Summary
Flowering is an indication of the transition from vegetative growth to reproductive growth and has considerable effects on the life cycle of soya bean (Glycine max). In this study, we employed the CRISPR/Cas9 system to specifically induce targeted mutagenesis of GmFT2a, an integrator in the photoperiod flowering pathway in soya bean. The soya bean cultivar Jack was transformed with three sgRNA/Cas9 vectors targeting different sites of endogenous GmFT2a via Agrobacterium tumefaciens‐mediated transformation. Site‐directed mutations were observed at all targeted sites by DNA sequencing analysis. T1‐generation soya bean plants homozygous for null alleles of GmFT2a frameshift mutated by a 1‐bp insertion or short deletion exhibited late flowering under natural conditions (summer) in Beijing, China (N39°58′, E116°20′). We also found that the targeted mutagenesis was stably heritable in the following T2 generation, and the homozygous GmFT2a mutants exhibited late flowering under both long‐day and short‐day conditions. We identified some ‘transgene‐clean’ soya bean plants that were homozygous for null alleles of endogenous GmFT2a and without any transgenic element from the T1 and T2 generations. These ‘transgene‐clean’ mutants of GmFT2a may provide materials for more in‐depth research of GmFT2a functions and the molecular mechanism of photoperiod responses in soya bean. They will also contribute to soya bean breeding and regional introduction.
Keywords: soya bean, GmFT2a, Agrobacterium tumefaciens‐mediated transformation, CRISPR/Cas9, genome editing, flowering time
Introduction
Soya bean (Glycine max (L.) Merr.) is an important legume crop with great economic value that provides abundant protein and oil for food production and animal feed. It is also a short‐day dicotyledon and is sensitive to seasonal changes in day length. This photoperiod sensitivity limits its geographical range of cultivation, and thus investigating the photoperiod response in flowering induction has great significance in soya bean regional introduction and domestication (Wang et al., 2016; Xu et al., 2013). In recent years, molecular biological studies in Arabidopsis thaliana have shown that FLOWER LOCUS T (FT) encodes florigen and plays an important role in flowering pathways as an integrator (Corbesier and Coupland, 2006; Turck et al., 2008). FT protein is a florigen that moves through the phloem to the shoot apex and functions as a long‐distance signal that induces floral initiation of Arabidopsis (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Notaguchi et al., 2008). Loss of FT function in A. thaliana results in a late‐flowering phenotype, whereas overexpression of FT causes precocious flowering independent of the transcription factor CONSTANS (CO) or photoperiod (Kobayashi et al., 1999; Koornneef et al., 1991). In soya bean, some homologous genes of FT have similar functions. Ten FT homologs in soya bean have been identified, and two, GmFT2a (Glyma16g26660) and GmFT5a (Glyma16g04830), have been confirmed to have functions similar to those of FT based on ectopic expression analysis in Arabidopsis. These homologs also coordinately control flowering in soya bean (Kong et al., 2010). Analysis of GmFT2a transcripts revealed that the expression of GmFT2a is regulated by photoperiod and is associated with flowering induction and maintenance (Sun et al., 2011). The GmFT2a promoter region harbours rich polymorphisms among different soya bean cultivars, but its coding sequence is highly conserved, and the polymorphisms in GmFT2a are not responsible for maturity diversity in soya bean (Jiang et al., 2013a). Subsequent studies have shown that GmFT2a and GmFT5a promote early flowering in soya bean upon overexpression of these two genes in the soya bean cultivar Williams 82 under long‐day (LD) conditions (Nan et al., 2014). Several flowering‐related genes in soya bean, such as GmAP1, GmSOC1 and GmLFY, were significantly up‐regulated by GmFT2a and GmFT5a on the basis of a redundant and differential pattern. Yeast two‐hybrid and bimolecular fluorescence complementation (BiFC) demonstrated that both GmFT2a and GmFT5a interact with the bZIP transcription factor GmFDL19, which can also cause early flowering (Nan et al., 2014). Fine mapping, sequencing and expression analysis revealed that the soya bean maturity gene E9 is FT2a, and that its recessive allele causes late flowering as a result of attenuated transcript abundance induced by allele‐specific transcriptional repression due to the Ty1/copia‐like retrotransposon SORE‐1 inserted in the first intron. SORE‐1 was highly methylated and did not generate damage to FT2a RNA processing (Zhao et al., 2016a). Obviously, GmFT2a plays an important role in flowering induction and maintenance in soya bean. However, previous studies of GmFT2a have mainly involved overexpression or ectopic expression analysis, and studies of soya bean endogenous gene GmFT2a mutants with loss of gene function remain relatively limited.
The recently developed CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (CRISPR‐associated) system has provided a robust and effective tool for targeted genome editing and more choice for gene functional researches. The main characteristic of the CRISPR/Cas9 system is the Cas9 protein, which comprises of two nuclease domains: the RuvC‐like domain and HNH domain (Cong et al., 2013). The Cas9 protein can form a complex with a synthetic sgRNA, which guides it to recognize target sequences and generate double‐strand breaks (DSBs) at expected target sites (Jinek et al., 2012). The DSBs subsequently induce DNA self‐repair mechanisms in the cell, which mainly include nonhomologous end‐joining (NHEJ) and homology‐directed repair (HDR). The NHEJ pathway is error‐prone and usually introduces some base insertions or deletions (indels) at the DNA break sites (Gorbunova and Levy, 1999). When these indels generate a frameshift mutation or disrupt important functional domains, the functions of the target genes will be damaged (Shan et al., 2013). The HDR pathway is a precise DNA repair mechanism that can be utilized to introduce specific point mutations or insertions of desired sequences at the target sites (Shan et al., 2013; Svitashev et al., 2015). The sequence of a fragment could also be replaced with desired sequences through the HDR pathway in the presence of an exogenous donor template (Gratz et al., 2014; Zhao et al., 2016b). The earliest reports of CRISPR/Cas9‐mediated genome editing in plants date from 2013 (Jiang et al., 2013b; Li et al., 2013). This new system has since been successfully applied to generate and estimate genome editing in many major crops, such as rice (Shan et al., 2013), wheat (Upadhyay et al., 2013), sorghum (Jiang et al., 2013b) and maize (Liang et al., 2014). CRISPR/Cas9‐mediated genome editing in soya bean was first successfully achieved in 2015. The CRISPR/Cas9 system has been successfully utilized to generate and estimate targeted mutations in both endogenous and exogenous genes in soya bean hairy roots (Cai et al., 2015; Du et al., 2016; Jacobs et al., 2015; Michno et al., 2015; Sun et al., 2015; Tang et al., 2016) and whole plants from embryonic calluses transformed by particle bombardment (Li et al., 2015). Targeted gene integrations through HDR were also detected by border‐specific polymerase chain reaction analysis at the callus stage, which revealed that one HDR event was transmitted to the T1 generation (Li et al., 2015). Rj4, not the gene previously reported, was confirmed to be the gene controlling nodulation specificity in soya bean through both complementation tests and CRISPR/Cas9‐mediated gene knockout experiments (Tang et al., 2016). These works have shown that CRISPR/Cas9 is a simple, efficient and highly specific genome editing tool in soya bean, although valuable phenotypic alterations have not been reported.
In this study, we employed the CRISPR/Cas9 system to specifically induce targeted mutagenesis of the GmFT2a gene in soya bean. A variety of homozygous ft2a mutants were generated from a sufficient number of stable transgenic soya bean events developed via Agrobacterium‐mediated transformation. In the results of GmFT2a‐CRISPR/Cas9, T1‐generation soya bean plants homozygous for null alleles of GmFT2a frameshift mutated by a 1‐bp insertion or short deletions exhibited late flowering under natural conditions (summer) in Beijing, China (N39°58′, E116°20′). The targeted mutations were stably inherited and maintained consistent mutation types from the T1 to T2 generation, and the homozygous T2 ft2a mutants exhibited late flowering under both long‐day (LD, 16 h light/8 h dark) and short‐day (SD, 12 h light/12 h dark) conditions. These results may contribute to soya bean regional introduction. The use of the CRISPR/Cas9 system to generate a phenotype of important agronomic traits in soya bean stable transformation has not been reported previously. We also obtained some ‘transgene‐clean’ targeted genome‐modified soya bean plants that were homozygous for the null alleles of endogenous GmFT2a and without any transgenic element. These mutants of GmFT2a that we obtained will provide materials for more in‐depth research on GmFT2a functions and the molecular mechanism of photoperiod responses in soya bean. These mutants will also contribute to soya bean breeding. More ‘transgene‐clean’ mutants of desired genes could be generated using the same method, and then be cross‐fertilized to stack favourable genes. The genome‐editing machinery will significantly increase breeding efficiency and speed up breeding process.
Results
Targeted mutagenesis of GmFT2a induced by CRISPR/Cas9
The CRISPR/Cas9‐mediated genome‐editing tool was utilized to knockout the soya bean endogenous gene GmFT2a. Three target sites (named GmFT2a‐SP1, GmFT2a‐SP2 and GmFT2a‐SP3) in the first exon of GmFT2a were chosen (Figure 1), and the corresponding sgRNA/Cas9 vectors were transformed into the soya bean cultivar Jack via Agrobacterium tumefaciens‐mediated transformation. In this study, the mutants of GmFT2a induced by CRISPR/Cas9 at the three target sites were separately named ft2a‐SP1, ft2a‐SP2 and ft2a‐SP3. DNA extracted from leaf tissue was used to examine CRISPR/Cas9‐induced mutations at the target sites using PCR and DNA sequencing analysis. The T0 transgenic lines harbouring the T‐DNA of the sgRNA/Cas9 vectors were identified, and we determined that 48% (12 of 25), 53% (19 of 36) and 37% (11 of 30) T‐DNA‐positive T0 lines at the three target sites had heterozygous‐targeted mutations of GmFT2a, respectively. Subsequently, all seeds collected from these self‐pollinated T0 lines were planted under natural conditions (summer) in Beijing, China (N39°58′, E116°20′). Site‐directed mutagenesis of GmFT2a was also observed at these three target sites in the T1 generation (Table 1). We identified a total of 64 T1 plants (18 ft2a‐SP1, 35 ft2a‐SP2 and 11 ft2a‐SP3) that were homozygous for null alleles of GmFT2a induced by CRISPR/Cas9 and detected two types of mutations at target site GmFT2a‐SP1 (8‐bp deletion and 1‐bp insertion). The 1‐bp insertion type was most frequently identified (Figure 2a,b). Simultaneously, two types of mutations were found at target site GmFT2a‐SP2 (4‐bp deletion and 1‐bp insertion), and the 1‐bp insertion type was most frequently identified (Figure 2a,c). Two types of mutations were also found at target site GmFT2a‐SP3 (14‐bp deletion and 1‐bp insertion), and the 14‐bp deletion type was most frequently identified (Figure 2a,d). All six types of frameshift mutations induced by CRISPR/Cas9 at three target sites of GmFT2a generated premature translation termination codons (PTCs) (Text S1).
Figure 1.
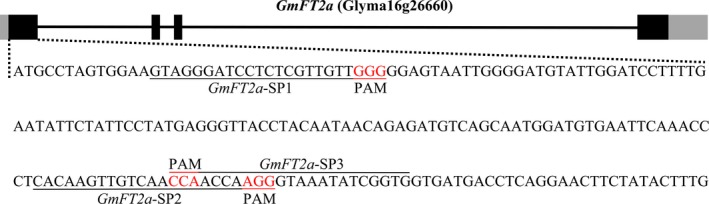
Gene structures of GmFT2a with target sites of CRISPR/Cas9 designed in the first exon. Black stripe, exon. Black line, intron. Grey stripe, UTR (untranslated regions). The underlined nucleotides indicate the target sites (named GmFT2a‐SP1, GmFT2a‐SP2 and GmFT2a‐SP3). Nucleotides in red represent PAM sequences. PAM, protospacer adjacent motif.
Table 1.
CRISPR/Cas9‐mediated targeted mutagenesis of GmFT2a in the T1 generation
| T1 lines with ft2a mutations | No. of plants sequenced | No. of homozygous ft2a mutants | No. of heterozygous ft2a mutants | No. of plants with no mutation |
|---|---|---|---|---|
| ft2a‐SP1‐T1#5 | 17 | 0 | 11 | 6 |
| ft2a‐SP1‐T1#10 | 14 | 2 | 10 | 2 |
| ft2a‐SP1‐T1#11 | 53 | 16 | 25 | 12 |
| ft2a‐SP1‐T1#16 | 24 | 0 | 18 | 6 |
| ft2a‐SP1‐T1#23 | 8 | 0 | 8 | 0 |
| ft2a‐SP2‐T1#8 | 12 | 8 | 2 | 2 |
| ft2a‐SP2‐T1#10 | 11 | 1 | 8 | 2 |
| ft2a‐SP2‐T1#13 | 13 | 0 | 10 | 3 |
| ft2a‐SP2‐T1#14 | 5 | 0 | 1 | 4 |
| ft2a‐SP2‐T1#15 | 5 | 3 | 0 | 2 |
| ft2a‐SP2‐T1#16 | 12 | 12 | 0 | 0 |
| ft2a‐SP2‐T1#22 | 36 | 10 | 20 | 6 |
| ft2a‐SP2‐T1#36 | 27 | 1 | 9 | 17 |
| ft2a‐SP3‐T1#2 | 8 | 0 | 2 | 6 |
| ft2a‐SP3‐T1#4 | 6 | 0 | 3 | 3 |
| ft2a‐SP3‐T1#10 | 3 | 0 | 2 | 1 |
| ft2a‐SP3‐T1#12 | 8 | 0 | 5 | 3 |
| ft2a‐SP3‐T1#30 | 29 | 11 | 18 | 0 |
Figure 2.
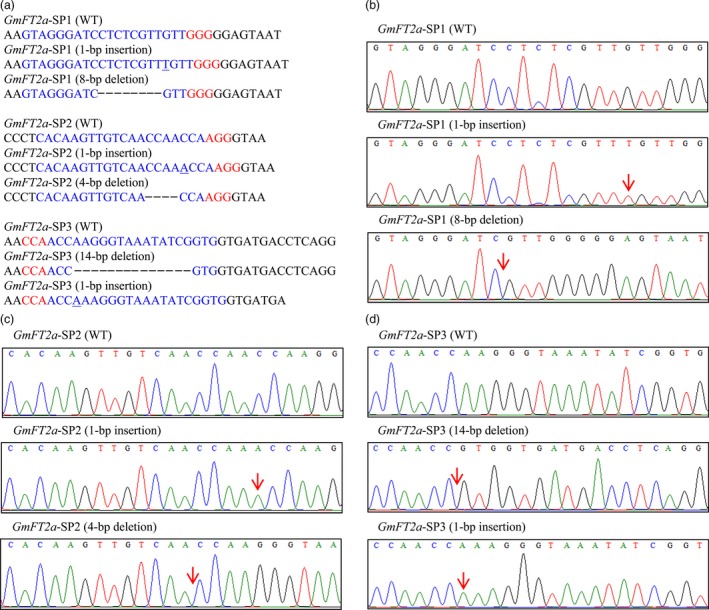
Homozygous targeted mutagenesis of GmFT2a induced by CRISPR/Cas9. (a) Sequences of wild type and representative mutation types induced at target sites GmFT2a‐SP1, GmFT2a‐SP2 and GmFT2a‐SP3 are presented, respectively. Underline, insertions. Dashes, deletions. (b), (c) and (d) are sequence peaks of wild type and representative mutation types at target sites GmFT2a‐SP1, GmFT2a‐SP2 and GmFT2a‐SP3, respectively. The red arrowheads indicate the location of mutations.
Potential off‐target analysis
To examine the specificity of CRISPR/Cas9 in soya bean and avoid affecting phenotype statistics by including off‐target sites, we analysed the potential off‐target effects of the editing of GmFT2a based on the predictions of the web tool CRISPR‐P (http://cbi.hzau.edu.cn/crispr/). The two most likely off‐target sites of these three target sites of GmFT2a were selected and examined by site‐specific genomic PCR and sequencing in the 64 T1 plants identified as homozygous ft2a mutants. All of the examined potential off‐target sites only possessed mismatches of 2–4 bp compared with the on‐target guide sequences. In this study, no mutations were observed in the examined potential off‐target sites (Table S1).
Stable inheritance of induced mutations and phenotypes of the mutants
In the T1 generation, we compared the flowering time between homozygous ft2a mutants induced by CRISPR/Cas9 at the three target sites with wild‐type (WT) plants under natural conditions (summer) in Beijing, China (N39°58′, E116°20′). We found that homozygous mutagenesis of GmFT2a at the three target sites delayed flowering time (Figure S1). To determine whether the ft2a mutants can transmit the induced mutations and phenotypes to their progenies, the T2 progeny of homozygous ft2a‐SP1‐T1#11 (1‐bp insertion), ft2a‐SP2‐T1#16 (1‐bp insertion) and ft2a‐SP3‐T1#30 (14‐bp deletion) lines were grown under both long‐day (LD, 16 h light/8 h dark) and short‐day (SD, 12 h light/12 h dark) photoperiodic conditions. The T2 plants of each mutation type were derived from three individuals of corresponding T1 ft2a mutant lines, respectively (34 ft2a‐SP1‐T2 plants were derived from ft2a‐SP1‐T1#11.14, ft2a‐SP1‐T1#11.15 and ft2a‐SP1‐T1#11.16 plants; 45 ft2a‐SP2‐T2 plants were derived from ft2a‐SP2‐T1#16.04, ft2a‐SP2‐T1#16.11 and ft2a‐SP2‐T1#16.12 plants; 30 ft2a‐SP3‐T2 plants were derived from ft2a‐SP3‐T1#30.02, ft2a‐SP3‐T1#30.08 and ft2a‐SP3‐T1#30.12 plants). Exact numbers of individuals are listed in Table S2. We subsequently examined the genotypes of the 109 T2‐generation plants and found that targeted mutagenesis of GmFT2a induced by CRISPR/Cas9 was stably inherited and maintained consistent mutation types from the T1 generation to T2 generation. Under SD conditions, T2 ft2a mutants did not have floral buds when WT plants were flowering, and when T2 ft2a mutants began to flower, the WT plants had obvious pods (Figure 3a). Similarly, under LD conditions, T2 ft2a mutants did not have floral buds when WT plants were flowering, and when T2 ft2a mutants came into flower, the flowers of WT plants fell off and began to produce pods (Figure 3b). We then compared the flowering time between T2 homozygous ft2a mutants at three target sites with WT plants under both SD and LD conditions via box‐plots (Figure 3c,d) and histograms (Figure 3e,f). These results exhibited that the T2 homozygous ft2a mutant plants showed a delayed flowering phenotype under both SD and LD conditions (Figure 3).
Figure 3.
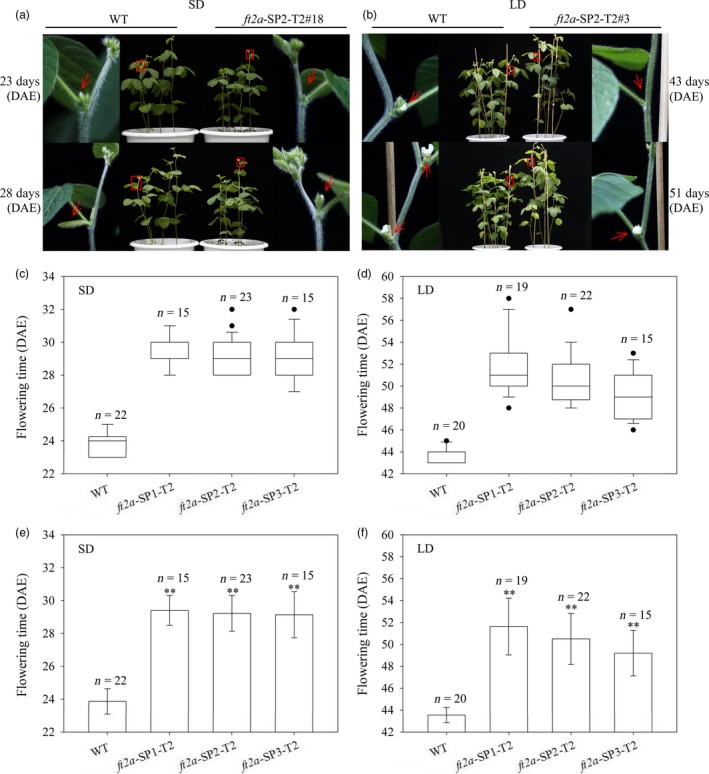
CRISPR/Cas9‐induced ft2a mutants exhibited late flowering in the T2 generation under both long‐day (LD) and short‐day (SD) conditions. (a) and (b) Phenotypes of wild‐type plants (WT, Jack) and homozygous T2 ft2a‐SP2 mutants under SD and LD condition, respectively. Top panel, ft2a‐SP2‐T2#18 and ft2a‐SP2‐T2#3 mutants did not have floral buds when WT plants were flowering. Bottom panel, flowers of WT fell off and produced the pods when ft2a‐SP2‐T2#18 and ft2a‐SP2‐T2#3 mutants were flowering. Red box, magnified view. (c) and (e) Flowering time of WT and homozygous T2 ft2a mutants at three target sites under SD conditions. (d) and (f) Flowering time of WT and homozygous T2 ft2a mutants at three target sites under LD conditions. n, exact numbers of individual plants identified. **, homozygous T2 ft2a mutants exhibit highly significant late flowering (P < 0.01). DAE, days after emergence. The flowering time is shown as the mean values ± standard deviation.
Expression patterns of GmFT2a in WT plants and T2 homozygous ft2a mutants
In this study, we compared the time course‐dependent expression patterns of GmFT2a between WT plants and T2 homozygous ft2a mutants grown under LD and SD photoperiodic conditions using RNAs extracted from trifoliate leaves sampled at 4 h after light every 5 days beginning at 10 DAE (Days after emergence). Under SD conditions, the expression levels of GmFT2a in WT plants increased to their maximum levels at 15 DAE (about 1 week before flowering) and thereafter decreased. The expression levels of GmFT2a in T2 ft2a mutants exhibited a similar pattern; however, they showed extremely low levels comparing with WT plants (Figure 4a). Under LD conditions, the expression of GmFT2a was relatively low. The transcript levels of GmFT2a in WT plants increased to their maximum levels at 30 DAE (About 2 weeks before flowering) and decreased thereafter. The transcript levels of GmFT2a in T2 ft2a mutants were significantly lower than that in WT plants, although they also increased slightly at 30 DAE (Figure 4b).
Figure 4.
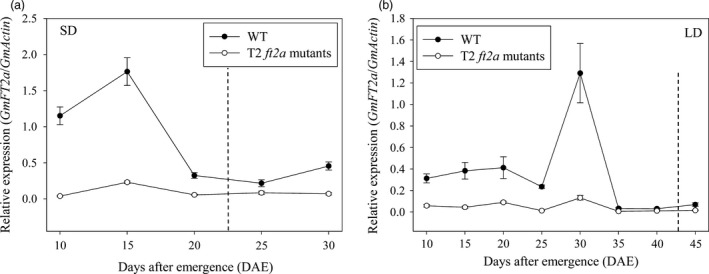
Expression patterns of GmFT2a in WT plants and T2 homozygous ft2a mutants under SD and LD conditions. (a) Expression analysis of GmFT2a under SD conditions. (b) Expression analysis of GmFT2a under LD conditions. Dotted lines indicate the flowering time of WT plants. DAE, days after emergence. RNAs were extracted from trifoliate leaves sampled at 4 h after light every 5 days beginning at 10 DAE. Relative transcript levels of GmFT2a were analysed by qRT‐PCR and normalized to GmActin. Average and SE (standard error) values for three replications are shown for each data point.
Generation of ‘transgene‐clean’ mutant soya bean lines
To obtain soya bean lines homozygous for endogenous GmFT2a mutations but without any transgenic element of the GmFT2a‐sgRNA/Cas9 vectors, ‘transgene‐clean’ plants were sought via a PCR strategy that used two sets of primer pairs spanning two distinct regions in the T‐DNA of sgRNA/Cas9 vectors, that is, part of the Cas9 coding sequence and region from the AtU6 promoter to the downstream vector sequence spanning the sgRNA (Figure 5a). The selectable marker gene bar was examined by test strip (Figure 5b). In this assay, we found that four of 64 T1 homozygous ft2a mutants were ‘transgene‐clean’ and that their 46 offspring plants were all ‘transgene‐clean’ homozygous ft2a mutants. The 63 progeny plants of five T‐DNA‐positive T1 homozygous mutants of GmFT2a were also examined, and we obtained 13 ‘transgene‐clean’ homozygous ft2a mutants (Table 2).
Figure 5.
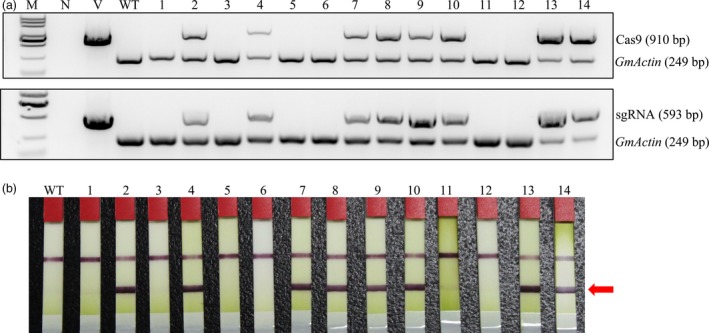
Identifying ‘transgene‐clean’ mutant soya bean lines of GmFT2a. (a) Gel image of PCR products obtained with primer sets for two T‐DNA regions of sgRNA/Cas9 vectors. Cas9 (910 bp), part of the Cas9 coding sequence. sgRNA (593 bp), region from the AtU6 promoter to the downstream vector sequence spanning the sgRNA. GmActin was used as a normalization control. M, DL2000 ladder DNA marker. N, negative control (water as template). V, plasmid of the vector used in transformation as template. WT, DNA of wild‐type soya bean plant as template. Labels above the gel: 1–14, individual mutant lines. (b) Detection of the selectable marker gene bar by test strip. WT, wild‐type soya bean plants. Labels 1–14, individual mutant lines. The bands at red arrowhead indicate that bar is positive.
Table 2.
Identifying ‘transgene‐clean’ homozygous ft2a mutants from T1 and T2 generations
| T1 homozygous ft2a mutants | T‐DNA in the T1 homozygous ft2a mutants | No. of the progeny plants identified | No. of the T2 ‘transgene‐clean’ homozygous ft2a mutants |
|---|---|---|---|
| ft2a‐SP1‐T1#11.14 | T‐DNA‐free | 7 | 7 |
| ft2a‐SP2‐T1#16.04 | T‐DNA‐free | 15 | 15 |
| ft2a‐SP3‐T1#30.08 | T‐DNA‐free | 12 | 12 |
| ft2a‐SP3‐T1#30.12 | T‐DNA‐free | 12 | 12 |
| ft2a‐SP1‐T1#11.15 | T‐DNA‐positive | 14 | 3 |
| ft2a‐SP1‐T1#11.16 | T‐DNA‐positive | 13 | 1 |
| ft2a‐SP2‐T1#16.11 | T‐DNA‐positive | 15 | 6 |
| ft2a‐SP2‐T1#16.12 | T‐DNA‐positive | 15 | 2 |
| ft2a‐SP3‐T1#30.02 | T‐DNA‐positive | 6 | 1 |
Discussion
We previously demonstrated that CRISPR/Cas9 system shared the same efficiency for both endogenous and exogenous genes in soya bean hairy roots (Cai et al., 2015), but the heritability of CRISPR/Cas9‐induced mutations cannot be studied in soya bean hairy roots because they cannot regenerate. In this study, we describe a rapid and highly specific method for targeted mutagenesis of soya bean endogenous genes by CRISPR/Cas9 in whole‐plant transformation. We determined that the CRISPR/Cas9‐induced mutations were inherited from the T0 to T1 generation. However, not all T0 lines could transmit the targeted mutations to the T1 generation. Targeted mutagenesis of GmFT2a in only 5 of 12, 8 of 19 and 5 of 11 T0 heterozygous ft2a mutant lines induced by CRISPR/Cas9 at the three target sites, respectively, was transmitted to the T1 generation (Table 1). These results demonstrated that the heritability of targeted mutations generated by CRISPR/Cas9 in soya bean from the T0 to T1 generation is difficult to predict. It is possible that in the T0 generation, CRISPR/Cas9‐induced mutations exist only in a proportion of cells, which subsequently generate somatic sectors via cell division (Gaj et al., 2013). If somatic mutant sectors occur in the floral primordia, CRISPR/Cas9‐induced targeted mutations could be transmitted to the T1 generation through the gametes. In contrast, the targeted mutations of GmFT2a induced by CRISPR/Cas9 were stably inherited and maintained consistent mutation types from the T1 to T2 generation, indicating a standard germ‐line transmission pattern. Taken together, our results described a feasible strategy to introduce transmissible mutations induced by CRISPR/Cas9 using the propagation of stable transgenic plants and it may be widely used for reverse genetics in soya bean.
Off‐target events are notable in the application of CRISPR/Cas9. To examine the specificity of CRISPR/Cas9 in soya bean and avoid affecting phenotype statistics by including off‐target sites, we analysed the potential off‐target effects of the editing of GmFT2a. The two most likely off‐target sites of the three target sites of GmFT2a were respectively selected and examined. We observed no mutations at all of the putative off‐target sites. Previous reports have demonstrated that the length of the sgRNA and the homology between the sgRNA and candidate off‐target site are the main influencing factors (Zhang et al., 2014). A double Cas9 nickase approach has also been reported to effectively reduce off‐target effects (Fauser et al., 2014). Whole‐genome sequencing should be performed if off‐target effects are fatal, especially in clinical research. However, it may not be a fatal problem in plant basic research. The risk of off‐target events induced by CRISPR/Cas9 in plants may not be high on account of the lower somatic mutation frequency in tissue culture‐based transformation or other mutagenic treatments (Ma et al., 2015). Through careful target selection, potential off‐target mutations could be minimized (Xie et al., 2014; Xu et al., 2015). In addition, undesired off‐target mutations in plants could be eliminated by hybridization, and off‐target events could be utilized to knockout multiple genes simultaneously. In previous studies, we demonstrated the potential of CRISPR/Cas9 to simultaneously edit two endogenous soya bean genes using only one customized sgRNA based on the off‐target principle (Cai et al., 2015). In short, the adverse effects of off‐target events could be eliminated in plant basic research.
In previous studies, site‐directed mutagenesis of FLOWER LOCUS T (FT) generated by CRISPR/Cas9 delayed flowering time in A. thaliana (Hyun et al., 2015). FT encodes florigen and plays an important role in flowering pathways as an integrator (Corbesier and Coupland, 2006; Turck et al., 2008). The FT protein is a florigen that moves through the phloem to the shoot apex and works as a long‐distance signal to induce floral initiation (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Notaguchi et al., 2008). GmFT2a is a soya bean ortholog of FT and plays an important role in flowering induction in soya bean (Kong et al., 2010; Nan et al., 2014; Sun et al., 2011). In the present study, three target sites in the first exon of GmFT2a were chosen. These mutations induced by CRISPR/Cas9 in the coding region were expected to inactivate the GmFT2a protein function by inducing frameshift mutations. We compared the flowering time between homozygous ft2a mutants with WT plants and found that the homozygous ft2a mutants exhibited late‐flowering phenotype under natural, SD and LD conditions. We also compared the time course‐dependent expression patterns of GmFT2a between WT plants and T2 homozygous ft2a mutants grown under LD and SD photoperiodic conditions using RNAs extracted from trifoliate leaves sampled at 4 h after light every 5 days beginning at 10 DAE. The results showed that the expression levels of GmFT2a increased to their maximum levels and thereafter decreased before flowering under both LD and SD photoperiodic conditions. What's more, the transcript levels of GmFT2a in T2 ft2a mutants were significantly lower than that in WT plants. In eukaryotic cells, non‐sense‐mediated mRNA decay (NMD) is an evolutionarily well‐conserved and effective mRNA surveillance pathway that ensures the detection and rapid degradation of mRNAs containing premature translation termination codons (PTCs), thereby preventing the potentially deleterious effects of truncated proteins (Conti and Izaurralde, 2005; Maquat, 2005). In this study, all six types of frameshift mutations induced by CRISPR/Cas9 at three target sites of GmFT2a generated PTCs (Text S1). Therefore, we predicted that the mRNA of GmFT2a containing PTCs was degraded by the NMD surveillance mechanism and then showed extremely low expression levels. GmFT2a likely encodes florigen, and thus loss of GmFT2a gene function resulted in a late‐flowering phenotype.
We obtained some ‘transgene‐clean’ targeted genome‐modified soya bean plants that were homozygous for the null alleles of endogenous GmFT2a and without any transgenic element in the T1 and T2 generations using the CRISPR/Cas9 system. In recent years, transgenic technology has provided many important technical breakthroughs and progress to overcome the shortcomings of traditional breeding methods, such as long cycle, labour intensive and low efficiency. However, there are also some disadvantages to this technology. Foreign genes are typically randomly integrated into the plant genome after entering the nucleus, which may generate negative results, such as the disruption of plant endogenous genes or exogenous gene silencing (Napoli et al., 1990). Although there is no scientific evidence that genetically modified organism (GMO) products are harmful to human health, debates about GMO safety continue. Genome‐editing techniques, especially CRISPR/Cas9, provide a new approach to solve this problem. The CRISPR/Cas9 system could accurately generate site‐directed mutations in specific genes and exogenous elements such as sgRNA. Cas9 and the associated selectable marker can be removed in later generations via genetic segregation by a selfing or hybridization approach. The ‘transgene‐clean’ mutants have been generated by the CRISPR/Cas9 system in many plant species, such as A. thaliana (Gao et al., 2016; Pyott et al., 2016), rice (Li et al., 2016; Xu et al., 2015; Zhou et al., 2016), wheat (Zhang et al., 2016) and tomato (Soyk et al., 2017). The ‘transgene‐clean’ soya bean varieties with high oleic acid have been created by targeted mutagenesis with transcription activator‐like effector nucleases (TALENs) (Haun et al., 2014). Similarly, CRISPR/Cas9 could also be utilized to improve soya bean varieties, such as quality optimization, disease resistance and growth period traits. In the present study, we obtained ‘transgene‐clean’ soya bean plants which were delayed flowering time by homozygous targeted mutagenesis of endogenous GmFT2a with CRISPR/Cas9. These mutants also provide materials for further in‐depth research; for example, they can be utilized to explore the movement of GmFT2a by importing the full GmFT2a expression cassette in combination with GFP (green fluorescent protein) and studying functional complementation. The same method could be used to generate more ‘transgene‐clean’ mutants of GmFT2a‐relevant genes by CRISPR/Cas9, which could then be crossed to research gene function. Soya bean is very sensitive to photoperiod, which limits its geographical range of cultivation, and our studies may contribute to soya bean regional introduction and domestication.
Experimental procedures
Plant materials and growth conditions
The soya bean cultivar Jack was used for transformation in this study. Wild‐type Jack (as a control) and all seeds collected from ft2a‐CRISPR/Cas9 T0 plants were sown under natural conditions in Beijing, China (N39°58′, E116°20′) on 11 May 2016. The progeny of T1 homozygous ft2a mutants was separately grown under long‐day (16 h light/8 h dark) and short‐day (12 h light/12 h dark) photoperiodic conditions at 27 °C with 50% relative humidity.
SgRNA design and construction of the CRISPR/Cas9 expression vector
To construct a plasmid vector carrying both sgRNA and Cas9 cassettes, the sequence of Cas9 was codon‐optimized for dicotyledons and assembled downstream of the CaMV 2× 35S promoter together with a customized sgRNA driven by the Arabidopsis U6 promoter (Text S2). The bar gene driven by a CaMV 35S promoter was used as a screening marker. The schematic of GmFT2a‐CRISPR/Cas9 vector is shown in Figure S2. The sequences were synthesized by ViewSolid Biotech (Beijing). The sequence and other information for the analysed soya bean endogenous gene GmFT2a were downloaded from the Phytozome website (www.phytozome.net/). We designed these sgRNAs using the web tool CRISPR‐P (http://cbi.hzau.edu.cn/crispr/) (Lei et al., 2014), which displayed all optional sgRNA sequences (20 bp) immediately followed by 5′‐NGG (PAM, protospacer adjacent motif) in the forward or reverse strand. In this study, we selected three sgRNAs targeting GmFT2a, named as GmFT2a‐SP1, GmFT2a‐SP2 and GmFT2a‐SP3. The first base was a guanine (G) nucleotide in GmFT2a‐SP1 but not GmFT2a‐SP2 and GmFT2a‐SP3. Thus, an extra G was appended before the 5′ end of the sequences of these two sgRNAs (Ran et al., 2013). For each sgRNA, a pair of DNA oligonucleotides was synthesized by the Beijing Genomics Institute (BGI) and annealed to generate dimers, which were subsequently integrated upstream of the sgRNA scaffolds in the plasmid vector simultaneously expressing Cas9 and sgRNA. After transformation into E. coli DH5α for in vivo cloning, these CRISPR/Cas9 expression vectors were purified using the EasyPure Plasmid MiniPrep Kit (TransGen Biotech) for subsequent use in soya bean transformation.
Transformation of CRISPR/Cas9 in soya bean and screening for mutations by sequencing analysis
CRISPR/Cas9 expression vectors were individually transformed into Agrobacterium tumefaciens strain EHA105 via electroporation. The soya bean cultivar Jack was used for tissue culture and transformation according to the protocol previously reported (Song et al., 2013). Genomic DNA was extracted from the leaves of each individual plant in the T0 generation, and then the regions spanning the target sites were amplified by PCR using Phanta® Super Fidelity DNA Polymerase (Vazyme Biotech) with the GmFT2a forward primer (5′‐ATTCATAACAAAGCAAACGAG‐3′) and reverse primer (5′‐ACTTGACCTTCCCTTAAACAC‐3′), purified and sequenced. Different types of gene editing can be identified via sequence peaks. Short base insertions or deletions (not multiples of three) induced by CRISPR/Cas9 can lead to frameshift mutations. The heterozygous mutations showed overlapping peaks from the target sites to the end. The wild‐type and homozygous mutations had no overlapping peaks at the target sites. Then, the homozygous mutant types were identified by sequence alignment with the wild‐type sequence. This method was also used in the T1 and T2 generations.
Flowering time measurements and statistical analyses
The flowering time of each soya bean plant was recorded as days from emergence to the R1 stage (the first flower appears at any node in the main stem; Fehr et al., 1971). For quantitative analyses of flowering time, at least 11 individual soya bean plants were analysed per genotype, and exact numbers of individuals (n) are shown in Figure 3 and Figure S1. Statistical analyses were performed using Microsoft Excel. A one‐way analysis of variance least significant difference test (LSD) was used to compare the significance of differences between controls and treatments at the 0.01 probability level. SigmaPlot 10.0 was used for drawing box‐plots and histograms. The flowering time is shown as the mean values ± standard deviation.
Gene expression analysis by quantitative real‐time PCR (qRT‐PCR)
The wild‐type plants and T2 homozygous ft2a mutants grown under long‐day (16 h light/8 h dark) and short‐day (12 h light/12 h dark) photoperiodic conditions were used to compare the expression levels of GmFT2a. Pieces of fully developed trifoliate leaves at the upper part of 16 wild‐type plants (Every 8 individual plants grown under LD and SD conditions, respectively) and 24 T2 ft2a mutants (Every 12 individual plants grown under LD and SD conditions, respectively) were sampled at 4 h after light every 5 days beginning at 10 DAE. These leaves were immediately frozen in liquid nitrogen. Total RNA was isolated from frozen tissue using TransZol Up Plus RNA Kit (TransGen Biotech). For reverse transcription, 1 μg of total RNA was used to synthesize single‐stranded cDNA using TransScript One‐Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). For qRT‐PCR, each 10‐μL reaction contained 1 μL of 1 : 5 diluted cDNA with 0.2 μL of each primer (10 μm), 5 μL of 2× KAPA SYBR® FAST qPCR Master Mix, 0.2 μL of 50× ROX High Reference Dye (KAPA Biosystems), and water to a final volume of 10 μL. The ABI Prism 7900HT Real‐Time PCR System was used. The PCR cycling conditions started with a denaturing step for 3 min at 95 °C followed by 40 cycles of 5 s at 95 °C and a primer extension reaction at 60 °C for 30 s. All PCR reactions were run with three biological replicates each. Data were analysed using the 2−ΔΔCt method with the mRNA level of GmActin (Glyma18g52780) gene as an internal control.
Primer sequences used in the present study
The primer sequences used for amplifying the regions which span the target sites, potential off‐target analysis, identifying ‘transgene‐clean’ mutant soya bean lines and qRT‐PCR analysis are listed in Table S3.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Homozygous ft2a mutants at three target sites in the T1 generation delayed flowering time under natural conditions (summer) in Beijing, China (N39°58′, E116°20′).
Figure S2 Schematic illustrating the basic architecture of the constructs used for CRISPR/Cas9‐mediated genome editing.
Table S1 Potential off‐target analysis at the three target sites of GmFT2a in the T1 generation.
Table S2 T2 ft2a mutants under LD and SD conditions.
Table S3 Primer sequences used in the present study.
Text S1 Frameshift mutations at three target sites of GmFT2a generated premature translation termination codons (PTCs). CDS, coding sequence. Blue capital letter, target sequence. Red capital letter, protospacer adjacent motif. Underline, insertions. Dashes, deletions. Yellow rectangle, termination codon.
Text S2 The sequences of the Cas9 and Arabidopsis U6 promoter used in the present study.
Acknowledgements
This work was supported by Major Science and Technology Projects of China (2016ZX08010‐004), National Natural Science Foundation of China (31471571), and the CAAS (Chinese Academy of Agriculture Sciences) Agricultural Science and Technology Innovation Project.
References
- Cai, Y. , Chen, L. , Liu, X. , Sun, S. , Wu, C. , Jiang, B. , Han, T. et al (2015) CRISPR/Cas9‐mediated genome editing in soybean hairy roots. PLoS ONE 10, e0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, E. and Izaurralde, E. (2005) Nonsense‐mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325. [DOI] [PubMed] [Google Scholar]
- Corbesier, L. and Coupland, G. (2006) The quest for florigen: a review of recent progress. J. Exp. Bot. 57, 3395–3403. [DOI] [PubMed] [Google Scholar]
- Corbesier, L. , Vincent, C. , Jang, S. , Fornara, F. , Fan, Q. , Searle, I. , Giakountis, A. et al (2007) FT protein movement contributes to long‐distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Du, H. , Zeng, X. , Zhao, M. , Cui, X. , Wang, Q. , Yang, H. , Cheng, H. et al (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. [DOI] [PubMed] [Google Scholar]
- Fauser, F. , Schiml, S. and Puchta, H. (2014) Both CRISPR/Cas‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . Plant J. 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Fehr, W.R. , Caviness, C.E. , Burmood, D.T. and Pennington, J.S. (1971) Stage of development descriptions for soybeans, Glycine Max (L.) Merrill. Crop Sci. 11, 929–931. [Google Scholar]
- Gaj, T. , Gersbach, C.A. and Barbas, C.F. (2013) ZFN, TALEN and CRISPR/Cas‐based methods for genome engineering. Trends Biotechnol. 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Chen, J. , Dai, X. , Zhang, D. and Zhao, Y. (2016) An effective strategy for reliably isolating heritable and Cas9‐free Arabidopsis mutants generated by CRISPR/Cas9‐mediated genome editing. Plant Physiol. 171, 1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova, V. and Levy, A.A. (1999) How plants make ends meet: DNA double‐strand break repair. Trends Plant Sci. 4, 263–269. [DOI] [PubMed] [Google Scholar]
- Gratz, S.J. , Ukken, F.P. , Rubinstein, C.D. , Thiede, G. , Donohue, L.K. , Cummings, A.M. and O'Connor‐Giles, K.M. (2014) Highly specific and efficient CRISPR/Cas9‐catalyzed homology‐directed repair in Drosophila . Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun, W. , Coffman, A. , Clasen, B.M. , Demorest, Z.L. , Lowy, A. , Ray, E. , Retterath, A. et al (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. [DOI] [PubMed] [Google Scholar]
- Hyun, Y. , Kim, J. , Cho, S.W. , Choi, Y. , Kim, J.‐S. and Coupland, G. (2015) Site‐directed mutagenesis in Arabidopsis thaliana using dividing tissue‐targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T.B. , LaFayette, P.R. , Schmitz, R.J. and Parrott, W.A. (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, K.E. and Wigge, P.A. (2007) FT protein acts as a long‐range signal in Arabidopsis. Curr. Biol. 17, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jiang, B. , Yue, Y. , Gao, Y. , Ma, L. , Sun, S. , Wu, C. , Hou, W. et al (2013a) GmFT2a polymorphism and maturity diversity in soybeans. PLoS ONE 8, e77474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013b) Demonstration of CRISPR/Cas9/sgRNA‐mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y. , Kaya, H. , Goto, K. , Iwabuchi, M. and Araki, T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kong, F. , Liu, B. , Xia, Z. , Sato, S. , Kim, B.M. , Watanabe, S. , Yamada, T. et al (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M. , Hanhart, C.J. and van der Veen, J.H. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana . Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Aach, J. , Norville, J.E. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al (2013) Multiplex and homologous recombination‐mediated plant genome editing via guide RNA/Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Liu, Z.B. , Xing, A. , Moon, B.P. , Koellhoffer, J.P. , Huang, L. , Ward, R.T. et al (2015) Cas9‐guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Li, X. , Zhou, Z. , Wu, P. , Fang, M. , Pan, X. , Lin, Q. et al (2016) Reassessment of the four yield‐related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhang, K. , Chen, K. and Gao, C. (2014) Targeted mutagenesis in zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 41, 63–68. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. (2005) Nonsense‐mediated mRNA decay in mammals. J. Cell Sci. 118, 1773. [DOI] [PubMed] [Google Scholar]
- Mathieu, J. , Warthmann, N. , Küttner, F. and Schmid, M. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Michno, J.‐M. , Wang, X. , Liu, J. , Curtin, S.J. , Kono, T.J.Y. and Stupar, R.M. (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web‐tool and a modified Cas9 enzyme. GM Crops Food 6, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan, H. , Cao, D. , Zhang, D. , Li, Y. , Lu, S. , Tang, L. , Yuan, X. et al (2014) GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS ONE 9, e97669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli, C. , Lemieux, C. and Jorgensen, R. (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co‐suppression of homologous genes in trans. Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi, M. , Abe, M. , Kimura, T. , Daimon, Y. , Kobayashi, T. , Yamaguchi, A. , Tomita, Y. et al (2008) Long‐distance, graft‐transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658. [DOI] [PubMed] [Google Scholar]
- Pyott, D.E. , Sheehan, E. and Molnar, A. (2016) Engineering of CRISPR/Cas9‐mediated potyvirus resistance in transgene‐free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A. , Hsu, P.D. , Wright, J. , Agarwala, V. , Scott, D.A. and Zhang, F. (2013) Genome engineering using the CRISPR‐Cas9 system. Nat. Protoc. 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. , Zhang, Y. , Chen, K. , Liang, Z. , Zhang, K. et al (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Song, S. , Hou, W. , Godo, I. , Wu, C. , Yu, Y. , Matityahu, I. , Hacham, Y. et al (2013) Soybean seeds expressing feedback‐insensitive cystathionine gamma‐synthase exhibit a higher content of methionine. J. Exp. Bot. 64, 1917–1926. [DOI] [PubMed] [Google Scholar]
- Soyk, S. , Muller, N.A. , Park, S.J. , Schmalenbach, I. , Jiang, K. , Hayama, R. , Zhang, L. et al (2017) Variation in the flowering gene SELF PRUNING 5G promotes day‐neutrality and early yield in tomato. Nat. Genet. 49, 162–168. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Jia, Z. , Cao, D. , Jiang, B. , Wu, C. , Hou, W. , Liu, Y. et al (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6, e29238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Hu, Z. , Chen, R. , Jiang, Q. , Song, G. , Zhang, H. and Xi, Y. (2015) Targeted mutagenesis in soybean using the CRISPR‐Cas9 system. Sci. Rep. 5, 10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene editing, and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Yang, S. , Liu, J. and Zhu, H. (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin‐like protein but not the one previously reported. Plant Physiol. 170, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F. , Fornara, F. and Coupland, G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. [DOI] [PubMed] [Google Scholar]
- Upadhyay, S.K. , Kumar, J. , Alok, A. and Tuli, R. (2013) RNA‐guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 3, 2233–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Gu, Y. , Gao, H. , Qiu, L. , Chang, R. , Chen, S. and He, C. (2016) Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 16, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Zhang, J. and Yang, Y. (2014) Genome‐wide prediction of highly specific guide RNA spacers for CRISPR–Cas9‐mediated genome editing in model plants and major crops. Mol. Plant 7, 923–926. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Xu, Z. , Liu, B. , Kong, F. , Tsubokura, Y. , Watanabe, S. , Xia, Z. et al (2013) Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA‐regulated post‐flowering responses of soybean. BMC Evol. Biol. 13, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R.F. , Li, H. , Qin, R.Y. , Li, J. , Qiu, C.H. , Yang, Y.C. , Ma, H. et al (2015) Generation of inheritable and “transgene clean” targeted genome‐modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5, 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.L. et al (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Takeshima, R. , Zhu, J. , Xu, M. , Sato, M. , Watanabe, S. , Kanazawa, A. et al (2016a) A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhang, C. , Liu, W. , Gao, W. , Liu, C. , Song, G. , Li, W.‐X. et al (2016b) An alternative strategy for targeted gene replacement in plants using a dual‐sgRNA/Cas9 design. Sci. Rep. 6, 23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , He, M. , Li, J. , Chen, L. , Huang, Z. , Zheng, S. , Zhu, L. et al (2016) Development of commercial thermo‐sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9‐mediated TMS5 editing system. Sci. Rep. 6, 37395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Homozygous ft2a mutants at three target sites in the T1 generation delayed flowering time under natural conditions (summer) in Beijing, China (N39°58′, E116°20′).
Figure S2 Schematic illustrating the basic architecture of the constructs used for CRISPR/Cas9‐mediated genome editing.
Table S1 Potential off‐target analysis at the three target sites of GmFT2a in the T1 generation.
Table S2 T2 ft2a mutants under LD and SD conditions.
Table S3 Primer sequences used in the present study.
Text S1 Frameshift mutations at three target sites of GmFT2a generated premature translation termination codons (PTCs). CDS, coding sequence. Blue capital letter, target sequence. Red capital letter, protospacer adjacent motif. Underline, insertions. Dashes, deletions. Yellow rectangle, termination codon.
Text S2 The sequences of the Cas9 and Arabidopsis U6 promoter used in the present study.


