Abstract
Objective
To provide an alternative to propensity scoring (PS) for the common situation where there are interacting covariates.
Setting
We used 1.3 million assessments of residents of the United States Veterans Affairs nursing homes, collected from January 1, 2000, through October 9, 2012.
Design
In stratified covariate balancing (SCB), data are divided into naturally occurring strata, where each stratum is an observed combination of the covariates. Within each stratum, cases with, and controls without, the target event are counted; controls are weighted to be as frequent as cases. This weighting procedure guarantees that covariates, or combination of covariates, are balanced, meaning they occur at the same rate among cases and controls. Finally, impact of the target event is calculated in the weighted data. We compare the performance of SCB, logistic regression (LR), and propensity scoring (PS) in simulated and real data. We examined the calibration of SCB and PS in predicting 6‐month mortality from inability to eat, controlling for age, gender, and nine other disabilities for 296,051 residents in Veterans Affairs nursing homes. We also performed a simulation study, where outcomes were randomly generated from treatment, 10 covariates, and increasing number of covariate interactions. The accuracy of SCB, PS, and LR in recovering the simulated treatment effect was reported.
Findings
In simulated environment, as the number of interactions among the covariates increased, SCB and properly specified LR remained accurate but pairwise LR and pairwise PS, the most common applications of these tools, performed poorly. In real data, application of SCB was practical. SCB was better calibrated than linear PS, the most common method of PS.
Conclusions
In environments where covariates interact, SCB is practical and more accurate than common methods of applying LR and PS.
Keywords: Balancing databases, propensity scoring, confounding, causal impact, prognosis
This article calculates the impact of a target event on outcomes after removing the influence of other co‐occurring events (covariates). When two events co‐occur, their impact is confounded and statistical procedures can be used to separate out the effect of each event. Statisticians typically address confounding through randomization. In 1983, Rosenbaum and Rubin proposed methods for removing confounding in observational data. They proposed the use of propensity scoring (PS) to balance rates of occurrence of covariates among treated and untreated subjects (Rosenbaum and Rubin 1983). Since then, different methods of PS have been proposed, including methods for matching (Rosenbaum and Rubin 1985; Rosenbaum 1989; Abadie and Imbens 2006), subclassification (Rosenbaum and Rubin 1984; Rosenbaum 1991; Hansen 2004), weighting (Rosenbaum 1987; Robins, Hernan, and Brumback 2000; Hirano, Imbens, and Ridder 2003), regression (Heckman, Ichimura, and Todd 1998), likelihood (Imai and Ratkovic 2014), or combinations of approaches (Robins, Rotnitzky, and Zhao 1995; Ho et al. 2007; Abadie and Imbens 2011). On June 28, 2016, a search on PubMed identified 10,050 articles referencing propensity scoring. Analysis of year of publications showed that the number of articles that used PS has grown exponentially (number of articles = 0.43e0.35 Years Since1987, R 2 = 0.98).
Despite widespread use, misspecification of PS remains a concern. Simulation studies have shown that misspecification can have a large impact on study findings, in many situations reversing the conclusions of the study (Kang and Schafer 2007). When the number of covariates is large, as it is in many health care studies, the typical linear combination of covariates can fail to balance all of the individual confounders. Nonlinear models are needed. Austin and colleagues recommended that in these situations, one should use interactions terms to balance residual unbalanced covariates (Austin 2009b, 2011a,b). Few investigators do so. On June 28, 2016, we reviewed a sample of 50 of the most recent publications using PS. Among these, 39 (78 percent) used no interaction terms, 9 (18 percent) used pairwise interactions terms, and 2 (4 percent) used select quadratic interaction terms. Despite recommendations to use interaction terms, no investigators included all interaction terms, perhaps because the sheer effort to do so is prohibitive or because introducing new interaction terms could make other covariates go out of balance. Investigators’ selective approach to inclusion of interaction terms makes the effort haphazard. No clear analytical solution is available except to exhort investigators to put more effort into modeling interaction among covariates. Some investigators have designed computer programs that automatically search for a model that can balance all terms in PS (McCaffrey, Ridgeway, and Morral 2004). We propose an alternative approach that does not rely on the investigators’ effort and reduces chances for misspecification of the PS.
A good example of how misspecification of PS can occur can be seen in analysis of inpatient data. There are many interactions present among the comorbidities of hospitalized patients (Extermann 2007; Ferdinandy et al. 2014; Alemi et al. 2016). For typical inpatient data, at least five diagnoses from a possible list of 14,000 diagnoses are listed for each hospitalization. To estimate the efficacy of a particular treatment, the influence of comorbidities that affect (confound) the outcome must be accounted for. This is not easy to do as this situation creates a factorial design where all first‐, second‐, third‐, and fourth‐order interaction terms are missing and fifth or more interaction terms are present. In this environment, balancing the main effects of covariates is not sufficient as many higher order interacting diagnoses are also present. We propose a new way of balancing covariates that works well with extensive interactions among the covariates.
Proposed Method
Step One: Identify Naturally Occurring Strata
The proposed method of analysis is based on a growing literature on design of covariate balanced randomized clinical trials (Pocock and Simon 1975; Wei 1978; Atkinson 1982; Signorini et al. 1993; Frangakis and Rubin 2002; Scott et al. 2002; Heritier, Gebski, and Pillai 2005; Yuan, Huang, and Liu 2011) and stratification procedures available since the 1950s (Tripepi et al. 2010). Like these methods, the first step in our proposed procedure is to stratify the data. In any database, certain events co‐occur, creating naturally occurring strata. For example, in analysis of comparative effectiveness of medications, diagnoses are used as covariates, and many diagnoses (e.g., diabetes and renal disease) co‐occur, creating natural strata.
The procedure for finding the natural strata is not statistical in nature. A standard query language (SQL) code to search the data for naturally occurring strata and calculate weights for balancing the data (a concept discussed in a later section) is available in the Appendix S2 and follows this algorithm:
| % E = {A,B,…,R} and F represent events | |
| % S represents a sample | |
| For each E in S | % Analysis done for each combination |
| F ← E union {X = 0} | % Select control combinations, X = 0 |
| f0[E] ← count(F) | % Count of Controls |
| Y0[E] ← sum(y(F))/count(F) | % Prob Y for Controls |
| For each E in S | % Analysis done for each combination |
| F ← E union {X = 1} | % Select Case combinations, X = 1 |
| f1[E] ← count(F) | % Count of Cases |
| Y1[E] ← sum(y(F))/count(F) | % Prob Y for Cases |
| For each E in f0 | % Weights for Control combinations |
| if E in f1 | |
| then w0[E] ← f1[E]/f0[E] | % Matched |
| else w0[E] ← 0 | % Not matched |
| For each E in f1 | % Weights for Case combinations |
| if E in f0 | |
| then w1[E] ← 1 | % Matched |
| else w1[E] ← 0 | % Not matched |
Here, we provide an intuitive understanding of the procedure. The analyst searches within the data, for all combinations of occurrences of covariates and treatment. Each unique combination of covariates is considered one stratum or subgroup. Within each stratum, the levels of covariates are fixed and cases are treated and controls are not. This allows cases and controls to be contrasted while holding covariates constant and the average impact of treatment on outcome calculated. The procedure organizes the data into a partial factorial design where cases and controls are examined at different factorial combinations of covariates.
One immediate question is how practical it is to search for combination of covariates. Theoretically, the possible combination of k binary covariates is 2k, which depending on the size of k may be computationally hard to do. However, the observed combinations of covariates are typically significantly lower than the theoretically possible combinations. Most combinations do not occur in the data. Extensive research on Apriori algorithm shows that the number of possible strata, even in large multidimensional data, is relatively small (Agrawal, Imieliński, and Swami 1993; Agrawal and Srikant 1994; Bayardo 1998). Readers, for example, may be surprised to know the number of observed combinations in the data analyzed in this paper. There were 1.3 million records of 11 variables. Theoretically, there should be 211 = 2,048 possible strata, but we observed only 418 unique combinations of 10 covariates and the treatment variable. Nearly 79 percent of possible combinations of covariates never occurred in the data, despite the large size of the data.
Step Two: Matched Estimation of the Effect
Once natural strata have been organized, the estimation of impact follows statistical procedures known since the 1950s for analysis of stratified case–control design (Cochran 1950; Mantel and Haenszel 1959). The chi‐square test for homogeneity is used to see whether, across strata, a common odds ratio exists. If cases and controls are counted as in Table 1, then the test of homogeneity of treatment impact across strata is calculated as:
Table 1.
Top 20 Most Frequent Strata
| k | Age | Male | Disabilities | Cases Unable to Eat, X = 1 | Matched Controls Able to Eat, X = 0 | |||
|---|---|---|---|---|---|---|---|---|
| Total, a i + b i | # Dead, ∑Y | Total, c i + d i | # Dead, ∑Y | Weight, w i0 | ||||
| 1 | 65–85 | M | SGTBWDL | 36,677 | 12,831 | 17,862 | 4,253 | 2.053 |
| 2 | 40–65 | M | SGTBWDL | 19,317 | 9,787 | 10,739 | 3,512 | 1.79 |
| 3 | 65–85 | M | SGTBWD | 14,494 | 3,118 | 7,456 | 1,153 | 1.944 |
| 4 | 85+ | M | SGTBWDL | 11,336 | 3,951 | 22,220 | 5,436 | 0.51 |
| 5 | 40–65 | M | SGTBWD | 10,987 | 3,263 | 6,318 | 1,358 | 1.739 |
| 6 | 65–85 | M | GTBWD | 6,386 | 3,275 | 3,032 | 1,121 | 2.106 |
| 7 | 65–85 | M | GTBWDL | 5,101 | 2,192 | 9,524 | 2,544 | 0.536 |
| 8 | 40–65 | M | GTBWD | 4,592 | 982 | 7,283 | 1,226 | 0.631 |
| 9 | 40–65 | M | GTBWDL | 4,465 | 3,210 | 2,002 | 1,017 | 2.23 |
| 10 | 40–65 | M | 4,113 | 762 | 11,173 | 1,695 | 0.368 | |
| 11 | 85+ | M | GTBWDL | 4,035 | 2,016 | 26,189 | 7,938 | 0.154 |
| 12 | 85+ | M | GTBWD | 4,027 | 2,031 | 9,406 | 3,167 | 0.428 |
| 13 | 65–85 | M | GTBD | 3,362 | 871 | 6,647 | 1,209 | 0.506 |
| 14 | 85+ | M | SGTBWD | 2,475 | 1,196 | 5,033 | 1,749 | 0.492 |
| 15 | 65–85 | M | 2,305 | 918 | 13,097 | 2,990 | 0.176 | |
| 16 | 65–85 | M | GB | 1,744 | 607 | 9,122 | 1,897 | 0.191 |
| 17 | 40–65 | M | GB | 1,629 | 647 | 3,186 | 666 | 0.511 |
| 18 | 65–85 | M | GBW | 1,534 | 566 | 4,038 | 998 | 0.38 |
| 19 | 40–65 | M | GBW | 1,406 | 362 | 782 | 124 | 1.798 |
| 20 | 65–85 | M | B | 1,317 | 384 | 100,237 | 12,786 | 0.013 |
M = male; B = unable to bathe; W = unable to walk; G = unable to groom; D = unable to dress; T = unable to toilet; L = bowel incontinent; S = unable to transfer; U = urinary incontinent.
| Outcomes in ith stratum, i = 1,…, k |
|
||||||
| Y = 1 | Y= 0 | ||||||
| Cases (X = 1) | a i | b i | |||||
| Controls (X = 0) | c i | d i | |||||
If a homogenous common odds ratio, , exists, then its statistical significance is tested as:
| Mantel–Haenszel test of Significance of Impact of X on Y over Different Strata | Estimate of Common Odds Ratio of Impact of X on Y | |||||
|
n i = a i + b + c i + d i |
|
If the Y variable is continuous, then paired t‐test (Anderson, Kish, and Cornell 1980) can be used:
The average treatment effect, , has a student's t‐distribution with k‐1 degrees of freedom and the t‐statistic is calculated as follows:
Some investigators (e.g., Rosenbaum and Rubin 1985; Austin 2009a) recommend using difference in means in units of the pooled standard deviation. Such an approach is not influenced by sample size, which in PS applications can be arbitrarily set depending on whether 1 to 1 or 1 to many matches are made. We prefer the paired t‐statistic because it takes advantage of the natural strata within the data.
Step Three: Weighted Estimation of the Effect
In calculation of treatment effect, first weights are used to balance the data; then, the average weighted treatment effect is calculated. The weights are chosen so that the rate of occurrence of covariates stays the same when treatment is present or absent. In an implausibly fortunate scenario, for example, when a study has been carefully designed, the original sample would include all factorial combinations of the covariates, with equal observation of cases and controls within each combination of covariates. This will provide a full factorial design, from which the estimate of impact of treatment on outcome can be readily estimated, with no consideration of confounding. This is almost never the case in observational studies. One can imagine repeated sampling until the desired pattern occurs and by luck a balanced factorial design is obtained. Alternatively, if one thinks of resampling as weighting, then one can select weights that accomplish the needed study design, thus removing the need for luck. In resampled data, the new count of cases and controls are shown as a weighted product of the original sample counts. Thus, in the strata “i”, the resampled counts are as follows: w i1(c i + d i) for cases and w i0(a i + b i) for controls. Weights are chosen so that we can remove the effects of co‐occurring covariates A,…, R. In the stratified sample, this idea can be expressed as selecting weights w i0 and w i1 such that there are no differences in probability of occurrence of any particular combination of covariates:
| (1) |
Note that the above equation must hold for every strata or combination of covariates. Thus, equation (1) is not one equation but a large number of different equations. Let Z be an event defined as the combination of all mutually exclusive co‐occurring events, that is, the strata, excluding X, from some record in S. Let Z i be an indicator variable that equals 1 when a unique combination of covariates is present. Thus, if all combination of covariates must be equally frequent among cases and controls, then equation (1) can be written as:
| (2) |
Note also that equation (2) does not specify what the rate should be but that the two rates on the two sides of the equation should be the same. Equation (2) holds if, and only if, the following equation is true for all Z k:
| (3) |
In the stratified sample, any combination of events occurs at most once with X = 1 and once with X = 0. Equation (3) has at most one term in the numerator of the summation and holds if:
| (4) |
To find a choice of weights for equation (1), suppose that we wish to exclude from analysis any case or control that is not matched, then if cases are absent, that is, a i + b i = 0, then assign control weight of w i0 = 0. If controls are absent, that is, c i + d i = 0, then assign cases the weight of w i1 = 0. Further assume, as typically is the situation, that cases are fewer than controls, then to create as little distortion in the data as possible, we assign weight of 1 for every case, w i0 = 1, given that the weight is not already assigned a 0. A weight of 1 makes sure that all matched cases are included in the analysis without any change. The sample weight for controls should then be set so that the rates of co‐occurring events are the same. Then, weights can be assigned as follows:
In this fashion, one can identify weights that balance the data for all combination of covariates. As each stratum is mutually exclusive, the weights assigned in this fashion will not contradict each other. As cases and controls within any combination of covariates are set to have equal frequency, then any subset of these combinations including the subset with just one covariate will occur with equal frequency. Finally, for the situation where all cases are weighted as 1, the proposed set of weights for controls is optimal because if any combination of the covariates among the controls is weighted in another fashion, then in at least one combination of covariates, the two samples are not balanced.
Result
Test in Simulated Data
In the simulation, the outcome was calculated as a function of 1 treatment variable, 10 covariates, and 210 = 1,024 combinations of covariates. All variables were generated using random binomial distributions. Multiple datasets were simulated, each time including a different number of combinations of covariates. The initial simulation calculated the outcome as a linear combination of treatment and the 10 covariates. This simulated the situation where impacts of covariates on the outcome were independent. Thereafter, progressively more interaction terms were included, simulating the situation where the impact of one covariate depends on another. The last simulation indicated the situation where the outcome variable was simulated as a function of a full factorial combination of covariates.
In all simulations, the impact of treatment on the outcome was set to be 2 and the impact of the covariate or the combinations of covariates was randomly chosen to be uniformly between 5 and −5. In each simulation, 10,000 observations were randomly generated. Four approaches were used to recover the treatment effect from the simulated data:
In the first approach, called pairwise logistic regression (LR), the simulated outcome was regressed on covariates, pairwise interaction among covariates, and the treatment variable. The coefficient of the treatment variable was used to estimate the treatment effect.
In the second approach, called pairwise propensity scoring (PS), first treatment was regressed on covariates, and pairwise interaction among the covariates. Then, the inverse PS was used to weigh the outcome. Treatment effect was calculated in the weighted data.
In the third approach, called proper LR, the simulated outcome was regressed on covariates, a priori known number of interaction among covariates, and treatment variable. To clarify, suppose the outcome was simulated from main effects, all two‐way, all three‐way, and a number of four‐way interactions. Then, the LR would include all main effects, all two‐way, all three‐way, and all four‐way interactions among the covariates. As before, the coefficient for treatment variable was used to estimate the treatment effect.
In the fourth approach, the naturally occurring strata within the data were identified and the method proposed in this article was used to balance covariates. No effort was made to match interactions among the covariates.
The findings of the simulation study are summarized in Figure 1. The Y‐axis shows the percent of error in estimating the impact of treatment on outcome. For simplicity, the percent of error is calculated as absolute value of difference of the actual and estimated effect divided by the actual estimate. The X‐axis shows the number of interaction terms included in generating the outcome. Initially, when the outcome was generated from a linear combination of the covariates, or when the outcome was based on pairwise interaction among the covariates, all four methods (pairwise LR, LR, pairwise PS, and SCB) perform similarly. In these initial comparisons, both the PS function and the logistic function were properly specified; they reflected the way the outcome was generated. As higher order interaction terms were used to generate the outcome, the SCB method and proper LR maintained their accuracy but both pairwise LR and the pairwise PS methods had increasing error. The simulation confirmed that SCB maintains its accuracy, even when the outcome was generated from higher order interaction terms. No matter how many and what type of interaction terms were used to generate the outcome, SCB was able to relatively accurately estimate the impact of treatment. LR and PS were not able to do so. When there was a mismatch between how the outcome was generated and how the variables were modeled, both PS and LR had higher error rates. In both LR and PS, one has to rely on the investigator's effort to incorporate the interaction terms. Often, they cannot incorporate all interaction terms and at best have been including only pairwise interactions. SCB does not rely on the investigator's effort and removes the confounding that results from pairwise or higher order interaction terms.
Figure 1.
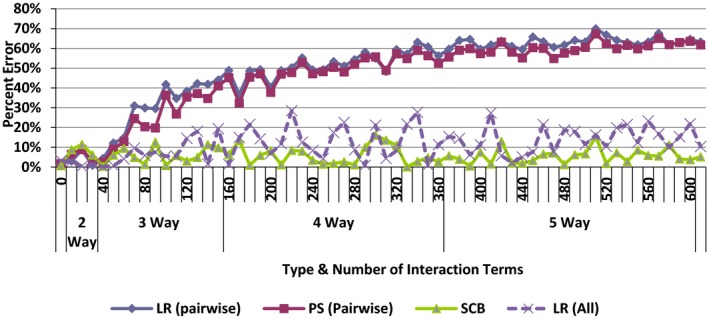
Percent Error in Estimating Impact of Treatment [Color figure can be viewed at wileyonlinelibrary.com]
Notes. Percent of error = |Estimated−Actual|/Actual. LR, logistic regression; PS, propensity scoring; SCB, stratified covariate balancing.
Test in Real Data
To demonstrate the use of SCB in a large dataset, we examined the relationship between feeding disability and 6‐month mortality for nursing home residents. The sample included 296,051 residents in Veterans Affairs nursing homes examined from January 1, 2000, through October 9, 2012. The mean age of the residents was 74.36 years (SD = 11.44) at time of first assessment. The majority were white (79.88 percent) and male (96.34 percent), typical of VA studies. An average resident had 6.15 (SD = 8.76) assessments that included information on nine disabilities. These included Bathing (B), Walking (W), Grooming (G), Dressing (D), Toileting (T), Bowel Continence (L), Transfer (S), Urinary continence (U), and Feeding (F) disabilities. The dependent variable was 6‐month mortality. Across the residents, there were a total of 1,329,260 assessments. After excluding negative values, missing age value, or age <40 years, 1,039,080 assessments were left.
Table 1 describes the top 20 most frequent cases that were matched. The Appendix S3 provides the full 418 matched strata. Each row in the table shows one naturally occurring stratum. These strata describe the types of residents within the data. Note that within each row, cases and controls occur with different frequency; the weighting procedure resets these frequencies so that the cases and controls have the same frequencies. Within the row, the mortality rate for cases and controls can be compared, as these values are calculated for the same type of resident, indicating same age, same gender, and same disabilities.
Figure 2 shows the odds of various events co‐occurring with “unable to eat.” When the sample was not weighted, these odds varied and were seldom 1 to 1. After the sample was weighted, the odds of all the co‐occurring events were 1 to 1, and they were all balanced. For example, before weighting, residents who were unable to eat were more likely to also have transfer disabilities than residents who were able to eat. The weighting procedure removed the differences in transfer disabilities. After weighting, both residents who were able and those who were not able to eat had the same rate of transfer disabilities, yielding an odds ratio of 1 to 1. Even though Figure 2 does not show it, we also examined other combinations of the listed covariates and these combinations also had odds ratio of 1 to 1. The SCB procedure had removed confounding from not only the main effects of covariates but also from any combination of the covariates and SCB weights had balanced all variations in the covariates and interactions of the covariates.
Figure 2.
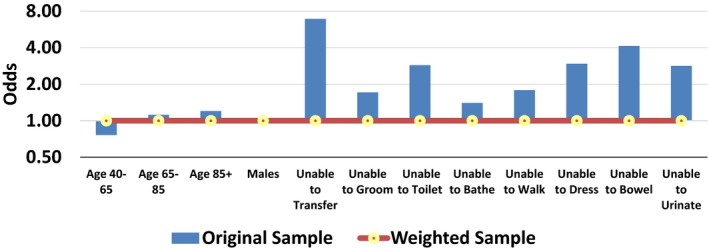
Odds of Occurrence of the Covariate for Able and Unable to Eat Residents [Color figure can be viewed at wileyonlinelibrary.com]
Given that the data were balanced, the next step was to calculate the unconfounded odds of mortality for residents who were unable to eat. Note that in the estimation section, the weighting procedure does not change the calculation of the common odds ratio, as the weights in the denominator and the numerator cancel each other out. But these weights do change the calculation of the confidence interval. The results indicate that in the original sample, the odds for mortality in 6 months for residents who were “unable to eat” was 2.56 to 1. After weighting the sample, so that confounded effects of age, gender, and other disabilities were removed, the odds of mortality was reduced to 1.86 to 1.
Calibration of Propensity Scores in Real Data
The stratification of the data allows us to examine whether the PS is well calibrated and not poorly specified. First, we estimated the PS by regressing “unable to eat” on the linear combination of the covariates, the most common way PS is used. The following equation was estimated:
As we are working with a large amount of data, it was not surprising that all coefficients in the logistic regression were statistically significant (alpha levels <0.001).
Second, for each stratum with at least 10 data points, we also calculated the observed probability of being unable to eat. This allowed us to compare the calibration of the PS and the observed probability of feeding disability. Figure 3 shows the results. The PS is shown as the dotted line (Observed = 0.3342 Predicted + 0.1846). If the PS was well calibrated, all points would fall around the 45 degree line (Observed = 0.45 Predicted), shown as the solid line. This is not the case. The PS is overestimating probability of being unable to eat in low probability strata and underestimating it in high probability strata. It is only well calibrated in strata with approximately 0.3 probability of being disabled. We conclude that the linear combination of the covariates used in balancing these data leads to a misspecified PS. In contrast, for each stratum, SCB used the weights calculated from the observed frequency of events; thus, it was perfectly calibrated. Our simulation presented earlier and simulation studies by others (Pirracchio, Resche‐Rigon, and Chevret 2012) have shown that misspecification of PS matters and could lead to wrong study conclusions.
Figure 3.
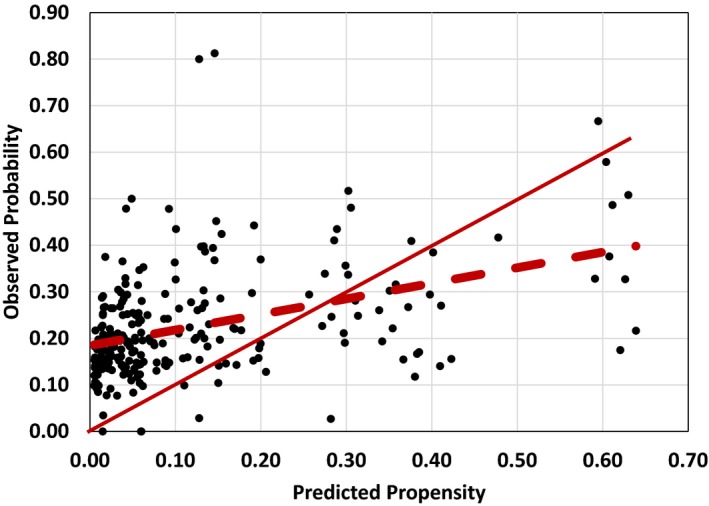
Calibration of Propensity Scores [Color figure can be viewed at wileyonlinelibrary.com]
Partial Matches and Sensitivity Analysis
At the end of the covariate balancing procedure, 164,003 of 164,017 cases were matched to 865,849 of 875,063 controls. This shows that on average, 99.99 percent of cases were matched and 98.94 percent of controls were used. A total of 9,214 controls were not used in the analysis. In this example, the dataset was large and the number of covariates relatively small. So, most of the data were used. This may not be the situation in other datasets. As the number of covariates increases, the number of cases per strata decreases, and combinations of covariates become quite rare. In these circumstances, it is possible that a large portion of the cases may not have matching controls, and therefore not used, reducing the generalizability of the findings. The findings will still be accurate for cases that were matched but perhaps not valid for other types of cases. In these situations, the analyst should progressively drop covariates from the analysis and examine the sensitivity of the estimates. Each time a covariate is dropped, a partial match is made. In partial matches, a larger number of patients fall within each strata. The important analytical issue is to examine whether the reduction in covariates changes the study finding, for example, by removing the statistical significance of the impact of the treatment. This approach allows one to test the sensitivity of the study conclusions to the percent of cases matched.
Table 2 provides the estimated odds, when one of the covariates is left unmatched. The first row in the table shows the estimated unconfounded odds ratio using all covariates. The remaining rows remove one covariate at a time. The number of cases matched increases when fewer covariates are used. For example, exclusion of “unable to urinate” increases the number of cases matched to 100 percent. At the same time, the exclusion of “unable to urinate” does not change the estimated odds of mortality by much. After dropping any of the covariates, the odds of mortality for residents who are unable to eat ranges from 1.79 to 1.87. The conclusion that residents who are unable to feed themselves are at increased risk of mortality does not change despite dropping one of the covariates from the analysis. This sensitivity analysis shows that (1) partial matches can increase the number of cases matched and (2) address sensitivity of conclusions to dropping any one of the covariates.
Table 2.
Sensitivity of Odds of Mortality for Residents Unable to Eat
| Covariate Removed | Cases Unable to Eat n (%) | Controls Able to Eat n (%) | Odds of Mortality |
|---|---|---|---|
| None | 164,003 (99.9%) | 865,849 (98.9%) | 1.86 |
| Age | 164,009 (99.9%) | 868,818 (99.2%) | 1.87 |
| Gender | 164,016 (99.9%) | 873,160 (99.7%) | 1.85 |
| Unable to bathe | 164,003 (99.9%) | 865,849 (98.9%) | 1.86 |
| Unable to walk | 164,009 (99.9%) | 868,818 (99.2%) | 1.87 |
| Unable to dress | 164,016 (99.9%) | 873,160 (99.7%) | 1.85 |
| Unable to bowel | 164,017 (100%) | 873,954 (99.8%) | 1.79 |
| Unable to urinate | 164,017 (100%) | 874,624 (99.9%) | 1.81 |
| Unable to groom | 164,017 (100%) | 874,624 (99.9%) | 1.81 |
| Unable to toilet | 164,017 (100%) | 875,063 (100%) | 1.83 |
| Unable to sit | 164,017 (100%) | 873,954 (99.8%) | 1.79 |
Discussion
SCB removes the impact of covariates by dividing the data into strata, where all patients falling into the strata share the same levels of the covariates. Within the strata, the covariates are held constant. Scientists refer to variables that are held constant as ceteris paribus events. Ceteris paribus events play important roles in scientific theories and in analysis of data to verify these theories. The approach could be used to evaluate the comparative effectiveness of treatment from observation data. In such an application, patients’ medical histories are held ceteris paribus and the impact of treatment on outcome is calculated. In our discussions with clinicians, we have found that most clinicians understand the concept of stratification, perhaps easier than regression analysis used for PS, which to some clinicians is a black box. Stratification seems to make intuitive sense to clinicians. Furthermore, they may value information SCB provides about treatment effectiveness in subsets of patients who fall within different strata.
By way of an example, the paper showed that SCB is computationally practical. This paper showed that SCB was able to remove confounding between feeding disability, age, gender, and other disabilities in a large dataset. A plot of odds of the covariates showed that we had successfully balanced the data; in addition, we reported that confounding caused by combination of covariates was also removed. After weighting the data, all co‐occurring covariates were equally likely to occur among residents with and without feeding disability. These data led us to conclude that SCB was practical and can balance the data without extensive search for a properly specified statistical model for PS.
Besides being easy to understand and computationally practical, when interaction terms were present, SCB was also more accurate than pairwise PS or pairwise LR. Typically, the PS or LR is calculated from linear combinations of the covariates, occasionally from pairwise combination of covariates. In simulated data, pairwise PS and pairwise LR performed poorly when higher order interaction terms were present; SCB consistently performed well even when higher order interaction terms were present. This simulation pointed out that correct specification of the PS is important in accurate estimation of impact of treatment. The misspecification of PS was also observed in real data, where PS was poorly calibrated.
One way to improve calibration of PS models is to include more interaction terms (Pirracchio et al. 2013). However, investigators may arrive at different set of weights depending on the extent of their effort to include interaction terms. In contrast, SCB does not depend on the effort of investigators. Using the proposed SCB method, all investigators will arrive at the same set of weights, whether they are concerned with interaction terms.
R Package
We recommend the use of SCB because of ease of understanding, computation considerations, and accuracy. To assist the use of SCB, on July 15, 2016, we provided a free R package online (search online for StratifiedBalancing R Package).
Limitations and Future Research
The procedure described here ignores cases that do not match to any controls. Failure to match all cases might affect the ability to generalize the findings to relatively rare subsets of the population. Investigators may wish to make partial matches between cases and controls so that their findings are relevant to a larger subset of the population. One way to make partial matches is to reduce the number of covariates used in constructing the strata. Analysts should always conduct sensitivity analysis of their findings to partial matches; one can do so by progressively dropping covariates from the analysis, examining treatment impact, and checking whether the study conclusions change. Besides progressively dropping covariates, two other strategies are also available for partial matching:
Analysts could use the Apriori algorithm (Bayardo 1998) to identify combinations with significant “support” (e.g., more than 29 cases) in the data.
Analysts could use classification and regression trees to predict participation in treatment from a subset of covariates. Such an approach would construct strata from smaller set of covariates that are most predictive of treatment.
All three approaches use fewer covariates (in other words make partial matches), provide a smaller number of strata, include more cases per strata, and are more likely to find a matching control for every case. The relative value of these three methods is not clearly understood, and more research is needed on optimal methods of constructing strata, especially in large multidimensional problems.
This paper compared the performance of SCB to inverse weighting of PS. Other methods of PS, reviewed earlier, are also available. The relative performance of SCB and these variants of PS is not known. Theoretically, at the core of all the variants of PS is a parametric model that may be misspecified. Therefore, we suspect that SCB will remain more accurate than variants of PS. Exceptions may exist, especially in recently proposed approach, where the PS model and the prediction of outcomes are carried out in one step (Imai and Ratkovic 2014). Future research can clarify conditions under which SCB and variants of PS perform best.
Supporting information
Appendix SA1: Author Matrix.
Appendix SA2: Standard Query Language for Stratified Covariate Balancing.
Appendix SA3: Strata for Predicting Impact of “Unable to Feed” on Mortality.
Acknowledgments
Joint Acknowledgement/Disclosure Statement: This study was initiated in Ithaca Greece, in conversations with Mastaneh Badii and Yara Alemi. This paper has benefitted from comment by Arthur Williams, Ph.D., Janusz Wojtusiak, Ph.D., and other colleagues. This project was funded by appropriation #3620160 from VA Office of Geriatrics and Extended Care. The contents of this paper do not represent the views of the Department of Veterans Affairs or the United States Government.
Disclosure: None.
Disclaimer: None.
References
- Abadie, A. , and Imbens G. W.. 2006. “Large Sample Properties of Matching Estimators for Average Treatment Effects.” Econometrica 74: 235–67. [Google Scholar]
- Abadie, A. , and Imbens G. W.. 2011. “Bias‐Corrected Matching Estimators for Average Treatment Effects.” Journal of Business and Economic Statistics 29: 1–11. [Google Scholar]
- Agrawal, R. , Imieliński T., and Swami A.. 1993. “Mining Association Rules between Sets of Items in Large Databases.” Proceedings of the 1993 ACM SIGMOD international conference on Management of Data — SIGMOD ‘93. p. 207.
- Agrawal, R. , and Srikant R.. 1994. “Fast Algorithms for Mining Association Rules in Large Databases.” Proceedings of the 20th International Conference on Very Large Data Bases, VLDB, pp. 487–499, Santiago, Chile, September 1994.
- Alemi, F. , Levy C., Citron B. A., Williams A. R., Pracht E., and Williams A.. 2016. “Improving Prognostic Web Calculators: Violation of Preferential Risk Independence.” Journal of Palliative Medicine ahead of print. doi:10.1089/jpm.2016.0126. [DOI] [PubMed] [Google Scholar]
- Anderson, D. W. , Kish L., and Cornell R. G.. 1980. “On Stratification, Grouping and Matching.” Scandinavian Journal of Statistics 7 (2): 61–6. [Google Scholar]
- Atkinson, A. C. 1982. “Optimum Biased Coin Designs for Sequential Clinical Trials with Prognostic Factors.” Biometrika 69 (1): 61–7. [Google Scholar]
- Austin, P. C. 2009a. “Balance Diagnostics for Comparing the Distribution of Baseline Covariates between Treatment Groups in Propensity‐Score Matched Samples.” Statistics in Medicine 28 (25): 3083–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, P. C. 2009b. “The Relative Ability of Different Propensity Score Methods to Balance Measured Covariates between Treated and Untreated Subjects in Observational Studies.” Medical Decision Making 29 (6): 661–77. [DOI] [PubMed] [Google Scholar]
- Austin, P. C. 2011a. “An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies.” Multivariate Behavioral Research 46 (3): 399–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, P. C. 2011b. “A Tutorial and Case Study in Propensity Score Analysis: An Application to Estimating the Effect of In‐Hospital Smoking Cessation Counseling on Mortality.” Multivariate Behavioral Research 46 (1): 119–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayardo, R. J. 1998. “Efficiently Mining Long Patterns from Databases (PDF).” ACM Sigmod Record 27 (2): 85–93. [Google Scholar]
- Cochran, W. G. 1950. “The Comparison of Percentages in Matched Samples.” Biometrika 37 (3/4): 256–66. [PubMed] [Google Scholar]
- Extermann, M. 2007. “Interaction between Comorbidity and Cancer.” Cancer Control 14 (1): 13–22. [DOI] [PubMed] [Google Scholar]
- Ferdinandy, P. , Hausenloy D. J., Heusch G., Baxter G. F., and Schulz R.. 2014. “Interaction of Risk Factors, Comorbidities, and Comedications with Ischemia/Reperfusion Injury and Cardioprotection by Preconditioning, Postconditioning, and Remote Conditioning.” Pharmacological Reviews 66 (4): 1142–74. [DOI] [PubMed] [Google Scholar]
- Frangakis, C. E. , and Rubin D. B.. 2002. “Principal Stratification in Causal Inference.” Biometrics 58: 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, B. B. 2004. “Full Matching in an Observational Study of Coaching for the SAT.” Journal of the American Statistical Association 99: 609–18. [Google Scholar]
- Heckman, J. J. , Ichimura H., and Todd P.. 1998. “Matching as an Econometric Evaluation Estimator.” Review of Economic Studies 65: 261–94. [Google Scholar]
- Heritier, S. , Gebski V., and Pillai A.. 2005. “Dynamic Balancing Randomization in Controlled Clinical Trials.” Statistics in Medicine 24 (24): 3729–41. [DOI] [PubMed] [Google Scholar]
- Hirano, K. , Imbens G., and Ridder G.. 2003. “Efficient Estimation of Average Treatment Effects using the Estimated Propensity Score.” Econometrica 71: 1307–38. [Google Scholar]
- Ho, D. E. , Imai K., King G., and Stuart E. A.. 2007. “Matching as Nonparametric Preprocessing for Reducing Model Dependence in Parametric Causal Inference.” Political Analysis 15: 199–236. [Google Scholar]
- Imai, K. , and Ratkovic M.. 2014. “Covariate Balancing Propensity Score.” Journal of the Royal Statistical Society: Series B 76 (Part 1): 243–63. [Google Scholar]
- Kang, J. D. , and Schafer J. L.. 2007. “Demystifying Double Robustness: A Comparison of Alternative Strategies for Estimating A Population Mean from Incomplete Data (with Discussions).” Statistical Science 22: 523–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel, N. , and Haenszel W.. 1959. “Statistical Aspects of the Analysis of Data from Retrospective Studies of Disease.” Journal of the National Cancer Institute 22 (4): 719–48. [PubMed] [Google Scholar]
- McCaffrey, D. F. , Ridgeway G., and Morral A. R.. 2004. “Propensity Score Estimation with Boosted Regression for Evaluating Causal Effects in Observational Studies.” Psychological Methods 9 (4): 403–25. [DOI] [PubMed] [Google Scholar]
- Pirracchio, R. , Resche‐Rigon M., and Chevret S.. 2012. “Evaluation of the Propensity Score Methods for Estimating Marginal Odds Ratios in Case of Small Sample Size.” BMC Medical Research Methodology 30 (12): 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirracchio, R. , Carone M., Rigon M. R., Caruana E., Mebazaa A., and Chevret S.. 2013. “Propensity Score Estimators for the Average Treatment Effect and the Average Treatment Effect on the Treated May Yield Very Different Estimates.” Statistical Methods in Medical Research 25 (5): 1938–54. [DOI] [PubMed] [Google Scholar]
- Pocock, S. J. , and Simon R.. 1975. “Sequential Treatment Assignment with Balancing for Prognostic Factors in the Controlled Clinical Trial.” Biometrics 31 (1): 103–15. [PubMed] [Google Scholar]
- Robins, J. M. , Hernan M. A., and Brumback B.. 2000. “Marginal Structural Models and Causal Inference in Epidemiology.” Epidemiology 11: 550–60. [DOI] [PubMed] [Google Scholar]
- Robins, J. M. , Rotnitzky A., and Zhao L. P.. 1995. “Analysis of Semiparametric Regression Models for Repeated Outcomes in the Presence of Missing Data.” Journal of the American Statistical Association 90: 106–21. [Google Scholar]
- Rosenbaum, P. R. 1987. “Model‐Based Direct Adjustment.” Journal of the American Statistical Association 82: 387–94. [Google Scholar]
- Rosenbaum, P. R. 1989. “Optimal Matching for Observational Studies.” Journal of the American Statistical Association 84: 1024–32. [Google Scholar]
- Rosenbaum, P. R. 1991. “A Characterization of Optimal Designs for Observational Studies.” Journal of the Royal Statistical Society: Series B 53: 597–610. [Google Scholar]
- Rosenbaum, P. R. , and Rubin D. B.. 1983. “The Central Role of the Propensity Score in Observational Studies for Causal Effects.” Biometrika 70: 41–55. [Google Scholar]
- Rosenbaum, P. R. , and Rubin D. B.. 1984. “Reducing Bias in Observational Studies Using Sub‐Classification on the Propensity Score.” Journal of the American Statistical Association 79: 516–24. [Google Scholar]
- Rosenbaum, P. R. , and Rubin D. B.. 1985. “Constructing a Control Group Using Multivariate Matched Sampling Methods that Incorporate the Propensity Score.” The American Statistician 39: 33–8. [Google Scholar]
- Scott, N. W. , McPherson G. C., Ramsay C. R., and Campbell M. K.. 2002. “The Method of Minimization for Allocation to Clinical Trials: A Review.” Controlled Clinical Trials 23 (6): 662–74. [DOI] [PubMed] [Google Scholar]
- Signorini, D. F. , Leung O., Simes R. J., Beller E., Gebski V. J., and Callaghan T.. 1993. “Dynamic Balanced Randomization for Clinical Trials.” Statistics in Medicine 12 (24): 2343–50. [DOI] [PubMed] [Google Scholar]
- Tripepi, G. , Jager K. J., Dekker F. W., and Zoccali C.. 2010. “Stratification for Confounding–Part 1: The Mantel‐Haenszel Formula.” Nephron Clinical Practice 116 (4): c317–21. [DOI] [PubMed] [Google Scholar]
- Wei, L. J. 1978. “Application of an Urn Model to Design of Sequential Controlled Clinical‐Trials.” Journal of American Statistical Association 73 (363): 559–63. [Google Scholar]
- Yuan, Y. , Huang X. L., and Liu S. Y.. 2011. “A Bayesian Response‐Adaptive Covariate‐Balanced Randomization Design with Application to a Leukemia Clinical Trial.” Statistics in Medicine 30 (11): 1218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix SA1: Author Matrix.
Appendix SA2: Standard Query Language for Stratified Covariate Balancing.
Appendix SA3: Strata for Predicting Impact of “Unable to Feed” on Mortality.


