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Dcdc2 knockout mice display exacerbated developmental disruptions following knockdown of doublecortin. Neuroscience. 2011;190:398–408. doi: 10.1016/j.neuroscience.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, Rosiene J, Che A, Becker A, LoTurco J. Tracking and transforming neocortical progenitors by CRISPR/Cas9 gene targeting and piggyBac transposase lineage labeling. Development. 2015;142(20):3601–3611. doi: 10.1242/dev.118836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones SL, Svitkina TM. Axon initial segment cytoskeleton: architecture, development, and role in neuron polarity. Neural Plast. 2016;2016:6808293. doi: 10.1155/2016/6808293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Ogawa Y, Hedstrom KL, Rasband MN. βIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J Cell Biol. 2007;176(4):509–519. doi: 10.1083/jcb.200610128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arellano JI, Guadiana SM, Breunig JJ, Rakic P, Sarkisian MR. Development and distribution of neuronal cilia in mouse neocortex. J Comp Neurol. 2012;520(4):848–873. doi: 10.1002/cne.22793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross GG, et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78(6):971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz DA, Lovallo EM, Stockton S, Rasband M, Lewis DA. Postnatal development of synaptic structure proteins in pyramidal neuron axon initial segments in monkey prefrontal cortex. J Comp Neurol. 2009;514(4):353–367. doi: 10.1002/cne.22006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng WC, Jenkins PM, Tanaka M, Mooney R, Bennett V. Giant ankyrin-G stabilizes somatodendritic GABAergic synapses through opposing endocytosis of GABAA receptors. Proc Natl Acad Sci U S A. 2015;112(4):1214–1219. doi: 10.1073/pnas.1417989112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci Rep. 2016;6:20611. doi: 10.1038/srep20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83(5):1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One. 2014;9(8):e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33(2):175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 30.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis JQ, Bennett V. Brain ankyrin. A membrane-associated protein with binding sites for spectrin, tubulin, and the cytoplasmic domain of the erythrocyte anion channel. J Biol Chem. 1984;259(21):13550–13559. [PubMed] [Google Scholar]
- 33.Ayalon G, Hostettler JD, Hoffman J, Kizhatil K, Davis JQ, Bennett V. Ankyrin-B interactions with spectrin and dynactin-4 are required for dystrophin-based protection of skeletal muscle from exercise injury. J Biol Chem. 2011;286(9):7370–7378. doi: 10.1074/jbc.M110.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenzo DN, et al. A PIK3C3-ankyrin-B-dynactin pathway promotes axonal growth and multiorganelle transport. J Cell Biol. 2014;207(6):735–752. doi: 10.1083/jcb.201407063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins E, Suminaite D, Jackson M. Cerebellar ataxias: β-III spectrin’s interactions suggest common pathogenic pathways. J Physiol (Lond) 2016;594(16):4661–4676. doi: 10.1113/JP271195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarkson YL, Perkins EM, Cairncross CJ, Lyndon AR, Skehel PA, Jackson M. β-III spectrin underpins ankyrin R function in Purkinje cell dendritic trees: protein complex critical for sodium channel activity is impaired by SCA5-associated mutations. Hum Mol Genet. 2014;23(14):3875–3882. doi: 10.1093/hmg/ddu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyagarajan SK, Fritschy JM. Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci. 2014;15(3):141–156. doi: 10.1038/nrn3670. [DOI] [PubMed] [Google Scholar]
- 38.Fritschy JM, Harvey RJ, Schwarz G. Gephyrin: where do we stand, where do we go? Trends Neurosci. 2008;31(5):257–264. doi: 10.1016/j.tins.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Shen D, et al. De novo GABRG2 mutations associated with epileptic encephalopathies. Brain. 2017;140(1):49–67. doi: 10.1093/brain/aww272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janve VS, Hernandez CC, Verdier KM, Hu N, Macdonald RL. Epileptic encephalopathy de novo GABRB mutations impair GABAA receptor function. Ann Neurol. 2016;79(5):806–825. doi: 10.1002/ana.24631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González MI. The possible role of GABAA receptors and gephyrin in epileptogenesis. Front Cell Neurosci. 2013;7:113. doi: 10.3389/fncel.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dejanovic B, et al. Exonic microdeletions of the gephyrin gene impair GABAergic synaptic inhibition in patients with idiopathic generalized epilepsy. Neurobiol Dis. 2014;67:88–96. doi: 10.1016/j.nbd.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Lionel AC, et al. Rare exonic deletions implicate the synaptic organizer Gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum Mol Genet. 2013;22(10):2055–2066. doi: 10.1093/hmg/ddt056. [DOI] [PubMed] [Google Scholar]
- 44.Dejanovic B, et al. Simultaneous impairment of neuronal and metabolic function of mutated gephyrin in a patient with epileptic encephalopathy. EMBO Mol Med. 2015;7(12):1580–1594. doi: 10.15252/emmm.201505323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papadopoulos T, Schemm R, Grubmüller H, Brose N. Lipid binding defects and perturbed synaptogenic activity of a Collybistin R290H mutant that causes epilepsy and intellectual disability. J Biol Chem. 2015;290(13):8256–8270. doi: 10.1074/jbc.M114.633024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimojima K, et al. Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet. 2011;56(8):561–565. doi: 10.1038/jhg.2011.58. [DOI] [PubMed] [Google Scholar]
- 47.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manent JB, Wang Y, Chang Y, Paramasivam M, LoTurco JJ. Dcx reexpression reduces subcortical band heterotopia and seizure threshold in an animal model of neuronal migration disorder. Nat Med. 2009;15(1):84–90. doi: 10.1038/nm.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(3):477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenkins PM, et al. Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc Natl Acad Sci U S A. 2015;112(4):957–964. doi: 10.1073/pnas.1416544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Althaus AL, Sagher O, Parent JM, Murphy GG. Intrinsic neurophysiological properties of hilar ectopic and normotopic dentate granule cells in human temporal lobe epilepsy and a rat model. J Neurophysiol. 2015;113(4):1184–1194. doi: 10.1152/jn.00835.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.