Summary
Background
Antibiotic use in human medicine, veterinary medicine, and agriculture has been linked to the rise of antibiotic resistance globally. We did a systematic review and meta-analysis to summarise the effect that interventions to reduce antibiotic use in food-producing animals have on the presence of antibiotic-resistant bacteria in animals and in humans.
Methods
On July 14, 2016, we searched electronic databases (Agricola, AGRIS, BIOSIS Previews, CAB Abstracts, MEDLINE, Embase, Global Index Medicus, ProQuest Dissertations, Science Citation Index) and the grey literature. The search was updated on Jan 27, 2017. Inclusion criteria were original studies that reported on interventions to reduce antibiotic use in food-producing animals and compared presence of antibiotic-resistant bacteria between intervention and comparator groups in animals or in human beings. We extracted data from included studies and did meta-analyses using random effects models. The main outcome assessed was the risk difference in the proportion of antibiotic-resistant bacteria.
Findings
A total of 181 studies met inclusion criteria. Of these, 179 (99%) described antibiotic resistance outcomes in animals, and 81 (45%) of these studies were included in the meta-analysis. 21 studies described antibiotic resistance outcomes in humans, and 13 (62%) of these studies were included in the meta-analysis. The pooled absolute risk reduction of the prevalence of antibiotic resistance in animals with interventions that restricted antibiotic use commonly ranged between 10 and 15% (total range 0–39), depending on the antibiotic class, sample type, and bacteria under assessment. Similarly, in the human studies, the pooled prevalence of antibiotic resistance reported was 24% lower in the intervention groups compared with control groups, with a stronger association seen for humans with direct contact with food-producing animals.
Interpretation
Interventions that restrict antibiotic use in food-producing animals are associated with a reduction in the presence of antibiotic-resistant bacteria in these animals. A smaller body of evidence suggests a similar association in the studied human populations, particularly those with direct exposure to food-producing animals. The implications for the general human population are less clear, given the low number of studies. The overall findings have directly informed the development of WHO guidelines on the use of antibiotics in food-producing animals.
Funding
World Health Organization.
Introduction
Infections with antibiotic-resistant bacteria result in increased mortality, morbidity, and social and economic costs.1, 2, 3, 4 By 2050, an estimated 10 million deaths per year globally will be attributable to antimicrobial resistance, with a cumulative economic cost of US$100 trillion.5 Governments around the world have mobilised to address this pressing public health concern at the recent G20 Summit and the meeting of the UN General Assembly.6, 7 Further, WHO has created a set of strategies to combat rising antibiotic resistance, which include improving sanitation and hygiene to reduce overall infection rates, and optimising the use (and preventing the overuse) of antibiotics in both humans and animals.8
Research in context.
Evidence before this study
Rising rates of antimicrobial resistance are a significant global concern. The widespread use of antibiotics in human beings and animals contributes to this problem. Antibiotics used in food-producing animals are closely related to those used in human medicine and can select for resistance in these animals. Cross-species transmission of resistant bacteria or resistant genetic elements from animals to humans can and does occur. As a result, many jurisdictions have implemented restrictions on the use of antibiotics in agriculture, in an effort to combat rising antibiotic resistance in humans. It is unclear, however, whether these restrictions are effective. There are a large number of original studies exploring effects of such interventions on antibiotic resistance. However, these studies have not yet been fully systematically reviewed or meta-analysed, nor have they been thoroughly explored to understand the heterogeneity of findings.
Added value of this study
We have done a large-scale systematic review of the global literature, seeking to fully summarise and distil existing knowledge. To our knowledge, this is the first review to comprehensively examine the associations between interventions that restrict antibiotic use in food-producing animals and antibiotic resistance in both animals and humans. We searched electronic databases (Agricola, AGRIS, BIOSIS Previews, CAB Abstracts, MEDLINE, Embase, Global Index Medicus, ProQuest Dissertations, Science Citation Index) and grey literature on July 14, 2016, with an update on Jan 27, 2017. Any interventions that aimed to reduce antibiotic use in food-producing animals (including but not limited to measures to improve antimicrobial stewardship, setting national antibiotic reduction targets, mandatory or voluntary bans, and antibiotic-free farming systems) were considered. We found a large body of literature (179 eligible studies, 81 of which were included in the meta-analysis) showing that interventions that reduce antibiotic use in food-producing animals are associated with a reduction in prevalence of antibiotic resistance in these animals. Our findings also suggest a similar association of reduced antibiotic resistance in human beings (21 studies, 13 of which were included in the meta-analysis), particularly those in direct contact with food-producing animals.
Implications of all available evidence
These findings are informative in the development of forthcoming guidelines from different sectors such as governmental bodies and WHO. Reducing antibiotic use in agriculture is associated with reductions in antibiotic resistance in food-producing animals. At minimum, the benefit appears to extend to farmers and those in direct contact with food-producing animals. The evidence of benefit for the general human population is less clear, but has potential for broad-reaching effects. The wide implementation of strategies to reduce antibiotic use will require balanced consideration of the benefits of reducing antibiotic resistance against any potential unintended consequences.
There is increasing recognition that widespread antibiotic use in agriculture and aquaculture might contribute to the development of resistance to antibiotics commonly used in human medicine,9, 10, 11 especially given the overlap of antibiotics used for these different purposes.10, 12, 13, 14 For example, bacteria in animals that are treated with antibiotics can develop antibiotic resistance, and these bacteria, which might carry resistance genes, can then be transmitted from animals to humans.15 This cross-species transmission can occur through food, direct contact between humans and animals, or shared environmental sources such as contaminated water.15
There is currently no consensus regarding the effect that antibiotic use in food-producing animals has on antibiotic resistance in the human population. Furthermore, the effect of interventions that restrict antibiotics in food-producing animals on antibiotic resistance in both animals and humans is somewhat unclear. Despite this evidence gap, many countries (particularly in the European Union) have made substantial efforts to reduce the overall use of antibiotics in food-producing animals, through the creation of national reduction targets, implementation of mandatory bans of antimicrobial drugs in the feed of food-producing animals, benchmarking antibiotic use at the farm level, and encouraging antibiotic stewardship such as by requiring susceptibility testing before the use of some high-priority antibiotics.16 Though it is plausible that such interventions will result in lower antibiotic resistance in animals, the environment, and in human beings, sound evidence is required to shape global policies regarding antibiotic use in the food animal industry.
We were commissioned by the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (WHO AGISAR) to do a systematic review and meta-analysis, to inform the development of WHO guidelines on the use of antibiotics in food-producing animals. The objective of our systematic review was to assess whether a restriction of antibiotic use in food-producing animals (specifically within the classifications of avian, swine, bovine, caprine, camel, equine, rabbit, ovine, fish, bees, molluscs, and crustaceans) reduces the presence of antibiotic-resistant bacteria in food-producing animals and in human beings.
Methods
Search strategy and selection criteria
We did this systematic review and meta-analysis using a predetermined protocol and in accordance with PRISMA reporting standards.17 On July 14, 2016, we identified potentially relevant articles by searching Agricola, AGRIS, BIOSIS Previews, CAB Abstracts, MEDLINE, Embase, Global Index Medicus ProQuest Dissertations, and Science Citation Index. No limits were placed based on publication date. We updated the literature search on Jan 27, 2017.
Our multidisciplinary team, which included experts in veterinary medicine and animal agriculture (HWB, SCC), infectious diseases (JDK), systematic review methodology (PER, WAG), and library sciences (HG), developed a search strategy consisting of controlled vocabulary and keywords (appendix pp 2–5) that described three comprehensive search themes: the animal populations of interest (theme 1), resistance to the antibiotics on the WHO list of Critically Important Antimicrobials for Human Medicine and the World Organisation for Animal Health (OIE) List of Antimicrobial Agents of Veterinary Importance (non-proprietary names searched only; theme 2),18, 19 and interventions to restrict antibiotic use (theme 3). Resources from the Food and Agriculture Organization of the UN informed the terms used for theme 1, specifically for terms relating to aquaculture.20 The three broad themes were combined using the Boolean operator “AND”. The search strategy was peer-reviewed by research librarians within the University of Calgary and at WHO. Though the terms used in the search strategy were in English, no limits were placed on publication language. Foreign-language articles were therefore still captured by this search strategy (appendix pp 2–5). Database-specific subject headings and controlled vocabulary were adapted when necessary from the original MEDLINE search.
The electronic database search was enhanced by scanning reference lists of relevant review articles and articles included in this systematic review that were published from 2010 onward, and by reviewing conference proceedings from major scientific meetings. Grey literature was identified by searching websites of relevant health agencies, professional associations, and other specialised databases (appendix pp 6–15). Finally, experts in surveillance and epidemiology of antimicrobial use and resistance, veterinary medicine, and animal health policy were contacted regarding potential missed, ongoing, or unpublished studies. The WHO Guideline Development Group also provided feedback with regards to relevant new or missing studies.
Two authors (KLT and NPC) independently reviewed all identified abstracts for eligibility. Abstracts that reported original research, described an intervention that aimed to reduce antibiotic use in animals, and described antibiotic resistance in animals or humans were selected for full-text review. Disagreements were resolved by consensus or third-party consultation (SCC) when consensus could not be achieved.
The same reviewers then performed a full-text review of articles meeting eligibility criteria for the systematic review. Articles were retained if the population studied included food-producing animals (within the previously mentioned classifications); the interventions aimed to reduce the use of one or more antibiotics in food-producing animals (these interventions may be of any type [including but not limited to mandatory or voluntary bans, antibiotic-free or organic farming systems, the creation of national reduction targets, requiring culture and sensitivity testing prior to antibiotic use] and at any level [including restrictions for only certain indications such as growth promotion to complete restrictions that do not permit any antibiotic use for any reason]); there was a presence of a comparator for which there was no restriction of antibiotic use in food-producing animals (historical comparators were considered eligible); the outcomes included the presence of genotypic or phenotypic antibiotic resistant bacteria or changes to antibiotic susceptibility in food-producing animals or in humans; and if the studies used were original research.
To be as inclusive and comprehensive as possible, there were no limits placed based on study quality, with study quality issues handled through sensitivity analyses instead. The classifications of food-producing animals included in the population of interest were identified through consensus within our study team and WHO AGISAR, and include the most commonly consumed food-producing animals globally.21 No limits were placed on the bacterial species studied. Inter-rater agreement between the two reviewers for inclusion into the systematic review at the full-text review stage was good (κ=0·74).
Data analysis
KLT and NPC extracted data from individual studies using a predesigned form obtaining data on the author, year, study design (eg, randomised trial, non-randomised trial, cross-sectional design, or pre-post study), country, species and age of animal, number of farms or animals used in analysis, sample type (eg, faeces and meat), sampling point (eg, farm, slaughter, or retail), description of intervention, description of comparator, antibiotic panels, laboratory procedure, bacteria investigated, and prevalence of antibiotic-resistant bacteria in intervention and control groups. The same two reviewers independently, and in duplicate, assessed the methodological quality of each individual study based on prespecified study quality indicators adapted from the Downs and Black checklist.22
Absolute risk differences within individual studies were calculated for each individual antibiotic by subtracting the proportion of isolates resistant to that antibiotic in the control group from the proportion in the intervention group. Resistance was considered a dichotomous outcome, as classified by the individual primary studies. Isolates with intermediate susceptibility were classified as susceptible.
Meta-analyses were done separately for animal and human studies. Meta-analysis was only done if there were six or more studies, as between-study variance cannot be estimated accurately with fewer than this number and might have resulted in biased pooled estimates when the meta-analysis was done.23 Studies that did not report absolute risk differences in antibiotic resistance or did not provide sufficient data to allow its calculation were not included in meta-analyses.
We pooled absolute risk differences, by antibiotic class, using DerSimonian and Laird random effects models.24 A pooled negative risk difference would indicate a lower prevalence of resistance to that antibiotic class in the intervention group compared to the control group. A random effects model was felt necessary a priori regardless of actual heterogeneity of findings because the studies were known to be clinically heterogeneous, assessing various interventions across different regions globally. The most common unit of analysis in individual studies was at the isolate level. Studies using a different unit of analysis were not included in meta-analysis.
For the animal studies, we made an a priori decision to pool analyses within five groups: Enterobacteriaceae in meat samples, Enterococcus spp in faecal samples, Campylobacter spp in faecal samples, Campylobacter spp in meat samples, and Staphylococcus spp in milk samples. These divisions were made because of fundamental differences in these bacterial groups in terms of their microbiological characteristics, innate antibiotic resistance, and potential for pathogenicity.25, 26, 27, 28 Within each of these groups, separate meta-analyses were done for each different antibiotic class, and also for multidrug resistance. If this was not defined by study authors, we considered multidrug resistance to be resistance to two or more antibiotics, as this was the most commonly used definition within the primary research studies. Heterogeneity across studies was assessed using the Q statistic (significance level of p≤0·10) and the I2 statistic.29, 30
If a study provided multiple point estimates (ie, multiple antibiotics that fell within the same antibiotic class, multiple bacterial species that fell within the same genus, or multiple measures across different geographical regions), we used a fixed effects model with inverse variance weighting to generate a single point estimate (risk difference) per antibiotic class, per analytic group, for each study. If studies examined more than one bacterial group (eg, Staphylococcus aureus and Enterococcus), then a separate set of risk differences (by antibiotic class) were generated for each group. For longitudinal study designs with repeated measures of antibiotic resistance, the first and last datapoints were used to calculate risk differences.
For animal studies that could not be included in meta-analysis because of inadequate reporting or where fewer than six studies could be pooled, a semi-quantitative analysis was completed to visualise trends in phenotypic antibiotic resistance across bacterial categories and antibiotic classes. Results from individual studies were colour-coded within a table to illustrate whether prevalence of resistance to each antibiotic class in intervention groups was lower, higher, or not significantly different compared with the control groups.
Because of the low numbers of human studies, we chose to do a single meta-analysis, rather than a set of meta-analyses based on the five analytic groupings as for the animal studies. This approach was justified as the meta-analysis results from animal studies suggested that pooled effect estimates were not modified by sample type, bacteria isolated, and antibiotics tested. The unit of analysis was the sample rather than the isolate, as most human studies reported sample-level data. Random effects models were used, and heterogeneity was assessed in the same manner as for animal studies.
For animal studies, we also did a so-called bundled meta-analysis, which included all studies amenable to meta-analysis, ignoring specific bacterial species, sample types, units of analysis, and antibiotic classes, for the sole purposes of stratifying analysis and assessing publication bias. A single effect estimate (absolute risk difference) was generated for each study by doing a within-study meta-analysis using random effects models. This bundled meta-analysis was done to allow meaningful and statistically powered stratified analyses. This approach was felt to be appropriate based on preceding meta-analytic results showing generally consistent findings across constituent elements.
We did stratified analyses for human and animal studies for different study populations, sample types, intervention characteristics, and study quality criteria. Meta-regression was used to identify whether these factors were significant predictors of the underlying heterogeneity.
Publication bias was assessed separately for animal and human studies, including all studies for which we did meta-analyses. This was done using Begg's test in addition to the visual inspection of a funnel plot.31 Sensitivity analysis using the Duval and Tweedie non-parametric trim and fill procedure was also implemented if there was any suggestion of visual asymmetry on the funnel plots.32 Data were analysed in Stata version 14.
Role of the funding source
WHO was involved in the development of the research question, the study design, and the study protocol; they also peer-reviewed the final search strategy. They had no involvement in data extraction or interpretation of findings. The authors have been given permission by WHO to publish this article. We presented the findings to WHO AGISAR in October, 2016, and to the WHO Guideline Development Group in March, 2017. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
The initial search strategy identified a total of 9008 citations, with an additional 56 identified by contacting experts in the field of antibiotic use and resistance, and another 82 through searching reference lists of included studies and relevant review articles. From these, 3201 duplicates were removed, and 5945 records were screened for eligibility through title and abstract review. After removal of the 5559 records that were not relevant to the research objectives, 386 full-text articles were reviewed. Of these, 181 studies were included in the systematic review (figure 1; references for all included studies are in the appendix pp 52–64). A total of 179 studies reported on the outcome of antibiotic resistance in animals, and 21 reported on the outcome of antibiotic resistance in humans (with an overlap of 19 studies that reported outcomes for both animals and humans).
Figure 1.
Flow diagram of the study selection process
*160 studies were exclusively in animals; the other 19 studied antibiotic resistance in both human beings and animals and so are counted in both animal studies and human studies. †Two studies were exclusively in the human population; the other 19 studied antibiotic resistance in both human beings and animals and so are counted in both animal studies and human studies.
81 studies were from the USA and 78 were from Europe (appendix p 16). Six study populations were from Asia and one was from Africa. Five studies did not specify the countries in which their study populations were located.
Of the 179 animal studies, 148 were journal articles, 20 were meeting abstracts or conference proceedings without accompanying full-text articles, nine were dissertations, and two were government or organisation reports (table 1). Most studies had a cross-sectional design (n=126). There were 50 longitudinal studies, and three non-randomised interventional trials. Poultry was the most commonly studied animal population, followed by swine and dairy cattle (appendix pp 17–32). Of the 21 human studies, 19 were journal articles, one was a meeting abstract, and one was an organisation report. 13 studies had a longitudinal design; eight were cross-sectional. The human population studied was most commonly farm workers and their household contacts (appendix pp 33–35).
Table 1.
Summary of study characteristics
| Number of animal studies (n=179) | Number of human studies (n=21) | ||
|---|---|---|---|
| Article type | |||
| Journal article | 148 (83%) | 19 (90%) | |
| Abstract only | 20 (11%) | 1 (5%) | |
| Dissertation | 9 (5%) | ·· | |
| Government or organisation report | 2 (1%) | 1 (5%) | |
| Study design | |||
| Non-randomised controlled trial | 3 (2%) | ·· | |
| Cross-sectional | 126 (70%) | 8 (38%) | |
| Longitudinal | 50 (28%) | 13 (62%) | |
| Population studied* | |||
| Beef cattle | 20 (11%) | ·· | |
| Dairy cattle | 36 (20%) | ·· | |
| Poultry: broilers, turkeys | 87 (49%) | ·· | |
| Poultry: egg layers | 10 (6%) | ·· | |
| Swine | 61 (34%) | ·· | |
| Goats | 1 (1%) | ·· | |
| Salmon | 1 (1%) | ·· | |
| Farm workers and household members | ·· | 12 (57%) | |
| Healthy adults | ·· | 5 (24%) | |
| Patients or cases | ·· | 6 (29%) | |
| Intervention studied* | |||
| Externally imposed bans and reductions | 36 (20%) | 9 (43%) | |
| Organic interventions | 87 (49%) | 2 (10%) | |
| Self-labelled antibiotic-free, pasture, or free-range | 38 (21%) | 5 (24%) | |
| Voluntary reduction or withdrawal in antibiotic use | 29 (16%) | 5 (24%) | |
| Sample studied* | |||
| Faecal/cloacal swabs/caecum | 106 (59%) | 12 (57%) | |
| Meat or carcass | 53 (30%) | ·· | |
| Milk | 20 (11%) | ·· | |
| Eggs | 7 (4%) | ·· | |
| Nasal swabs | 11 (6%) | 8 (38%) | |
| Urine | ·· | 1 (5%) | |
| Blood | ·· | 1 (5%) | |
| Unknown | 4 (2%) | 2 (10%) | |
| Bacteria studied* | |||
| Campylobacter spp. | 30 (17%) | 2 (10%) | |
| Enterococcus spp. | 39 (22%) | 8 (38%) | |
| Enterobacteriaceae | ·· | ·· | |
| Escherichia coli | 58 (32%) | 3 (14%) | |
| Salmonella spp. | 31 (17%) | 1 (5%) | |
| Yersinia enterocolitica | 1 (1%) | ·· | |
| Unspecified Enterobacteriaceae | 2 (1%) | ·· | |
| Staphylococcus spp. | 31 (17%) | 8 (38%) | |
| Other | ·· | ·· | |
| Listeria monocytogenes | 3 (2%) | ·· | |
| Lactobacillus spp | 1 (1%) | ·· | |
| Unspecified | 7 (4%) | ·· | |
Data are n (%).
Categories are not mutually exclusive and studies can be included in more than one category.
Interventions were classified into four categories: externally imposed bans or restrictions of antibiotic use (36 animal studies, nine human studies); organic interventions, as defined by the study and the country-specific regulations for organic certification (87 animal studies and two human studies); self-labelled antibiotic-free, free-range, or pasture systems (38 animal studies, five human studies); and voluntary reduction of antibiotic use (29 animal studies, five human studies). These intervention categories were not mutually exclusive. For example, studies that reported antibiotic resistance in antibiotic-free versus organic versus conventional meats were considered to include both organic and antibiotic-free interventions.
Study quality was assessed for all studies (table 2; appendix pp 36–46). Nearly all studies had a clearly defined research question or objective. For both animal and human studies, an area of weaker study quality was the poor description of the study populations and management practices on farms and in slaughterhouses. Without a clear description of sample characteristics, it was difficult to assess whether intervention and control groups were similar and whether they were representative of the source population. There was also poor description of the interventions, specifically in animal studies that sampled at the retail level, such as those comparing retail meats labelled as organic or antibiotic-free with conventional meats. As a result, many studies had insufficient detail regarding their farming and production systems and degree of antibiotic use (eg, whether antibiotics are allowed therapeutically). Finally, few animal or human studies adjusted for potential confounders, despite the many variables that are likely to confound the complex association between antibiotic resistance and antibiotic use in animals.
Table 2.
Proportion of studies meeting individual study quality criteria
| Animal studies (n=179) | Human studies (n=21) | |
|---|---|---|
| Hypothesis, aim, and objective definition | ||
| Clear | 167 (93%) | 18 (86%) |
| Unclear | 10 (6%) | 3 (14%) |
| Unknown | 2 (1%) | 0 |
| Description of animal and human participants | ||
| Good | 57 (32%) | 9 (43%) |
| Poor | 118 (66%) | 10 (48%) |
| Unknown | 4 (2%) | 2 (10%) |
| Intervention descriptions | ||
| Good | 112 (63%) | 14 (67%) |
| Poor | 64 (35%) | 6 (29%) |
| Unknown | 3 (2%) | 1 (5%) |
| Description of main outcomes | ||
| Good quality | 157 (88%) | 16 (76%) |
| Poor quality | 18 (10%) | 3 (14%) |
| Unknown | 4 (2%) | 2 (10%) |
| Provision of estimates of variability | ||
| Yes | 44 (25%) | 7 (33%) |
| No | 133 (74%) | 13 (62%) |
| Unknown | 2 (1%) | 1 (5%) |
| Representative study population | ||
| Yes | 37 (21%) | 3 (14%) |
| No | 82 (47%) | 7 (33%) |
| Unknown | 58 (32%) | 11 (52%) |
| Recruitment of intervention and control group populations | ||
| Same source | 78 (44%) | 7 (33%) |
| Alternate sources | 46 (26%) | 7 (33%) |
| Unknown | 53 (30%) | 7 (33%) |
| Recruitment window of intervention and control group populations | ||
| Same time period | 137 (77%) | 7 (44%) |
| Different time periods | 10 (5%) | 4 (25%) |
| Unknown | 32 (18%) | 5 (31%) |
| Adjustment for confounders | ||
| Adequate adjustment | 27 (14%) | 4 (19%) |
| Inadequate adjustment | 69 (70%) | 11 (52%) |
| Unknown | 28 (16%) | 6 (29%) |
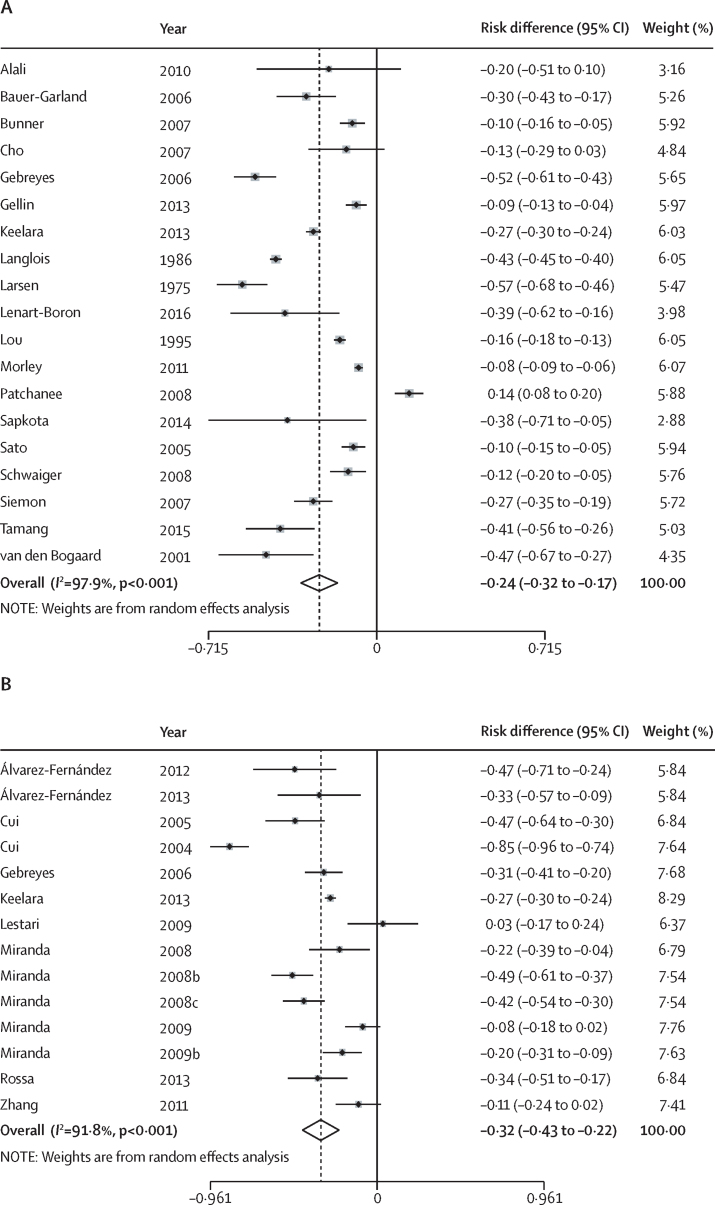
Of the 179 studies reporting antibiotic resistance in animals, 81 were included in meta-analyses within the five analytic groups (table 3). Overall, all pooled estimates were less than zero except for amphenicol and tetracycline resistance in Campylobacter spp in faecal (relative difference 0·00, 95% CI −0·02 to 0·02) and meat samples (0·01, 95% CI −0·19 to 0·21). For nearly all antibiotic classes, sample types, and bacterial groups, the pooled risk of antibiotic resistance in the intervention group was lower than in the control group. Of the meta-analyses that resulted in a negative pooled estimate, the pooled absolute risk differences ranged between −0·01 for cephalosporin resistance in Enterobacteriaceae in faecal samples (95% CI −0·04 to 0·01) and −0·39 for macrolide resistance in Enterococcus spp in faecal samples (95% CI −0·56 to −0·23). This corresponds to a reduction in the proportion of isolates that were antibiotic resistant in the intervention group compared with the control group by 1–39%. Most absolute risk differences were in the −0·15 to −0·10 range, indicating a 10–15% reduction in the proportion of antibiotic-resistant isolates in the intervention groups compared with the control groups. The pooled effect estimates appeared greatest for Enterococcus spp (range relative difference [RD] −0·39 to −0·13), and least for Campylobacter (range RD −0·15 to 0·01). Meta-analysis of the absolute risk difference of multidrug resistance was done for animal studies testing Enterobacteriaceae in faecal (figure 2A) and meat samples (figure 2B). This showed an absolute risk reduction of 24–32% in the proportion of isolates that were multidrug resistant in intervention groups compared with control groups.
Table 3.
Pooled absolute risk differences of antibiotic resistance from meta-analysis of animal studies, by antiobiotic drug class
| Number of studies | Pooled absolute risk difference (95% CI) | |
|---|---|---|
| Enterobacteriaceae in faecal samples | ||
| Aminoglycosides | 21 | −0·12 (−0·17 to −0·07) |
| Amphenicols | 16 | −0·04 (−0·06 to −0·03) |
| Cephalosporins | 17 | −0·01 (−0·04 to 0·01) |
| Penicillins | 20 | −0·12 (−0·18 to −0·07) |
| Quinolones | 17 | −0·01 (−0·02 to 0·00) |
| Sulfonamides | 20 | −0·06 (−0·09 to −0·02) |
| Tetracyclines | 21 | −0·16 (−0·27 to −0·05) |
| Enterobacteriaceae in meat samples | ||
| Aminoglycosides | 12 | −0·07 (−0·12 to −0·02) |
| Amphenicols | 11 | −0·08 (−0·14 to −0·03) |
| Cephalosporins | 11 | −0·07 (−0·14 to 0·01) |
| Penicillins | 11 | −0·16 (−0·25 to −0·08) |
| Quinolones | 12 | −0·09 (−0·17 to −0·02) |
| Sulfonamides | 13 | −0·23 (−0·32 to −0·13) |
| Tetracyclines | 13 | −0·20 (−0·36 to −0·03) |
| Enterococcus spp. in faecal samples | ||
| Aminoglycosides | 7 | −0·13 (−0·23 to −0·02) |
| Glycopeptides | 12 | −0·22 (−0·32 to −0·12) |
| Macrolides | 10 | −0·39 (−0·56 to −0·23) |
| Penicillins | 7 | −0·10 (−0·18 to −0·02) |
| Streptogramins | 8 | −0·31 (−0·46 to −0·17) |
| Tetracyclines | 7 | −0·30 (−0·48 to −0·13) |
| Campylobacter spp. in faecal samples | ||
| Aminoglycosides | 8 | −0·02 (−0·03 to 0·00) |
| Amphenicols | 7 | 0·00 (−0·02 to 0·02) |
| Macrolides | 11 | −0·15 (−0·26 to −0·04) |
| Penicillins | 8 | −0·03 (−0·08 to 0·02) |
| Quinolones | 11 | −0·06 (−0·16 to 0·05) |
| Tetracyclines | 10 | −0·12 (−0·20 to −0·03) |
| Campylobacter spp. in meat samples | ||
| Macrolides | 7 | −0·04 (−0·17 to 0·09) |
| Quinolones | 9 | −0·08 (−0·17 to 0·01) |
| Tetracyclines | 7 | 0·01 (−0·19 to 0·21) |
| Staphylococcus spp. in milk samples | ||
| Aminoglycosides | 6 | −0·04 (−0·13 to 0·05) |
| Lincosamides | 7 | −0·09 (−0·16 to −0·02) |
| Macrolides | 8 | −0·06 (−0·10 to −0·01) |
| Penicillins | 10 | −0·07 (−0·11 to −0·02) |
| Sulfonamides | 6 | −0·04 (−0·07 to 0·00) |
| Tetracyclines | 9 | −0·06 (−0·10 to −0·01) |
Figure 2.
Forest plot of absolute risk differences of multi-drug resistance
Differences are shown for Enterobacteriaceae isolates in (A) faecal and (B) meat samples. The references used have been provided in the appendix.
63 studies that reported on phenotypic resistance could not be included in either the main meta-analyses (within the five analytic groupings) or the bundled meta-analyses (for stratified analyses), because of insufficient data to calculate a risk difference in antibiotic resistance, or because sample sizes were not reported. Results of these studies are qualitatively described in the appendix (pp 47–50), and corroborated the results of the meta-analysis. Most studies showed lower antibiotic resistance in intervention compared with control groups, although a smaller number of studies showed no difference in antibiotic resistance between the two groups.
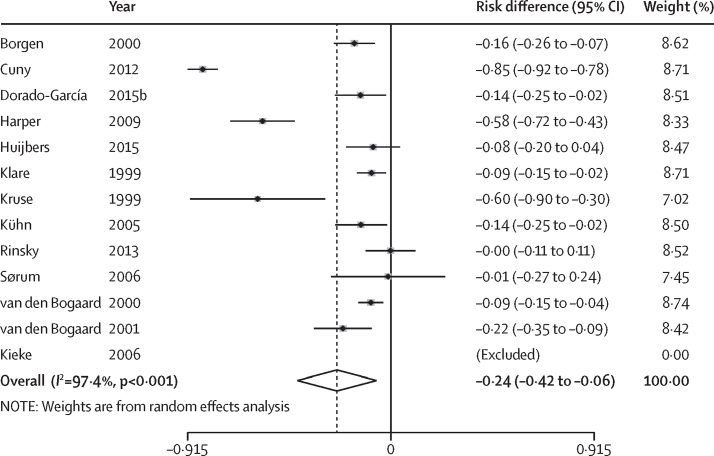
Of the 21 studies that reported antibiotic resistance in humans, 13 were included in the meta-analysis (figure 3). The other eight studies were not pooled because the unit of analysis was at the isolate (rather than the human) level, there was insufficient data to calculate risk differences in antibiotic resistance, sample sizes of intervention and control groups were not reported, or a combination of the above. All 13 studies showed either no difference or a lower risk of antibiotic resistance in the intervention group compared with the control group. Results from nine of the 13 studies were significant, with absolute risk differences ranging from −0·85 to −0·09. The pooled estimate of the absolute risk difference of antibiotic resistance, across all classes of antibiotics, was −0·24 (95% CI −0·42 to −0·06). The pooled prevalence of antibiotic-resistant bacteria in humans was 24% lower in intervention groups in which there was reduced use of antibiotics in animals, compared with control groups.
Figure 3.
Forest plot of absolute risk differences of antibiotic resistance in humans
The references used have been provided in the appendix.
Due to the heterogeneity across studies, with an I2 generally between 51·3% and 99·4% for animal studies and an I2 of 97·4% for human studies, stratified analysis was done by study characteristics and quality criteria (table 4 for animal studies, table 5 for human studies). Overall, there were no significant differences in the stratified effect estimates based on the strength of intervention. Furthermore, effect estimates from studies where interventions restricted the use of all antibiotics for any indication did not differ from studies where interventions allowed the use of antibiotics for therapeutic purposes.
Table 4.
Stratified meta-analysis for animal studies, by stratification variable
| Number of studies (n=101) | Pooled absolute risk difference (95% CI) | Meta-regression p value | |
|---|---|---|---|
| Study population | |||
| Swine | 30 (30%) | −0·21 (−0·27 to −0·14) | 0·89 |
| Cattle | 24 (24%) | −0·04 (−0·06 to −0·03) | ·· |
| Poultry | 55 (54%) | −0·18 (−0·22 to −0·14) | ·· |
| Sample type | |||
| Retail | 25 (25%) | −0·15 (−0·19 to −0·10) | 0·99 |
| Non-retail | 79 (78%) | −0·14 (−0·16 to −0·12) | ·· |
| Strength of intervention | |||
| Stronger interventions* | 31 (31%) | −0·19 (−0·26 to −0·11) | 0·37 |
| Weaker interventions† | 70 (69%) | −0·13 (−0·15 to −0·11) | ·· |
| Level of antibiotic restriction | |||
| Complete restriction of all antibiotic use | 38 (38%) | −0·16 (−0·19 to −0·12) | 0·59 |
| Therapeutic antibiotic use allowed | 67 (66%) | −0·14 (−0·17 to −0·12) | ·· |
| Laboratory procedure to measure resistance | |||
| Disk diffusion | 32 (32%) | −0·15 (−0·19 to −0·10) | 0·16 |
| Broth or agar dilution | 53 (52%) | −0·11 (−0·13 to −0·09) | ·· |
| Use of selective media containing antibiotics | 11 (11%) | −0·16 (−0·31 to −0·01) | ·· |
| Other or undetermined | 6 (6%) | −0·30 (−0·50 to −0·11) | ·· |
| Recruitment of study populations | |||
| Same source population for intervention and comparator groups | 46 (46%) | −0·12 (−0·16 to −0·09) | 0·15 |
| Different source populations for intervention and comparator groups | 55 (54%) | −0·16 (−0·19 to −0·14) | ·· |
| Recruitment period | |||
| Intervention and comparator groups are recruited over the same time period | 75 (74%) | −0·12 (−0·14 to −0·10) | 0·21 |
| Intervention and comparator groups are not recruited over the same time period | 26 (26%) | −0·20 (−0·28 to −0·13) | ·· |
| Description of interventions | |||
| Well described interventions | 72 (71%) | −0·14 (−0·16 to −0·11) | 0·58 |
| Poorly described interventions | 29 (29%) | −0·17 (−0·21 to −0·12) | ·· |
| Adjustment for potential confounders | |||
| Adequate adjustment | 14 (14%) | −0·10 (−0·15 to −0·06) | 0·35 |
| Inadequate adjustment | 87 (86%) | −0·16 (−0·18 to −0·13) | ·· |
| Publications | |||
| Peer-reviewed publications | 90 (89%) | −0·15 (−0·17 to −0·13) | 0·33 |
| Non-peer reviewed publications‡ | 11 (11%) | −0·10 (−0·19 to −0·02) | ·· |
Studies might be included into more than one stratum if stratum-specific estimates were provided.
Externally imposed restrictions, and voluntary restrictions.
Self-reported organic, antibiotic-free, and related labels.
Includes meeting abstracts, reports, and dissertations.
Table 5.
Stratified meta-analysis for human studies, by stratification variable
| Number of studies*(n=13) | Pooled absolute risk difference (95% CI) | Meta-regression p value | |
|---|---|---|---|
| Population | |||
| Farm workers and household members | 9 (69%) | −0·29 (−0·54 to −0·04) | 0·32 |
| Non-farm workers | 3 (23%) | −0·09 (−0·13 to −0·05) | ·· |
| Strength of intervention | |||
| Stronger interventions† | 8 (62%) | −0·14 (−0·20 to −0·08) | 0·22 |
| Weaker interventions‡ | 4 (31%) | −0·38 (−0·84 to 0·08) | ·· |
| Level of antibiotic restriction | |||
| Complete restriction of antibiotic use | 2 (15%) | −0·43 (−1·00 to 0·40) | 0·29 |
| Therapeutic antibiotic use allowed | 10 (77%) | −0·19 (−0·27 to −0·10) | ·· |
| Laboratory procedure to measure resistance | |||
| Disk diffusion | 2 (15%) | −0·04 (−0·12 to 0·05) | 0·093 |
| Broth or agar dilution | 3 (23%) | −0·11 (−0·16 to −0·05) | ·· |
| Use of selective media containing antibiotics | 4 (31%) | −0·30 (−0·74 to 0·14) | ·· |
| Other or undetermined | 3 (23%) | −0·43 (−0·77 to −0·08) | ·· |
| Recruitment of populations | |||
| Same source population for intervention and comparator groups | 3 (23%) | −0·11 (−0·17, to −0·05) | 0·30 |
| Different source populations for intervention and comparator groups | 9 (69%) | −0·29 (−0·53 to −0·06) | ·· |
| Recruitment period | |||
| Intervention and comparator groups recruited over the same time period | 6 (46%) | −0·09 (−0·13 to −0·06) | 0·037 |
| Intervention and comparator groups not recruited over the same time period | 6 (46%) | −0·41 (−0·72 to −0·09) | ·· |
| Description of interventions | |||
| Well described interventions | 9 (69%) | −0·11 (−0·16 to −0·06) | 0·008 |
| Poorly described interventions | 3 (23%) | −0·55 (−0·94 to −0·16) | ·· |
| Adjustment for potential confounders | |||
| Adequate adjustment | 1 (8%) | −0·14 (−0·25 to −0·02) | 0·70 |
| Inadequate adjustment | 11 (85%) | −0·25 (−0·44 to −0·06) | ·· |
| Publications | |||
| Peer-reviewed publications | 11 (85%) | −0·21 (−0·40 to −0·03) | 0·98 |
| Non-peer reviewed publications§ | 1 (8%) | −0·58 (−0·71 to −0·43) | ·· |
Of the 13 studies that could be meta-analysed, one was excluded from stratified meta-analysis and meta-regression because of a standard error of the risk difference of 0.
Externally imposed restrictions, and voluntary restrictions.
Self-reported organic, antibiotic-free, and related labels.
Including meeting abstracts, reports, and dissertations.
Stratified meta-analysis of studies that met study quality criteria and studies that did not meet quality criteria both showed significant risk reductions in antibiotic resistance in intervention compared with control groups, although effects appeared to be smaller for higher quality compared with lower quality studies. This was true of both animal and human studies, though p values on meta-regression were significant for only two quality criteria in human studies (whether intervention and control groups were recruited over the same time period; and whether interventions were well described).
We further stratified the meta-analysis by study population. In animal studies, the pooled effect estimates appeared stronger when the intervention was targeted to swine and poultry than when the intervention was targeted to cattle, though this finding was not significant on meta-regression. The differences in pooled effect estimates might be because swine and poultry farming practices are generally intensive (with larger numbers of animals and potentially greater antibiotic use), and because antibiotic administration tends to be at the herd or flock level, rather than the individual animal level. Interventions in swine and poultry farming are therefore likely to result in greater overall reductions in antibiotic use. A similar dose-response effect of the interventions within human studies is suggested, with pooled effect estimates being stronger in farm workers (−0·29, 95% CI −0·54 to −0·04) compared with human beings without direct contact with livestock animals (−0·09 95% CI −0·13 to −0·05). Despite large differences in these pooled risk differences, the meta-regression did not result in a significant p value, which is likely to be because meta-regression analyses are often underpowered from few completed studies.
Separate funnel plots were produced for animal and human studies (appendix p 51). Visual inspection of both plots revealed asymmetry. Begg's test for funnel plot asymmetry was not significant for animal studies (p=0·16) but was for human studies (p=0·047). Because there was evidence of visual asymmetry, a trim and fill procedure was done for both sets of studies. There was no change in the risk reduction of antibiotic resistance in animals with interventions that reduced antibiotic use with and without imputation (both −0·13, 95% CI −0·15 to −0·11). Furthermore, there was no change in the risk reduction of antibiotic resistance in humans with imputation of potential missing studies (−0·27, 95% CI −0·43 to −0·10) and without imputation (−0·24, 95% CI −0·42 to −0·06). This suggests that publication bias, if present, was likely to have had a minimal effect on our findings.
Discussion
Our systematic review assessed interventions that reduce antibiotic use in farm animals and uncovered extensive literature and important findings. Of 179 identified studies, we found an association between interventions that restrict antibiotic use and reduction in the prevalence of antibiotic-resistant bacteria in animals and in different human subgroups. Overall, reducing antibiotic use decreased prevalence of antibiotic-resistant bacteria in animals by about 15% and multidrug-resistant bacteria by 24–32%. The evidence of effect on human beings was more limited and less robust, though meta-analysis of 13 studies showed similar results, with a 24% absolute reduction in the prevalence of antibiotic-resistant bacteria in humans with interventions that reduce antibiotic use in animals.
Three recent systematic reviews have explored antibiotic resistance in bacteria isolated from organically—versus conventionally—farmed animals.33, 34, 35 Their conclusions were similar to the ones we have reached, although all focused only on the single type of intervention, and assessed only a single bacterial species, a single sample type, or both. Our systematic review is, to our knowledge, the most comprehensive and the first to include studies that examine all types of interventions that aim to reduce antibiotic use in animals, with no limitation on the type of sample obtained, the animal species included, or the bacterial species tested. We also believe that this is the first systematic review of studies examining the association between interventions to reduce antibiotic use in food-producing animals and changes to antibiotic-resistant bacteria in human beings. This summary of evidence is an essential ingredient to creating evidence-informed global strategies on antibiotic use in food-producing animals.
The potential effect that interventions to reduce antibiotic use in food-producing animals have on human antibiotic resistance is complex. The results of our meta-analysis on the human studies included in our systematic review consistently suggest that antibiotic-resistant bacteria can be exchanged between livestock and farm workers, with the evidence being weaker and more indirect for transmission to other human populations. Transmission from food-producing animals to humans can occur through contaminated animal retail products,36 although the risk of this might be low if animal products are adequately prepared and cooked. Resistance can also be transmitted through the environment through animal faecal matter, waste water, and contaminated produce.37 Our systematic review has shown that reducing the level of antibiotic use in livestock populations is likely to be a beneficial strategy for both animals and human beings. Although we do not fully understand the mechanisms of cross-species transmission of resistant bacteria and their genetic elements, it seems clear that the health of humans, animals, and the ecosystem are intricately linked, and that an interdisciplinary and multi-sectoral approach will be required to address the problem of antimicrobial resistance.38
There are some caveats and limitations to our review. Despite considerable heterogeneity across studies of both food-producing animals and human beings, we chose to pool results through meta-analyses for three reasons. First, we anticipated a priori that there would be clinical heterogeneity across studies, given the wide variety of settings studied and interventions described and tested. Second, it would have been surprising to not find statistical heterogeneity, given the large number of samples in the constituent studies that would have resulted in high power when statistical testing for heterogeneity. Finally, even with this expected clinical and statistical heterogeneity, the overall findings were surprisingly homogeneous in their ultimate findings, with a consistent effect that spanned different populations, interventions, and bacterial groups. Having made the decision to pool study results, it was essential to have mitigating strategies to handle the heterogeneity and explore its sources. To this end, we used random effects models in all our analyses and used extensive stratified analyses and meta-regression to explore contributors to heterogeneity.
As with any systematic review, our study is limited by the varied quality and nature of the underlying studies. Although many studies had distinct strengths in clearly reporting their objectives, hypotheses, and outcomes, areas of deficiency in some studies included poor descriptions of study groups or interventions, the absence of a conrol group in longitudinal studies, and inadequate adjustment for potential confounders. Stratified analysis showed that studies not meeting quality criteria might overestimate the effects of interventions, which is likely to be because of the presence of selection bias and confounding. Most studies involved cross-sectional assessments of antibiotic resistance in intervention compared with control groups; in such studies, causal inferences between reduction in antibiotic use and reduction in the prevalence of antibiotic-resistant bacteria are less robust than for longitudinal studies. The issue of causality was especially challenging for human studies in which linkages between the intervention in food-producing animals and changes in bacterial resistance in humans were indirect and implied; the mode of transmission and level of contact with animal or animal products with antibiotic resistance were often not identified. Additionally, most studies were from North America and Europe, and only one study originated from India, China, or Brazil—three of the top five global users of antibiotics in livestock and in humans.39 This distribution is likely to reflect geographical areas in which there has been greatest focus on reduction of antibiotic use in animals. Furthermore, with only one study included in the review pertaining to aquaculture, the effect of interventions to restrict antibiotic use in this setting is unknown. Lastly, because primary research studies focused only on selected bacterial groups, the overall effects of reducing antibiotic use in food-producing animals on the resistomes of both animals and human beings remain unknown.
The caveats discussed are balanced by the considerable weight of findings compiled here. We have reviewed and meta-analysed a substantial body of evidence that suggests there is a reduction in the prevalence of antibiotic-resistant bacteria in food-producing animals when antibiotic use is reduced in this population. We have also summarised and analysed a smaller body of evidence suggesting that such interventions might also be associated with a reduction in prevalence of antibiotic-resistant bacteria in humans, particularly those with direct contact with food-producing animals. The evidence on the whole is not only substantial in its volume, but also in its consistency. The findings were consistent regardless of bacteria studied, food-producing animals in question, interventions implemented, samples studied, and regardless of the quality of the studies. Therefore, despite the limitations posed by the quality of studies and the methodological issues and assumptions that are made in them, it would be imprudent to entirely discount this body of evidence given its coherence and consistency.
Our systematic review and meta-analysis provides evidence for only a small piece of a large global problem, with many policy-relevant questions remaining unanswered. Specifically, our systematic review was not designed to identify which type or level of restriction on antibiotic use in food-producing animals confers the greatest benefit. Furthermore, we have not explored any potential harms arising from the restriction of antibiotic use in food-producing animals, though other studies have suggested only limited or temporary effects to productivity and animal health when antibiotic growth promoter bans have been implemented.40, 41, 42, 43, 44 Ultimately, policy decisions regarding restriction of antibiotic use will depend upon a careful weighing of the evidence of potential unintended consequences against the benefits we have shown in this review; this should be the focus of future research. There is also potential for the use of non-antibiotic alternatives for growth promotion and disease prevention. There is a need for additional research in this area to better understand their mechanisms of action and to more robustly show their effectiveness in field trials.16
This systematic review, commissioned by WHO, focuses on the fundamental question of whether interventions that restrict antibiotic use are effective in reducing antibiotic resistance. It appears that such interventions are indeed associated with a reduction in resistance in food-producing animals and in human beings that have direct contact with such animals, with a possible, but less clear, association in the more general population. These findings will be an important consideration as global policy options are being considered.
This online publication has been corrected. The corrected version first appeared at thelancet.com/planetary-health on November 15, 2017
Acknowledgments
Acknowledgments
This study was commissioned and paid for by WHO. Copyright in the original work on which this article is based belongs to the WHO. The authors have been given permission to publish this article. The authors alone are responsible for the views expressed in this publication and do not necessarily represent the views, decisions, or policies of WHO. KLT was supported by fellowship awards from the Canadian Institutes of Health Research and Alberta Innovates—Health Solutions. We would like to thank the Dr Carl Ribble (University of Calgary, AB, Canada), Dr John Prescott (University of Guelph, ON, Canada), Dr Sylvia Checkley (University of Calgary, AB, Canada), Dr Tim McAllister (Agriculture and Agri-Food, Canada), and Dr Vic Gannon (Public Health Agency of Canada) for their feedback and guidance regarding antimicrobial resistance. We thank Jay Son (Sunnybrook Research Institute), Jan Tomka (NAFC Research Institute for Animal Production Nitra, Slovakia), Julia Kupis (University of Calgary, AB, Canada), and Zoe Thomson (University of Calgary, AB, Canada), Dr Caroline Ritter (University of Calgary, AB, Canada), Dr David Hall (University of Calgary, AB, Canada), Dr Karin Lienhard (University of Calgary, AB, Canada), and Dr Lis Alban (Danish Agriculture and Food Council, Denmark) for their assistance with language translation of studies not available in English. We extend our gratitude to Jill de Grood and the W21C Research and Innovation Centre for project coordination and oversight.
Contributors
All authors were involved in the design of the study, development of the study protocol and the development of the data extraction form. HG created the search strategy and did the literature search in electronic databases. DBN did the grey literature search. KLT and NPC screened all studies for inclusion into the systematic review and did all study quality assessments. SCC, PER, and HWB provided input on studies where consensus could not be reached. KLT, NPC, DBN, AJP, and NS extracted data. All authors contributed to data interpretation and data analysis. KLT drafted the manuscript and all authors revised it critically for content. All authors had full access to all data and can take responsibility for the integrity of the data and accuracy of the data analysis. All authors have read and approved the manuscript.
Declaration of interests
JDK was the principal investigator on an unrestricted grant in aid to conduct an epidemiological study of invasive pneumococcal disease in humans, including impact of pneumococcal vaccines (Pfizer Canada). He is also the local co-investigator on contract of a clinical trial of a maternal pertussis vaccine (GSK Canada). All other authors declare no conflicts of interest other than WHO funding of this study.
Supplementary Material
References
- 1.de Kraker ME, Davey PG, Grundmann H. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011;8:e1001104. doi: 10.1371/journal.pmed.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Kraker ME, Wolkewitz M, Davey PG, et al. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother. 2011;66:398–407. doi: 10.1093/jac/dkq412. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006;42(suppl 2):S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove SE, Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin Infect Dis. 2003;36:1433–1437. doi: 10.1086/375081. [DOI] [PubMed] [Google Scholar]
- 5.The review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. UK. 2016. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed March 1, 2017).
- 6.G20 G20 Leaders' Communique Hangzhou Summit. 2016. http://g20.org/English/Dynamic/201609/t20160906_3396.html (accessed Nov 3, 2016).
- 7.United Nations General Assembly of the United Nations: President of the 71st Session. 2016. http://www.un.org/pga/71/event-latest/high-level-meeting-on-antimicrobial-resistance/ (accessed Nov 3, 2016).
- 8.WHO Antimicrobial resistance: global antimicrobial resistance surveillance system: manual for early implementation. 2015. http://www.who.int/antimicrobial-resistance/publications/surveillance-system-manual/en/ (accessed Oct 1, 2016).
- 9.WHO . WHO Global principles for the containment of antimicrobial resistance in animals intended for food: report of a WHO consultation with the participation of the Food and Agriculture Organization of the United Nations and the Office International des Epizooties. World Health Organization; Geneva: 2000. [Google Scholar]
- 10.WHO . Joint FAO/OIE/WHO expert workshop on non-human antimicrobial usage and antimicrobial resistance: scientific asssessment. World Health Organization; Geneva: 2003. [Google Scholar]
- 11.WHO . Critically important antibacterial agents for human medicine for risk management strategies of non-human use: report of a WHO working group consultation. World Health Organization; Canberra: 2005. [Google Scholar]
- 12.WHO . Antimicrobial use in aquaculture and antimicrobial resistance: report of a joint FAO/OIE/WHO expert consultation on antimicrobial use in aquaculture and antimicrobial resistance. World Health Organization; Seoul: 2006. [Google Scholar]
- 13.Tusevljak N, Dutil L, Rajic A, et al. Antimicrobial use and resistance in aquaculture: findings of a globally administered survey of aquaculture-allied professionals. Zoonoses Public Health. 2013;60:426–436. doi: 10.1111/zph.12017. [DOI] [PubMed] [Google Scholar]
- 14.UN FAO . Joint FAO/WHO/OIE expert meeting on critically important antimicrobials. UN Food and Agriculture Organization; Rome: 2008. [Google Scholar]
- 15.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Reports. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Medicines Agency and European Food Safety Authority EMA and EFSA joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety. EFSA J. 2017;15:1–245. doi: 10.2903/j.efsa.2017.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. W64. [DOI] [PubMed] [Google Scholar]
- 18.WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) Critically important antimicrobials for human medicine. World Health Organization; Geneva: 2012. [Google Scholar]
- 19.World Organization for Animal Health (OIE) OIE list of antimicrobial agents of veterinary importance. World Health Organization; Geneva: 2015. [Google Scholar]
- 20.UN FAO . List of animal species used in aquaculture. UN Food and Agriculture Organization; Rome: 1996. [Google Scholar]
- 21.UN Food and Agriculture Organization Sources of meat. 2014. http://www.fao.org/ag/againfo/themes/en/meat/backgr_sources.html (accessed June 13 2016).
- 22.Downs SH, Black N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J Epidemiol Commun Health. 1998;52:377–384. doi: 10.1136/jech.52.6.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Introduction to meta-analysis. John Wiley & Sons Ltd; Chichester: 2009. [Google Scholar]
- 24.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control. 2006;34(suppl 1):S3–S10. doi: 10.1016/j.ajic.2006.05.219. [DOI] [PubMed] [Google Scholar]
- 26.Brenner DJ, Staley JT, Krieg NR. In: Bergey's manual of systematic bacteriology (2nd edn): Volume two—the proteobacteria. Garrity G, editor. Springer; New York, NY: 2005. Classification of procaryotic organisms and the concept of bacterial speciation; pp. 27–32. [Google Scholar]
- 27.Vos P, Garrity G, Jones D, et al., editors. Bergey's manual of systematic bacteriology (2nd edn): Volume three—the firmicutes. Springer-Verlag; New York, NY: 2009. [Google Scholar]
- 28.Imhoff JF. In: Bergey's manual of systematic bacteriology (2nd edn): Volume two—the proteobacteria part B. Garrity G, editor. Springer-Verlag US; New York, NY: 2005. Enterobacteriales; pp. 587–850. [Google Scholar]
- 29.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 30.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 32.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith-Spangler C, Brandeau ML, Hunter GE, et al. Are organic foods safer or healthier than conventional alternatives?: a systematic review. Ann Intern Med. 2012;157:348–366. doi: 10.7326/0003-4819-157-5-201209040-00007. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelm B, Rajic A, Waddell L, et al. Prevalence of zoonotic or potentially zoonotic bacteria, antimicrobial resistance, and somatic cell counts in organic dairy production: current knowledge and research gaps. Foodborne Pathog Dis. 2009;6:525–539. doi: 10.1089/fpd.2008.0181. [DOI] [PubMed] [Google Scholar]
- 35.Young I, Rajic A, Wilhelm BJ, Waddell L, Parker S, McEwen SA. Comparison of the prevalence of bacterial enteropathogens, potentially zoonotic bacteria and bacterial resistance to antimicrobials in organic and conventional poultry, swine and beef production: a systematic review and meta-analysis. Epidemiol Infect. 2009;137:1217–1232. doi: 10.1017/S0950268809002635. [DOI] [PubMed] [Google Scholar]
- 36.Lazarus B, Paterson DL, Mollinger JL, Rogers BA. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. 2015;60:439–452. doi: 10.1093/cid/ciu785. [DOI] [PubMed] [Google Scholar]
- 37.Aarestrup FM. The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140085. doi: 10.1098/rstb.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Prevention One Health. 2013. http://www.cdc.gov/onehealth/ (accessed Sept 10, 2016).
- 39.Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aarestrup FM, Jensen VF, Emborg HD, Jacobsen E, Wegener HC. Changes in the use of antimicrobials and the effects on productivity of swine farms in Denmark. Am J Vet Res. 2010;71:726–733. doi: 10.2460/ajvr.71.7.726. [DOI] [PubMed] [Google Scholar]
- 41.Bengtsson B, Wierup M. Antimicrobial resistance in Scandinavia after ban of antimicrobial growth promoters. Anim Biotechnol. 2006;17:147–156. doi: 10.1080/10495390600956920. [DOI] [PubMed] [Google Scholar]
- 42.Collignon P, Wegener HC, Braam P, Butler CD. The routine use of antibiotics to promote animal growth does little to benefit protein undernutrition in the developing world. Clin Infect Dis. 2005;41:1007–1013. doi: 10.1086/433191. [DOI] [PubMed] [Google Scholar]
- 43.Emborg H, Ersboll AK, Heuer OE, Wegener HC. The effect of discontinuing the use of antimicrobial growth promoters on the productivity in the Danish broiler production. Prev Vet Med. 2001;50:53–70. doi: 10.1016/s0167-5877(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 44.Wierup M. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb Drug Resist. 2001;7:183–190. doi: 10.1089/10766290152045066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.