Summary
The NHX‐type cation/H+ transporters in plants have been shown to mediate Na+(K+)/H+ exchange for salinity tolerance and K+ homoeostasis. In this study, we identified and characterized two NHX homologues, HtNHX1 and HtNHX2 from an infertile and salinity tolerant species Helianthus tuberosus (cv. Nanyu No. 1). HtNHX1 and HtNHX2 share identical 5′‐ and 3′‐UTR and coding regions, except for a 342‐bp segment encoding 114 amino acids (L272 to Q385) which is absent in HtNHX2. Both hydroponics and soil culture experiments showed that the expression of HtNHX1 or HtNHX2 improved the rice tolerance to salinity. Expression of HtNHX2, but not HtNHX1, increased rice grain yield, harvest index, total nutrient uptake under K+‐limited salt‐stress or general nutrient deficiency conditions. The results provide a novel insight into NHX function in plant mineral nutrition.
Keywords: Helianthus tuberosus, NHX‐type transporters, nutrient deficiency, potassium, rice, salt stress
Introduction
Naturally occurred and fertilization‐induced soil salinity, and low nutrient use efficiency are significant constraints in modern agriculture production (Heuer et al., 2016; Horie et al., 2009; Munns and Gilliham, 2015; Munns and Tester, 2008; Xu et al., 2012). At the cellular level, the extrusion of Na+ ions at the cell plasma membrane and the compartmentation of Na+ into vacuoles are efficient mechanisms to avoid excessive cytosolic Na+ concentration and maintain an adequate cytosolic K+/Na+ ratio (Julkowska and Testerink, 2015; Munns and Tester, 2008). The plasma membrane electrical potential difference of root cells is usually maintained around −120 mV, while the tonoplast (vacuole membrane) potential is positive and around 20–40 mV (Shabala et al., 2016). These potentials allow the root to acquire sufficient K+ via high‐affinity transporter systems (HATS) in K+‐limited soils or via low‐affinity transporter systems (LATS) at normal external K+ supplies. The presence of high external Na+ concentrations suppresses the K+ conductance through LATS (Qi and Spalding, 2004) and competes with K+ uptake through HATS (Alemán et al., 2011; Leidi et al., 2010), causing a decrease in intracellular K+ with a concomitant [K+]/[Na+] imbalance.
The plant vacuolar Na+/H+ antiporters (NHXs) were shown originally to mediate the electroneutral Na+/H+ exchange, driving the excess cytosolic Na+ into the vacuole (Apse et al., 1999; Blumwald and Poole, 1985; Gaxiola et al., 1999). The NHX proteins belong to the large mono‐valent cation/proton transporters (CPA) family, showing three distinct functional clades (Chanroj et al., 2012). In Arabidopsis, in addition to the plasma membrane‐located NHX7 and NHX8, also known as SOS (salt‐over‐sensitive), intracellular NHXs sharing high sequence similarity are further divided into type‐I and type‐II, based on their subcellular location (Chanroj et al., 2012). Type‐I NHXs (AtNHX1‐AtNHX4) are vacuolar‐located, while type‐II NHXs (AtNHX5 and AtNHX6) are found at endosome, Trans‐Golgi Network (TGN)/Golgi and prevacuolar compartments (Andrés et al., 2014; Barragán et al., 2012; Bassil et al., 2011a,b; McCubbin et al., 2014). In the rice genome, there are at least five NHX members with OsNHX1‐OsNHX4 belonging to the type‐I and OsNHX5 to the type‐II (Fukuda et al., 2011). The operation of the NHXs also affects the pH gradients across the different luminal compartments (Bassil et al., 2012).
Both type‐I and type‐II NHXs have different subcellular localization; they may have different mechanisms in salt tolerance, and other not yet defined functions. Large efforts have been paid to increasing the plant salt tolerance by overexpressing NHX orthologue genes from different species. In rice, overexpression of the NHX1 homologue genes from Oryza sativa, Chenopodium glaucum and Atriplex dimorphostegia resulted in the enhanced tolerance to salt stress (Li et al., 2008). However, these NHX proteins actually mediate both Na+/H+ and K+/H+ exchange and their functions cannot be solely explained by accumulating Na+ into vacuole (Andrés et al., 2014; Barragán et al., 2012; Bassil et al., 2011b; Jiang et al., 2010; Leidi et al., 2010; Reguera et al., 2014).
The type‐II NHXs have also been shown to be involved in salt tolerance. AtNHX5 and AtNHX6 are the only two type‐II NHXs in Arabidopsis. Although their function appears to be redundant, the double knockout Atnhx5nhx6 displayed high sensitivity to salt stress (Bassil et al., 2011a). In tomato, the overexpression of LeNHX2, a type‐II NHX located in prevacuolar compartments and Golgi, enhanced salt tolerance at high external K+ levels (Venema et al., 2003).
Plants use a number of strategies to deal with high salinity (Munns and Tester, 2008). Halophytes rely heavily on the homoeostasis of three major inorganic ions (Na+, Cl−, and K+) to maintain their osmotic and turgor pressure under saline conditions, while glycophytes predominantly increase the synthesis of compatible solutes (Deinlein et al., 2014). For salt tolerant plants grown in high external saline conditions, the efficient compartmentation of Na+ into vacuole and other cell organs via intracellular NHXs is of importance, together with the extrusion of Na+ at the roots, via plasma membrane‐bound antiporters such as SOS (Deinlein et al., 2014; Munns and Tester, 2008).
Jerusalem artichoke (Helianthus tuberosus L.) belonging to the same Asteraceae family as sunflower (Helianthus annuus L.) is an herbaceous perennial plant that have potential as a biorefinery crop (Johansson et al., 2015). H. tuberosus is highly tolerant to infertile, drought and saline stresses and its variety Nanyu No. 1 (NY‐1) could fully emerge in the coastal region containing 1% salt at soil surface layer (0–15 cm) and grow well in soil containing 0.5%–0.6% salt or under irrigation with 50%–75% sea water (Long et al., 2008). In this study, we isolated two putative NHX genes, HtNHX1 and HtNHX2, from H. tuberosus cv. NY‐1 and examined their roles in enhancing the tolerance of salt stress and nutrient deficiency in rice, Arabidopsis and yeast. Our results indicate that HtNHX2 could function in enhancing the plant tolerance to salinity stress and improving plant nutrient efficiency.
Results
Cloning of salt‐stress up‐regulated HtNHX1 and HtNHX2 genes from Helianthus tuberosus
To clone NHX genes in H. tuberosus, we designed the primers (Table S1) based on the conserved regions of known NHXs in plants (Figure S1) for PCR amplification of cDNA isolated from the salt‐treated cv. NY‐1. The two different fragments were obtained, and their full lengths were further isolated by 5′ and 3′ RACE‐PCR. The two putative NHX genes with 2148‐ and 1806‐bp encoding regions were named HtNHX1 and HtNHX2, respectively.
Interestingly, HtNHX1 and HtNHX2 have identical 269 bp of 5′ UTR and 501 bp of 3′UTR. Furthermore, their coding sequences are also identical except a segment of 342 bp in HtNHX1 encoding 114 amino acids (L272 to Q385) which is absent in HtNHX2 (Figure S1A). By alignment of the amino acids sequences of HtNHX1 and HtNHX2 with more than forty NHX members from different plant species, both HtNHX1 and HtNHX2 are classified as IC type‐I NHXs (Figure S1B). Transmembrane topology predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) or TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/) indicated that HtNHX1 comprised 10 transmembrane domains, while HtNHX2 has seven to eight transmembrane domains (Figure S1CD).
As the expression of most known type‐I NHXs was up‐regulated by salt stress, we analysed the expression of HtNHX1 and HtNHX2 in H. tuberosus in responses to salt treatments (100 mm NaCl). The expression of HtNHX1 was determined by real‐time qRT‐PCR with the primers flanking the region absent in HtNHX2, while the expression level of HtNHX2 was distinguished from that of HtNHX1 by semi‐quantitative RT‐PCR with a product of HtNHX2 342 bp shorter than that of HtNHX1. As shown in Figure 1a,b, both HtNHX1 and HtNHX2 were up‐regulated by increasing the salinity levels. HtNHX1 expression was enhanced in the roots and stems, and to a lesser extent in leaves, while HtNHX2 expression was enhanced similarly in all three organs (Figure 1c,d).
Figure 1.
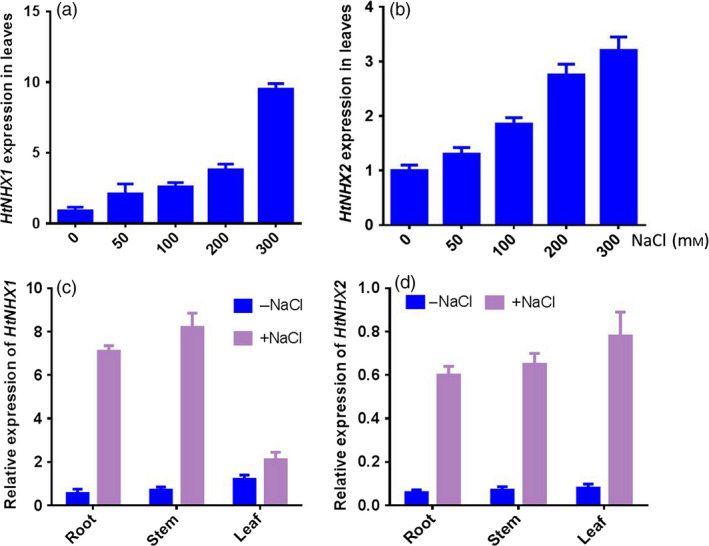
Effects of salt treatments on expression of HtNHX1 and HtNHX2 in entire shoot (a, b) and root, stem and leaf (c, d) of H. tuberosus grown in nutrient sufficient solution. The seedlings of H. tuberosus (cv. Nanyu no. 1) were grown in the nutrient solution for 14 days, and then different amount of NaCl solution was added into the culture solution for 24 h (a, b) and 100 mm NaCl for 12 h (c, d) before the sampling for RNA extraction. The expression of HtNHX1 and HtNHX2 was relative to HtActin abundance in the cultivar. The bar of each column is SE (standard error) with three biological replicates.
HtNHX1 and HtNHX2 expression conferred salt tolerance in rice
To assess HtNHX1 and HtNHX2 functional role(s), HtNHX1 and HtNHX2 were overexpressed in rice (cv. Nipponbare) driven by the ubiquitin promoter (see Materials and methods, Figure S2) and salt tolerance tested. Wild‐type (WT) and transgenic lines were grown under normal conditions for 2 weeks, and the seedlings were exposed to 100 mm NaCl for 3 weeks and growth measured (Figure 2). While the expression of HtNHX1 or HtNHX2 did not affect plant root or shoot biomass (Figure 2a,c,d), the presence of NaCl induced a reduction of growth in both shoot and roots, but the growth of the transgenic plants was less affected by salt stress (Figure 2b,e,f). When grown under nonsalinized conditions, the K+ contents of shoots and roots were similar in WT and transgenic plants, although shoots contained higher K+ amounts than roots (Figure 3a). Transgenic plants expressing HtNHX1 displayed higher shoot and root K+ contents (Figure 3b), while transgenic plants expressing HtNHX2 displayed higher K+ contents than WT but similar root K+ contents (Figure 3b). All transgenic lines displayed higher shoot and root Na+ amounts than WT, although Na+ contents were lower in shoots expressing HtNHX1 than plants expressing HtNHX2 (Figure 3c).
Figure 2.
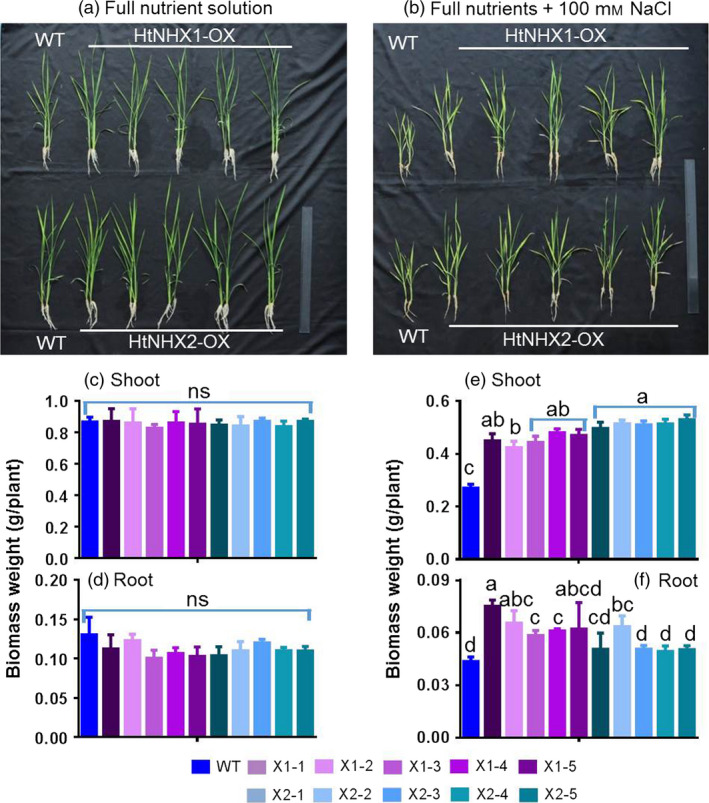
Effects of overexpression of HtNHX1 and HtNHX2 on rice growth under different nutrient supply and salt‐stress condition. The rice was grown for 21 days in the solution before the sampling. (a, c, d) IRRI solution with full supply of nutrients (see Materials and methods); (b, e, f) Full nutrient solution + 100 mm NaCl. WT: cv. Nipponbare; X1‐1, 1‐2, 1‐3, 1‐4 and 1‐5: five T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2, 2‐3, 2‐4 and 2‐5: five T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%. ns: no significant difference.
Figure 3.
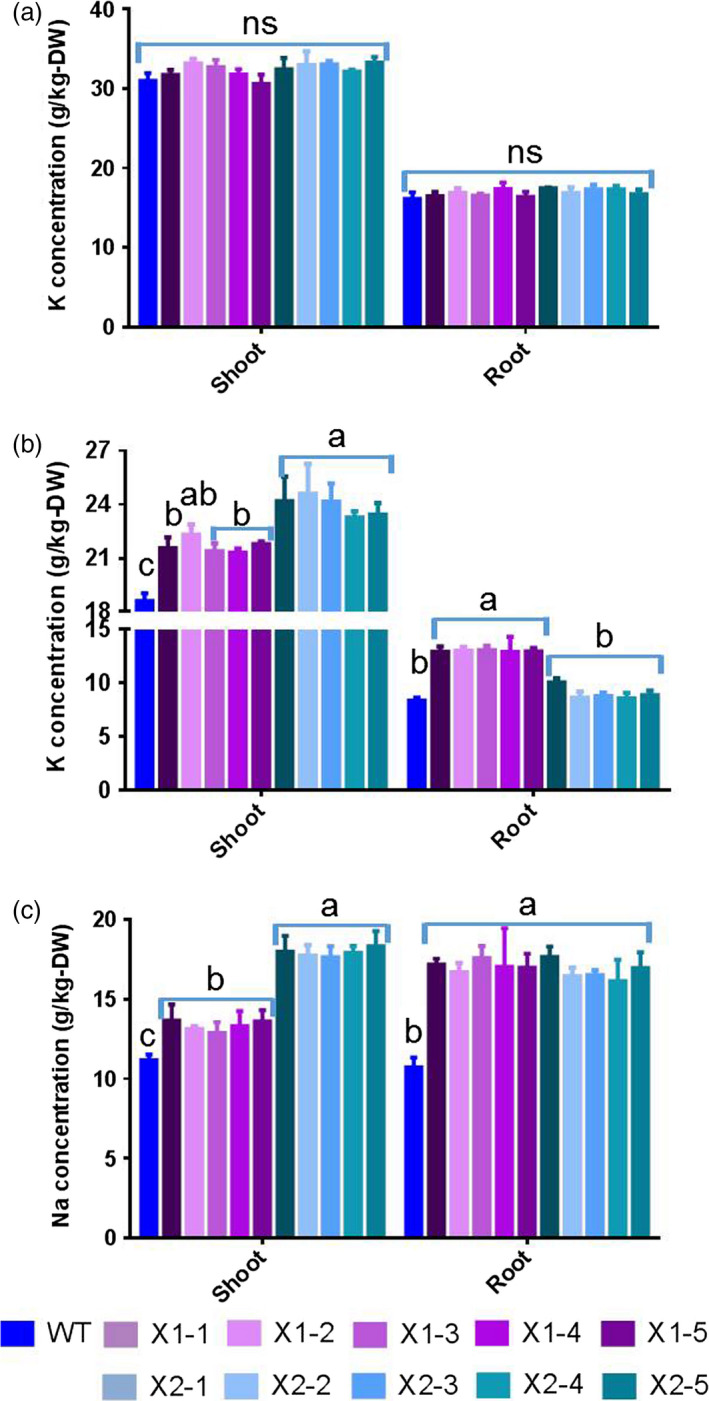
Effects of overexpression of HtNHX1 and HtNHX2 on uptake of K+ and Na+ under full nutrient supply and salt‐stress condition. (a, b) K+ concentration in shoot and root under full nutrient supply and nonsalinized condition (1 mm K+ + 0 mm Na+) (a) and under salinized condition (1 mm K+ + 100 mm Na+) (b); (c) Na+ concentration in shoot and root under salinized condition (1 mm K+ + 100 mm NaCl). WT: cv. Nipponbare; X1‐1, 1‐2, 1‐3, 1‐4 and 1‐5: five T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2, 2‐3, 2‐4 and 2‐5: five T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%. ns: no significant difference.
We also examined the effects of HtNHX1 and HtNHX2 on grain yields of plants grown under salinized soils (Figure 4). Plant biomass and harvest index (grain/straw ratio) was similar in WT and transgenic plants under control conditions (Figure 4a,b). Under salt treatments, WT plants displayed significant yield penalties, while the yield of the transgenic plants was only slightly affected (Figure 4a,b), confirming the role of HtNHX conferring salt tolerance.
Figure 4.
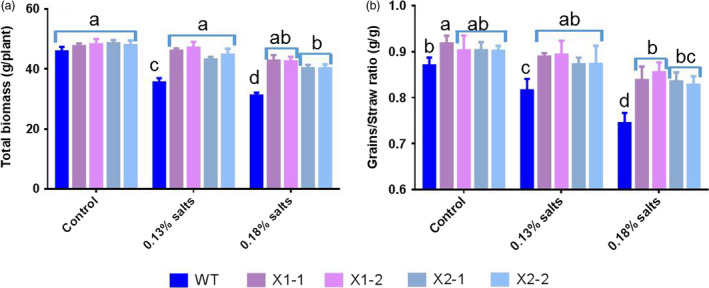
Effects of overexpression of HtNHX1 and HtNHX2 on rice growth, harvest index grown in the soil with different amount of additive salts (NaCl). Two‐week‐old seedlings were acclimated to the soil containing 0.1% (1‐g/kg soil) NaCl for 1 week before being transplanted to the pot filled with 40 kg of soil which had been fertilized and mixture with indicated extra amount of salts for 2 weeks. Control: no additive NaCl was provided into the fertile soil (see Table S2); 0.13% and 0.18% salts: adding 1.3 and 1.8 g NaCl per kg soil in the pot. WT: cv. Nipponbare; X1‐1, 1‐2: two T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2: two T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%. ns: no significant difference.
Roles of HtNHX1 and HtNHX2 in enhancing rice tolerance to K+ deficiency and salt tolerance
When transgenic rice plants were grown under low K+ concentrations (i.e. 0.25 mm), the expression of HtNHX1 did not affect either plant growth or K+ contents in the shoot (Figure 5a–d). On the other hand, the overexpression of HtNHX2 resulted in significant increases in shoot biomass (70%), root biomass (35%) and plant K+ contents (25%–55%) (Figure 5a–d).
Figure 5.
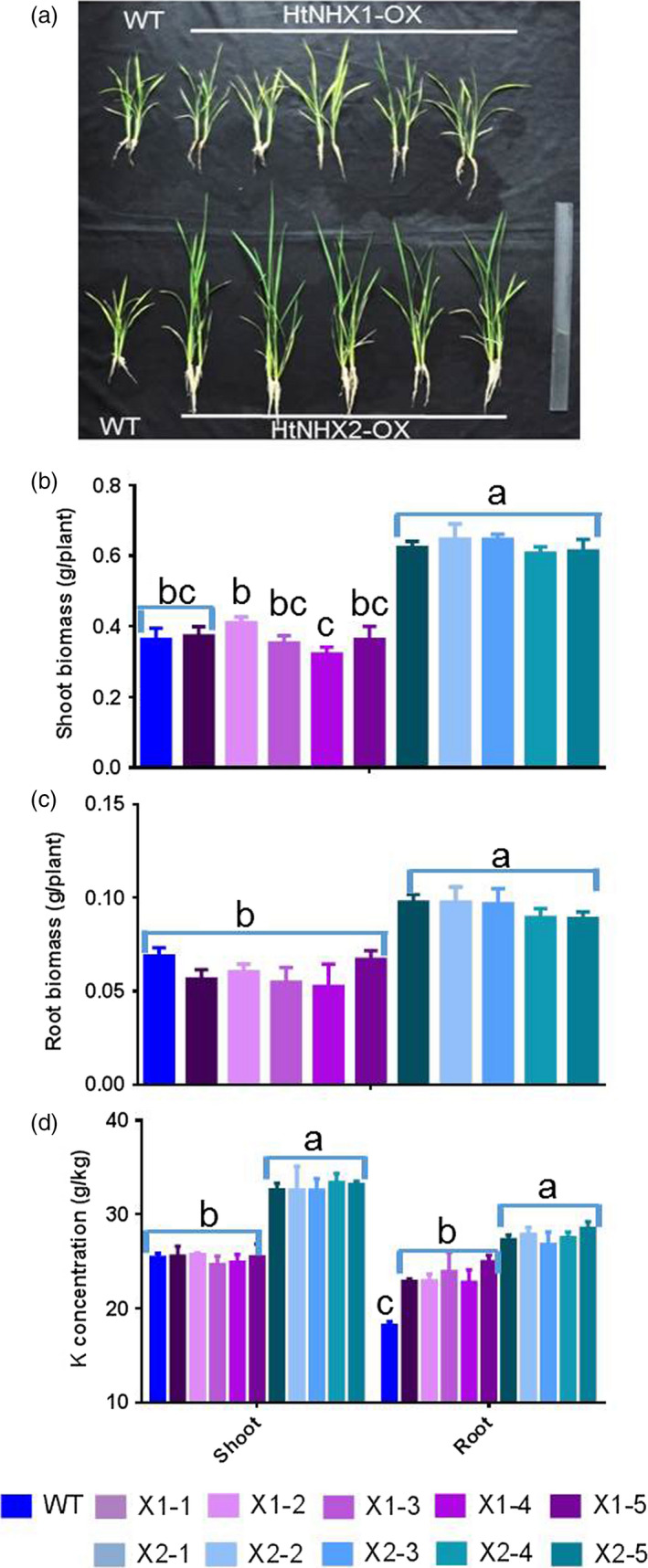
Effects of overexpression of HtNHX1 and HtNHX2 on rice growth and K+ accumulation under limited K+ supply condition. The rice was grown for 21 days in the solution containing 1/4 K+ (0.25 mm) of IRRI solution with sufficient supply of all other nutrients (see Materials and methods). (a) Growth phenotypes; (b and c) biomass of shoot (b) and root (c); (d) K+ concentration in shoot and root. WT: cv. Nipponbare; X1‐1, 1‐2, 1‐3, 1‐4 and 1‐5: five T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2, 2‐3, 2‐4 and 2‐5: five T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%. ns: no significant difference.
The functions of HtNHX1 and HtNHX2 in improving rice tolerance to K+ deficiency were also tested in low K+‐containing soil (Figure S3). When K+ fertilizer was applied, there were no differences between WT and transgenic plants expressing HtNHX1 or HtNHX2 (Figure S3A, B). However, in low K+‐containing soil, the transgenic lines expressing HtNHX2 displayed about 30% increase in grain yield (Figure S3A), increased harvest index (Figure S3B) and increased straw K+ contents (Figure S3C). In contrast, expression of HtNHX1 significantly decreased rice grain yield and grain to straw ratio by about 40%–50% even though the concentrations of N, P and K+ in the straw were not significantly affected (Figure S3A–C).
Interestingly, a large difference in the effects of expressing HtNHX1 or HtNHX2 in enhancing the tolerance to salt stress under limited K+ supply was noted (Figure 6). As compared to WT, the transgenic lines expressing HtNHX2 showed increased shoot biomass, K+ and Na+ contents of 95%, 40% and 90%, respectively, while a lesser increase was seen in lines expressing HtNHX1 (Figure 6a–c). Total shoot K+/Na+ remained unchanged in plants expressing HtNHX1, but decreased significantly in plants expressing HtNHX2 (Figure 6d). Although comparable increases in root growth and Na+ contents were obtained by expressing HtNHX1 and HtNHX2, K+ root contents were higher in plants expressing HtNHX1 than HtNHX2 (Figure 6a–c), resulting in higher root K+/Na+ ratio in HtNHX1 lines (Figure 6d).
Figure 6.
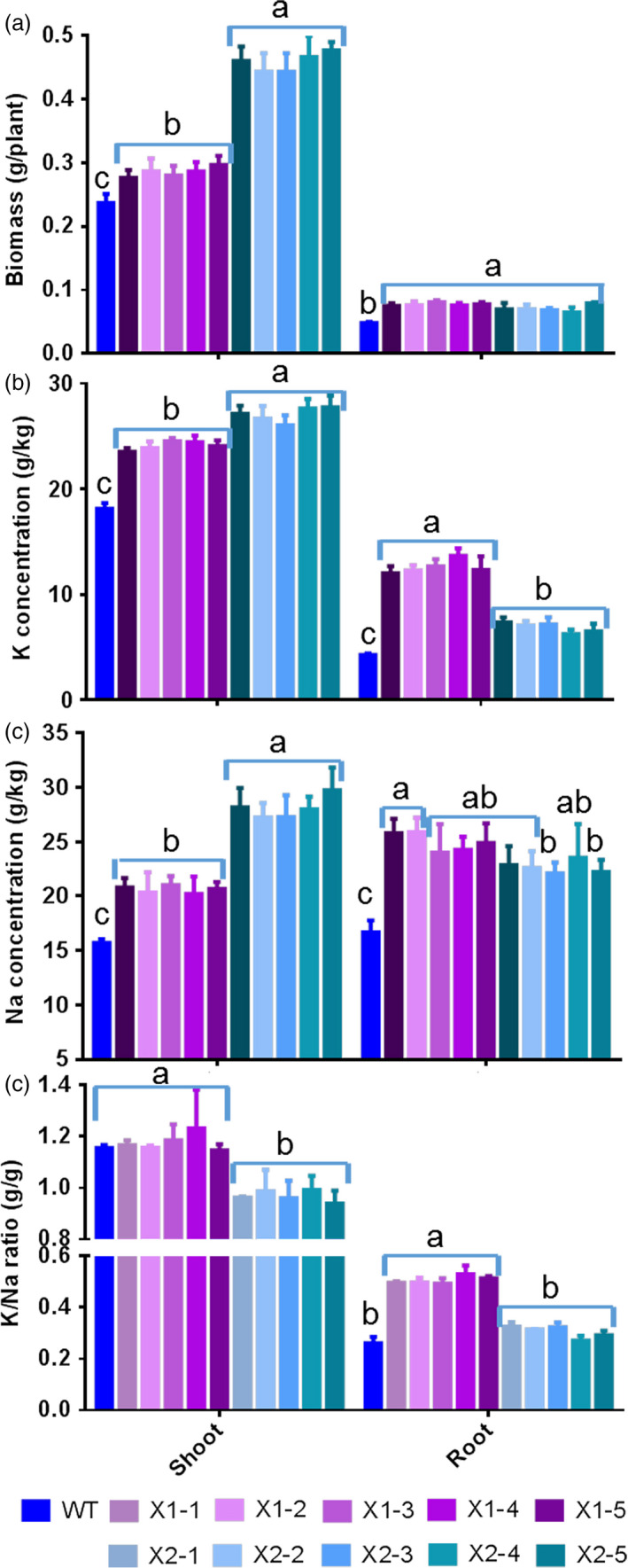
Effects of overexpression of HtNHX1 and HtNHX2 on rice biomass production and total K+ and Na+ accumulation under limited K+ supply and salt‐stress condition. The rice was grown for 21 days in the solution containing ¼ K+ (0.25 mm) of IRRI solution with sufficient supply of all other nutrients and 100 mm NaCl (see Materials and methods). (a) Biomass of shoot and root; (b, c) concentration of K (b) and Na (c) in shoot and root; (d) K+/Na+ ratio in shoot and root. WT: cv. Nipponbare; X1‐1, 1‐2, 1‐3, 1‐4 and 1‐5: five T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2, 2‐3, 2‐4 and 2‐5: five T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%. ns: no significant difference.
HtNHX2, but not HtNHX1, enhanced rice tolerance to limited supply of major nutrients
While the ectopic expression of HtNHX1 or HtNHX2 did not induce phenotypical differences in rice grown under control conditions, transgenic rice plants expressing HtNHX2 displayed remarkable higher growth than WT or transgenic plants expressing HtNHX1, when grown under low K supply (Figure 5). Considering the essential roles of K+ in balancing uptake and distribution of anions, particularly, nitrate and phosphate (Drew and Saker, 1984; Drew et al., 1990; Engels and Marschner, 1992) and improving plant growth, we characterized the effects of expressing HtNHX1 and HtNHX2 on enhancing the tolerance of rice to nutrient deficiency. The reduction in nutrient supply to 1/4 of its full strength decreased the growth and biomass of WT and HtNHX1‐expressing rice equally (Figure 7a,b) while did not affect the growth of HtNHX2‐expressing rice plants (Figure 7a,b). The HtNHX2 lines showed about 35%, 25% and 45% increase in total biomass, N and P contents, respectively, in comparison with WT (Figure 7b–d), indicating that HtNHX2 expression could remarkably enhance the root acquisition of N and P, thus improving plant growth under limited nutrient supply conditions. In contrast, N and P contents in the HtNHX1 lines were lower than those in WT. 15N quantification analysis of rice grown in low N and K conditions showed that HtNHX2 expression did not alter root N contents, but significantly increased shoot N contents (Figure S4), suggesting the enhanced N translocation from shoot to root, in addition to the increase in total N uptake.
Figure 7.
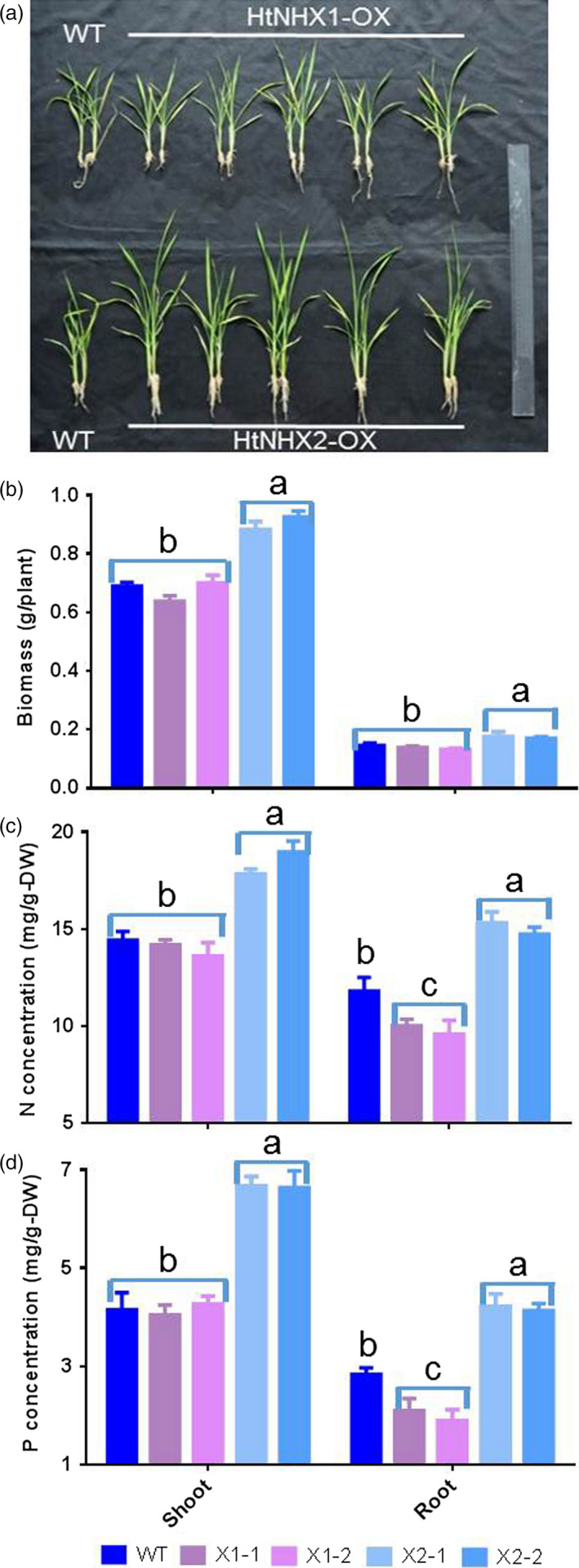
Effects of overexpression of HtNHX1 and HtNHX2 on rice growth and N and P concentrations under supply of ¼ concentrations of all the essential nutrients in IRRI culture solution. (a) The rice was grown in IRRI solution with ¼ concentrations of the full supply; (b) biomass of shoot and root; (c, d) concentration of N (c) and P (d) in shoot and root. WT: cv. Nipponbare; X1‐1, 1‐2: two T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2: two T2 individual transgenic lines of expressing HtNHX2. Different letter on the column indicates the significant difference at probability at 5%.
The notion of the role(s) of HtNHX2, but not HtNHX1, in enhancing K+ uptake and mediating N and P accumulation was further supported by the growth of rice plants in infertile paddy fields (low N and K contents) (see Table S2). Compared to WT, HtNHX1 lines showed 20% lower straw weight (biomass) and 40% lower grain yield, resulting in significant lower grain harvest index due to a larger portion of unfilled grains (Figure 8a–c). The HtNHX1 lines contained significant higher total N, similar P and lower K+ in the straw at mature stage (Figure 8d). Remarkably, the HtNHX2 lines showed 45% increase of total grain yield and 90%, 40% and 13% higher N, P and K+ contents in the straw (Figure 8c,d).
Figure 8.
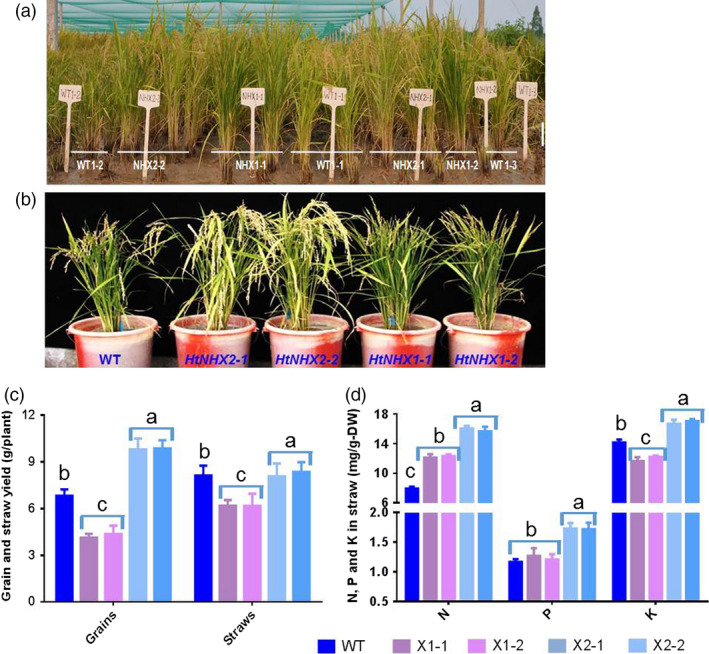
Effects of overexpression of HtNHX1 and HtNHX2 on grain yield and straw biomass, and concentrations of N, P, K+ in rice grown in nutrient deficient soil. (a, b) The rice was grown in Ledong experimental station, Hainan Province. Soil properties were described in Table S2, and no fertilizer was applied during the season. (c) Grain yield and straw biomass. (d) Concentrations of total N, P and K+ in the harvested straws. WT: cv. Nipponbare; X1‐1, 1‐2: two T2 individual transgenic lines of expressing HtNHX1; X2‐1, 2‐2: two T2 individual transgenic lines of expressing HtNHX2. Different letter(s) on the column indicates the significant difference at probability at 5%.
Both HtNHX1 and HtNHX2 could rescue the growth impairment of nhx5 and nhx6 double mutant of Arabidopsis
In Arabidopsis, AtNHX5 and AtNHX6 are located at TGN, Golgi and prevacuolar compartments and the double knockout Atnhx5nhx6 displayed growth impairment under normal growth condition and high sensitivity to salinity (Bassil et al., 2011a). We found that either HtNHX1 or HtNHX2 expression could rescue the growth defects and salt sensitivity of the Atnhx5nhx6 mutant similarly (Figure 9a,b), suggesting a functional similarity between HtNHX1, HtNHX2 and AtNHX5, AtNHX6. Interestingly, only HtNHX1, but not HtNHX2, could enhance the tolerance of nhx1 mutant yeast (AXT3 strains) to hygromycin (Figure S5). In addition, the transformation of the Atnhx5nhx6 double mutant with HtNHX1 or HtNHX2 resulted in increased hygromycin resistance, although HtNHX1‐expressing plants displayed much stronger hygromycin tolerance (Figure S5).
Figure 9.
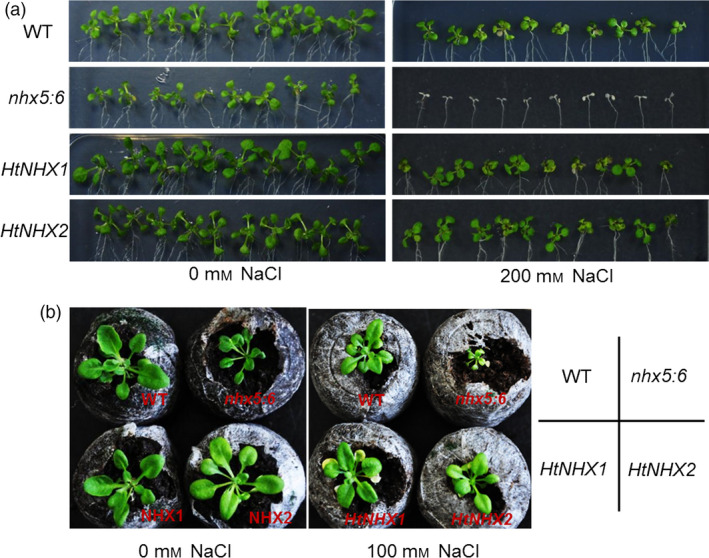
Either HtNHX1 or HtNHX2 rescued the growth defect of Arabidopsis nhx5 and nhx6 double mutant under both normal and salt‐stress conditions. (a) The growth in agar medium with or without additive NaCl; (b) The growth in the substrate with or without additive NaCl in the pots for 6 weeks.
HtNHX1 and HtNHX2 showed different subcellular localization
The differential response displayed by transgenic rice plants expressing HtNHX1 or HtNHX2 would suggest their dissimilar cellular localization. Therefore, we used rice protoplasts expressing HtNHX1‐GFP or HtNHX2‐GFP encoding chimeras and several Arabidopsis markers to detect their localization. HtNHX1‐GFP, but not HtNHX2‐GFP fluorescence colocalized with the RFP signal of the tonoplast marker AtTPK1 (Figure 10a). Either HtNHX1‐GFP or HtNHX2‐GFP fluorescence was largely separated from RFP signal of the endoplasmic reticulum (ER) marker AtHDEL (Figure 10b).
Figure 10.
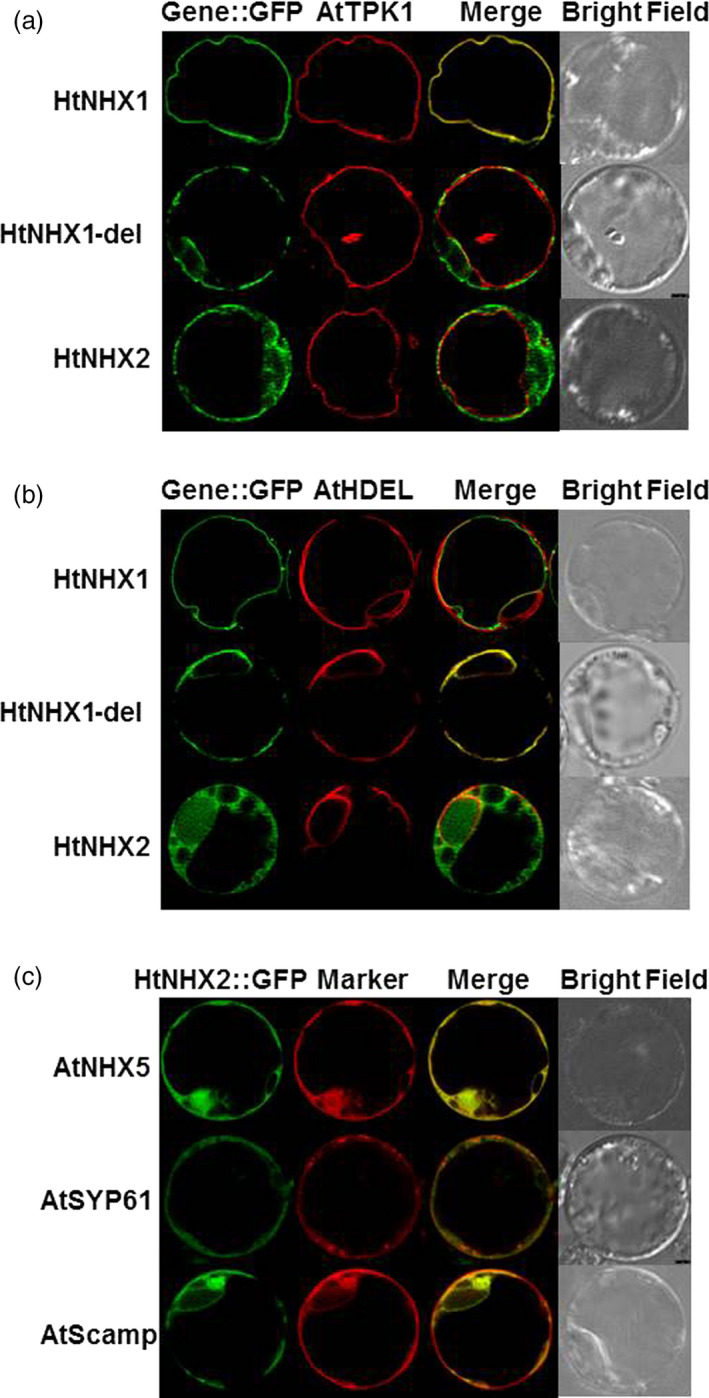
Subcellular localization of HtNHX1 and HtNHX2 in rice protoplast. Expression of HtNHX1, HtNHX2, HtNHX1del fused with GFP, and AtTPK1 (a), AtHDEL (b)?AtNHX5, AtSYP61, AtScamp (c) fused with RFP in rice protoplast cells.. HtNHX1del: expression of mutated HtNHX1 by deletion of conserved eight amino acids in NHX1 group from different plant species (see Figure S1). AtTPK1, AtHDEL, AtNHX5, AtSYP61 and AtScamp are the localization markers on tonoplast, endoplasmic reticulum, and endosome or trans‐Golgi network/Golgi, respectively, in Arabidopsis.
It has been addressed that the type‐I NHX members including AtNHX1‐AtNHX4 are located mainly at the vacuolar membranes while the type‐II NHX members including AtNHX5, AtNHX6, LeNHX2, HvNHX4 and OsNHX5 are at endosome membranes (Bassil et al., 2012). We aligned sequences of HtNHX1 and HtNHX2 with those of known plant NHXs and found a patch of conserved eight amino acids (S/T,P/F,G,X,S/T,Ø,X,V) in type‐I NHXs is missed in the sequences of type‐II NHXs (Figure S1C). Interestingly, this eight amino acids motif is also missed in HtNHX2 (Figure S1C). To gain insight on the effect of these eight amino acids on the cellular localization of type‐I NHX members, we generated the HtNHX1del‐GFP encoding chimera and expressed it in rice protoplasts. Interestingly, the mutated HtNHX1 was separated from AtTPK1 location (Figure 10a), but largely overlapped with the ER marker (AtHDEL) (Figure 10b). The results suggested that the eight amino acids patch was important for targeting HtNHX1 to the vacuolar membrane.
In the rice protoplasts, HtNHX2 localized similar to AtNHX5 and partially overlapped with AtSYP61 and AtScamp (Figure 10c), indicating that HtNHX2 was largely distributed at endosomal membranes.
Discussion
Plant NHX‐type antiporters play important roles in salt tolerance and the maintenance of cellular K+ and Na+ homoeostasis and the regulation of intracellular pH. In this study, we cloned two highly identical salt‐stress up‐regulated NHX orthologues genes, HtNHX1 and HtNHX2 from H. tuberosus (Figures 1 and S1). HtNHX1 and HtNHX2 share identical 5′‐UTR, 3′‐UTR and coding sequences, except for a fragment of consecutive 342 bp in HtNHX1 which is absent in HtNHX2. Whether the two genes were alternatively spliced or evolutionary independently generated is not clear. According to their sequences, both genes are predicted to belong to the type‐I family and to locate at the vacuole (Figure S1B). The ability of HtNHX1 but not HtNHX2 to confer resistance to hygromycin in yeast and Arabidopsis (Figure S5) and different cellular localization in rice (Figure 10) would suggest functional differences between the two isoforms. Notably, the role of HtNHX2 in conferring tolerance to nutrient deficiency, in addition to conferring tolerance to salinity, provides a novel insight on NHX functions (Figures 5, 6, 7, 8, S3 and S4).
The contribution of HtNHX1 and HtNHX2 to salt tolerance in rice depends on the K+‐supply levels
We observed that HtNHX1 or HtNHX2 improved rice tolerance to salt stress (comparison of Figures 2b,e,f and 3b with Figure 2a,c,d; Figure 4a,c), but they did not have significant influence on growth and uptake of K+ and Na+ at normal growth conditions (Figures 2a,c,d, 3a and 4a,c). At salinized conditions, HtNHX1 increased rice K+ and Na+ accumulation, that is keeping the same ratio of K+/Na+ as that in WT (Figure 3d). This indicated that HtNHX1 indistinguishably transported K+ and Na+, which is similar to that of AtNHX1 and AtNHX2 in Arabidopsis (Barragán et al., 2012; Bassil et al., 2011b). In addition, the K+ and Na+ contents of plant expressing HtNHX1 were higher in the roots (about 40%–50%) and the shoots (about 10%–25%) than in WT plants (Figure 3b,c), supporting the role of HtNHX1 as type‐I NHXs (Figure 10a) in sequestering Na+(K+) into the vacuoles (Barragán et al., 2012; Bassil et al., 2011b). Interestingly, the accumulation and distribution of K+ and Na+ in plants expressing HtNHX2 differed from that seen in HtNHX1 plants, as HtNHX2 plants accumulated relatively more Na+ and K+ (Figure 3c). These results were in agreement with the localization of HtNHX2 to intracellular compartments other than the vacuole (Figure 10a).
Endosomal trafficking and the vesicle fusion to the vacuole are important components of the response of plant cells to salinity. Transporters playing roles in cation sequestration, such as NHX1 (Apse et al., 1999) and H+‐pumps (Gaxiola et al., 2001), depend on vesicular trafficking for their delivery to the tonoplast. Thus, the expression of endosomal NHXs may influence protein trafficking from the Golgi/TGN to the vacuoles (Bassil et al., 2011), necessary for the response to high salinity. The up‐regulation of endosomal NHXs, such as AtNHX5 in response to salt stress, but not to osmotic shock, supported the role of endosomal AtNHXs (Bassil et al., 2011a; Yokoi et al., 2002). In addition, salinity induced bulk endocytosis, promoted the rapid increase in vacuolar volume and the accumulation of sodium into the vacuole (Baral et al., 2015; Hamaji et al., 2009; Leshem et al., 2006; Mimura et al., 2003). These results suggest that the overexpression of endosomal NHXs, such as HtNHX2, contributed to salt tolerance through several mechanisms: increasing vesicle fusion to the vacuole, contributing to the accumulation of Na+ and by increasing the trafficking of transporters that could explain the improved nutrient uptake in the transgenic plants.
HtNHX1 and HtNHX2 rescued the salt sensitivity of the Arabidopsis Atnhx5 nhx6 double mutant at the same extent (Figure 9a,b), but HtNHX1 was more effective than HtNHX2 in conferring tolerance of Arabidopsis to external hygromycin (Figure S5). It has been shown that AtNHX5 and AtNHX6 located at endosomes, TGN and prevacuolar compartments and nhx5 nhx6 double knockouts displayed abnormal vesicular trafficking and sensitivity to salinity (Bassil et al., 2011a). In rice, HtNHX2 showed similar cellular localization as AtNHX5 (Figure 10c); thus, it is plausible that HtNHX2 may play a similar role as AtNHX5 in the regulation of endosomal ion and pH homoeostasis. Nevertheless, these results should be taken with some caution as both HtNHX1 and HtNHX2 were expressed under the control of a constitutive promoter (ubiquitin) and abnormal localization due to unregulated expression cannot be ruled out.
The expression of HtNHX2, but not HtNHX1, improved rice growth and grain yield
Potassium is a plant essential nutrient, and vacuolar K+ plays roles in the regulation of cellular volume and tissue expansion. Although plants grown under different K+ supply can change their vacuolar K+ contents, cytosolic K+ is maintained at a relative constant level of around 60–100 mm (Cuin et al., 2003; Walker et al., 1996). The activity of NHXs plays a significant role in the transport of K+ into the vacuole (Bassil et al., 2012). The overexpression of AtNHX1 in tomato led to increased vacuolar K+ accumulation (Zhang and Blumwald, 2001) and higher tissue K+ contents. In the knockout Atnhx1, Apse et al. (2003) reported a reduction in K+/H+ and Na+/H+ exchange and reduced cell expansion. Bassil et al. (2011b) observed lower vacuole pH and K+ concentration in the nhx1 nhx2 mutant, further supporting the role of AtNHX1 and AtNHX2 in driving the uptake of K+ into the vacuole.
The expression of HtNHX2 improved plant growth and grain yield at low K+ concentrations (Figures 5, S3), and the tissue K+ contents of the transgenic plants were higher than the WT and HtNHX1‐expressing plants. Notably, HtNHX2 localization appeared to be similar to that of AtNHX5 (Figure 10), that is intracellular vesicles, TGN. (Bassil et al., 2011a), thus associated with vesicular trafficking and storage protein sorting (Reguera et al., 2014). In yeast, nhx1 mutants have been shown to have impaired vacuolar biogenesis and protein sorting (Qiu and Fratti, 2010). In tomato, LeNHX2 colocalized with prevacuolar and Golgi markers and appeared to be more selective to K+ than Na+ (Venema et al., 2003). Interestingly, HtNHX2 lacks a continuous 114 amino acids stretch at predicted six to eight transmembrane domains of HtNHX1 (Figure S1D). This stretch also comprises a segment of eight amino acids that is absent in type‐II NHXs (Figure S1C). Although the occurrence of a shorter HtNHX2 isoform retaining its transport function has not been reported before, shorter AtNHX1 isoforms have been reported (Xu et al., 2010). Using DNA shuffling mutagenesis, Xu et al. (2010) generated a 296 amino acids deleted AtNHX1, AtNHXS1, in which, several transmembrane domains and the C‐terminus hydrophilic tail containing the CaM‐binding domain (Yamaguchi et al., 2003, 2005) were deleted. Interestingly, such large fragment deletion did not alter its vacuolar localization but increased the Na+/K+ selectivity and Na+/H+ exchange activity, enhancing the NaCl tolerance of yeast expressing AtNHXS1 (Xu et al., 2010). Notably, plants expressing the endosomal HtNHX2 accumulated more K+ and Na+ than the transgenic rice expressing the vacuolar HtNHX1. Although it is possible to speculate that the deletion in HtNHX2 resulted in increased transport activity of the antiporter, a detailed structure/function analysis is needed to assess this point.
The increased tolerance to nutrient deficiency displayed by transgenic rice plants expressing HtNHX2 was associated with increased K+ contents and enhanced N assimilation as seen by the increase in 15N translocation from root to shoot (Figure S4) and the amounts of N, P and K+ in the straw of plants grown in infertile soil (Figure 8a,b). K+ is the major accompanying ion for the translocation of NO3 − and sucrose in plants (Drew and Saker, 1984; Drew et al., 1990). About 40%–90% of root acquired K+ could be retranslocated from the shoot via the phloem and recycled through the roots (Lu et al., 2005; Peuke, 2010), and K+ recycling in plants can act as an important signal for feedback control of nutrient uptake (Drew et al., 1990; Engels and Marschner, 1992). It could be speculated that the expression of HtNHX2 improved the synthesis/delivery of the transporters of N, P and K+ to plasma membranes by improving vesicular trafficking and/or protein targeting (Reguera et al., 2014).
The functions of NHX‐type transporters have been extensively studied (Bassil et al., 2012 and references therein), and the effects of the overexpression of NHXs on salt tolerance have been shown in different crop species (Bassil and Blumwald, 2014). Here, we show that although the expression of HtNHX1 conferred salt tolerance, only the expression of HtNHX2, a shorter isoform ot the known type‐II NHXs, conferred salt tolerance under nutrient stress conditions.
Experimental procedures
Plant materials and growth conditions
The tubers of H. tuberosus genotype NY‐1 after dormancy were germinated on moist sand in an incubator. Uniformly germinated slices of the buds were selected, sown in sands and then transplanted into 1/2 Hoagland nutrient solution in the greenhouse as described previously (Li et al., 2014). After emergence of the fourth leaf, the seedlings were treated with 50, 100, 200 and 300 mm NaCl for 24 h or with 100 mM NaCl for 12 h before the sampling for analysis of HtNHX1 and HtNHX2 expression. Each treatment was replicated three times.
Arabidopsis thaliana (ecotype Columbia (Col‐0) wild‐type (WT) and nhx5 nhx6 double knockout mutants (Bassil et al., 2011b) were provided by Professor Eduardo Blumwald at University of California (Davis, California). For the salt and hygromycin tolerance analysis in agar medium, the seeds were germinated and grown by following the method described by Bassil et al. (2011b). For the salt tolerance analysis in the pot filled with peat culture medium, the seedlings of uniform size after 2 weeks growth in a plate were selected and transplanted to the nutrient‐enriched medium in a growth chamber at 23 °C with a 14 : 10 h L : D photoperiod. For salt treatment, NaCl was added to final concentrations as indicated in the figure legends.
The rice hypotonic culture and soil pot culture were basically followed the protocols previously described by Li et al. (2014) and Yang et al. (2014). Briefly, for hypotonic culture, 2‐week‐old seedlings of similar size were transferred to IRRI nutrient solution with specific treatment (indicated in figure legends) for additional 3 weeks. The nutrient concentrations (mm) of IRRI solution are as follows: N 2.5, P 0.3, K 1, Ca 1, Mg 1, Si 0.5, Fe 0.02, B 0.02, Mn 0.009, Zn 0.00077, Cu 0.00032, Mo 0.00039. For salt‐stressed pot culture, 2‐week‐old seedlings were acclimated to the soil containing 0.1% (1‐g/kg soil) NaCl for 1 week before being transplanted to the pot filled with 40 kg of soil which had been fertilized and mixture with indicated extra amount of salts (0, 1.3 and 1.8‐g/kg soil, respectively) for 2 weeks. For either K+ deficient or N + P deficient soil culture, 4‐week‐old seedlings were transplanted to the respective isolated paddy field and no extra fertilizer was added during the growth season. The soil properties used for the plant tolerance to salt stress, K+ deficiency or N+P deficiency were described in Table S2.
Cloning of HtNHX1 and HtNHX2 sequences
Both cDNAs of HtNHX1 and HtNHX2 were reversely transcribed from the RNA extracted from the leaves of NY‐1. Degenerate primers (P1, P2) designed for cloning the Na+/H+ antiporters (Table S1) were based on alignment of the sequences of yeast, microbes, moss and plant Na+/H+ antiporters. The amplified fragments were ligated into the pEasy‐Blunt vector (Transgene, China). Based on the sequences of this RT‐PCR fragment, rapid‐amplification of cDNA ends (RACE) PCR was carried out using SMARTer RACE cDNA amplification kit (Clontech, China) to obtain the full‐length cDNA sequences according to the instruction. First strand of 5′‐ and 3′‐RACE‐ready cDNA was synthesized according to the kit manual with total RNA. The 3′ and 5′ RACE‐PCRs were carried out with each pair of gene‐specific and universal primers using the first strand cDNA as the template. The 3′ nested PCR was performed with the nested gene‐specific primer and nested universal primer A (provided with the kit) using diluted 3′ RACE‐PCR product as template because the level of background in the 3′‐RACES reaction was too high. The primers used in RACE‐PCR were designed based on the partial DNA sequence: P3: (for 5′ RACE‐PCR), P4: (for 5′ nested RACE‐PCR), P5: (for 3′ RACE‐PCR), and P6: (for 3′ nested RACE‐PCR). The PCR product was purified and cloned into pEasy‐Blunt vector and sequenced. By aligning the sequences of the 5′‐end 3′‐end PCR products and the previous partial DNA sequence, the full‐length cDNA sequence of HtNHX1 and HtNHX2 was deduced and obtained by RT‐PCR with KOD‐PLUS (TOYOBO, Japan) using the specific primers: P7 and P8 (Table S1).
HtNHX1 and HtNHX2 transcripts analysis
Total RNA was extracted from root or shoot of H. tuberosus genotype NY‐1 subjected to different salt stresses. Real‐time qRT‐PCR analysis of HtNHX1 was performed as described by Li et al. (2014). Semi‐quantitative RT‐PCR analysis for HtNHX2 was performed according to the protocol described previously (Tsuchiya et al., 2004; Yan et al., 2011). The target genes and actin standards in 1 : 10, 1 : 100 and 1 : 1000 dilutions were always present in the experiments. All the primers are listed in Table S1. All the PCR products were checked by electrophoresis and sequenced to confirm their identity.
Generating the transgenic rice expressing either HtNHX1 or HtNHX2
The open reading frames of HtNXH1 and HtNXH2 were amplified by gene‐specific primers (see Table S1). The fragment was treated with restriction enzymes and inserted into pTCK303 expression vectors under the control of the Ubi‐promoter. The final constructed vector was sequenced before transformation. The gene transformation into rice (Oryza sativa L. cv. Nipponbare) was performed by Agrobacterium‐mediated cocultivation method (Upadhyaya et al., 2010; Li et al., 2014). Total 13 lines of HtNHX1 and 17 lines of HtNHX2 expressed homozygotes were initially obtained from T1 generation. By Southern blot analysis, each five lines of HtNHX1 or HtNHX2 expressing rice plants with only one copy of the gene insertion (Figure S2A) were selected and propagated for getting the seeds of T2‐T4 generations. The exact integration sites of HtNHX1 or NtNHX2 in the rice genome for HtNHX1.1‐HtNHX1.2 and HtNHX2.1‐HtNHX2.2 lines were sequenced (Figure S2B), and their T3 generation was used for further experiments.
Expression of HtNH1 or HtNHX2 gene in Arabidopsis nhx5 nhx6 double mutant
The HtNHX1‐ or HtNHX2‐coding sequence was amplified and built into the BamHI/SacI sites of the pTCK303 vector driven by the ubiquitin promoter. The resulting constructs were introduced into nhx5‐2 nhx6‐2 for the functional complementation by Agrobacterium tumefaciens strain EHA105 using the floral dipping method (Clough and Bent, 1998).
Southern blot and TAIL‐PCR analysis
The independent transgenic lines with HtNHX1 or HtNHX2 expression lines, NHX1‐1, NHX1‐2 and NHX2‐1, NHX2‐2, were determined by Southern blot analysis following the procedures described previously (Yan et al., 2011). A TAIL‐PCR procedure was performed as described previously (Liu et al., 1995). The PCR products were purified and then cloned into pMD‐19 T vector (TaKaRa Biotechnology Co., Ltd., Dalian, China) for sequencing. Sequence identity was determined by blasting against the NCBI database. All of the primers used for TAIL‐PCR and procedures are listed in Table S1.
HtNHX1 and HtNHX2 subcellular localization assay
Rice protoplasts were isolated according to Yang et al. (2014), C‐terminal or N‐terminal GFP fused HtNHX1 or HtNHX2 was constructed in the vector and transiently expressed in the rice protoplasts and detected by laser scanning microscope (LSM410; Carl Zeiss) as described by Yang et al. (2014). The separation of the membrane fractions of BY2 cells was conducted according to the bio‐protocol of Membrane Preparation, Sucrose Density Gradients and Two‐phase Separation Fractionation from 5‐day‐old Arabidopsis seedlings (http://www.bio-protocol.org/e1014) by Yang et al. (2013).
Determination total Na+, K+, N and P content in rice
All the samples were dried at 105 °C for 30 min and then kept at 70 °C in an oven for 72 h before weighing. After grinding, the samples were extracted by 2 m HCl for 2 days before measuring K+ and Na+ by a flame emission spectrometry. For N and P, samples were digested completely with 98% H2SO4–30% H2O2 and measured as described by Tang et al. (2012) and Ai et al. (2009).
Statistical analysis
Data were analysed by ANOVA using the SPSS 10 program (SPSS). Different letters above columns in the figures indicate statistical differences (Probability at 0.05) between individual transgenic lines and wild‐type and/or between different treatments.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Bioinformatic prediction of HtNHX1 and HtNHX2 structures.
Figure S2 Isolation of single copy inserted rice lines of HtNHX1 or HtNHX2 expression.
Figure S3 Effects of overexpression of HtNHX1 and HtNHX2 on grain yield, harvest index, and NPK concentrations in the rice grown in low K supplied soil.
Figure S4 Effects of HtNHX1 and HtNHX2 expression on ammonium N acquisition and distribution under low N and K supply condition.
Figure S5 HtNHX1, but not HtNHX2, functioned in enhancing tolerance to external hygromycin in both nhx1 mutated yeast cells (A) and Arabidopsis (B).
Table S1 The primers used in the experiments.
Table S2 The properties of the soil used in the experiments.
Acknowledgements
This work was supported by the National R&D Program for Transgenic Crops (Grant no. 2016ZX08009003005‐008), National Natural Science Foundation of China (Grant no. 31272226), The National Key Program for Research and Development (Grant no. 2016YFD0100700) and International Potash Institute (IPI). We thank Prof. Huixia Shou and Dr. Yinhui Ying from Zhejiang University, Prof. Yiqun Bao for providing the localization markers, Dr. Francisco Javier Quintero from Consejo Superior de Investigaciones Cientificas for some critical comments of this work.
Contributor Information
Ling Yu, Email: lyu@njau.edu.cn.
Guohua Xu, Email: ghxu@njau.edu.cn.
References
- Ai, P. , Sun, S.B. , Zhao, J.N. , Fan, X.R. , Xin, W.J. , Guo, Q. , Yu, L. et al. (2009) Two rice phosphate transporters, ORYsa;Pht1;2 and ORYsa;Pht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. [DOI] [PubMed] [Google Scholar]
- Alemán, F. , Nieves‐Cordones, M. , Martínez, V. and Rubio, F. (2011) Root K(+) acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol. 52, 1603–1612. [DOI] [PubMed] [Google Scholar]
- Andrés, Z. , Pérez‐Hormaeche, J. , Leidi, E.O. , Schlücking, K. , Steinhorst, L. , McLachlan, D.H. , Schumacher, K. et al. (2014) Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl Acad. Sci. USA, 111, E1806–E1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse, M.P. , Aharon, G.S. , Snedden, W.A. and Blumwald, E. (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science, 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Apse, M.P. , Sottosanto, J.B. and Blumwald, E. (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T‐DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36, 229–239. [DOI] [PubMed] [Google Scholar]
- Baral, A. , Shruthi, K.S. and Mathew, M.K. (2015) Vesicular trafficking and salinity responses in plants. IUBMB Life, 67, 677–686. [DOI] [PubMed] [Google Scholar]
- Barragán, V. , Leidi, E.O. , Andrés, Z. , Rubio, L. , De Luca, A. , Fernández, J.A. , Cubero, B. et al. (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell, 24, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, E. and Blumwald, E. (2014) The ins and outs of intracellular ion homeostasis: NHX‐type cation/H+ antiporters. Curr. Opin. Plant Biol. 22, 1–6. [DOI] [PubMed] [Google Scholar]
- Bassil, E. , Ohto, M.A. , Esumi, T. , Tajima, H. , Zhu, Z. , Cagnac, O. , Belmonte, M. et al. (2011a) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell, 23, 224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, E. , Tajima, H. , Liang, Y. , Ohto, M. , Ushijima, K. , Nakano, R. , Esumi, T. et al. (2011b) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell, 23, 3482–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, E. , Coku, A. and Blumwald, E. (2012) Cellular homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 3, 5727–5740. [DOI] [PubMed] [Google Scholar]
- Blumwald, E. and Poole, R.J. (1985) Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris . Plant Physiol., 78, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanroj, S. , Wang, G. , Venema, K. , Zhang, M.W. , Delwiche, C.F. and Sze, H. (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 3, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cuin, T.A. , Miller, A.J. , Laurie, S.A. and Leigh, R.A. (2003) Potassium activities in cell compartments of salt‐grown barley leaves. J. Exp. Bot. 54, 657–661. [DOI] [PubMed] [Google Scholar]
- Deinlein, U. , Stephan, A.B. , Horie, T. , Luo, W. , Xu, G.H. and Schroeder, J.I. (2014) Plant salt‐tolerance mechanisms. Trends Plant Sci. 19, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, M.C. and Saker, L.R. (1984) Uptake and long‐distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: evidence of non‐allosteric regulation. Planta, 160, 500–507. [DOI] [PubMed] [Google Scholar]
- Drew, M.C. , Webb, J. and Saker, L.R. (1990) Regulation of K+ uptake and transport to the xylem in barley roots: K+ distribution determined by electron probe X‐ray microanalysis of frozen‐hydrated cells. J. Exp. Bot. 41, 815–825. [Google Scholar]
- Engels, C. and Marschner, H. (1992) Adaptation of potassium translocation into the shoot of maize (Zea mays) to shoot demand: evidence for xylem loading as a regulating step. Physiol. Plant. 86, 263–268. [Google Scholar]
- Fukuda, A. , Nakamura, A. , Hara, N. , Toki, S. and Tanaka, Y. (2011) Molecular and functional analyses of rice NHX‐type Na+/H+ antiporter genes. Planta, 233, 175–188. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A. , Rao, R. , Sherman, A. , Grisafi, P. , Alper, S.L. and Fink, G.R. (1999) The Arabidopsis thaliana transporters, AtNHX1 and Avp1, can function in cation detoxification in yeast. Proc. Natl Acad. Sci. USA, 96, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A. , Li, J. , Undurraga, S. , Dang, L.M. , Allen, G.J. , Alper, S.L. and Fink, G.R. (2001) Drought‐ and salt‐tolerant plants result from overexpression of the AVP1 H1‐pump. Proc. Natl Acad. Sci. USA, 98, 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji, K. , Nagira, M. , Yoshida, K. , Ohnishi, M. , Oda, Y. , Uemura, T. , Goh, T. et al. (2009) Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 50, 2023–2033. [DOI] [PubMed] [Google Scholar]
- Heuer, S. , Gaxiola, R. , Schilling, R. , Herrera‐Estrella, L. , López‐Arredondo, D. , Wissuwa, M. , Delhaize, E. et al. (2016) Improving phosphorus use efficiency—a complex trait with emerging opportunities. Plant J. 90, 868–885. 10.1111/tpj.13423. [DOI] [PubMed] [Google Scholar]
- Horie, T. , Hauser, F. and Schroeder, J.I. (2009) HKT transporter‐mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Leidi, E.O. and Pardo, J.M. (2010) How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav. 5, 792–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, E. , Prade, T. , Angelidaki, I. , Svensson, S.E. , Newson, W.R. , Gunnarsson, I.B. and Hovmalm, H.P. (2015) Economically viable components from Jerusalem artichoke (Helianthus tuberosus L.) in a biorefinery concept. Int. J. Mol. Sci. 16, 8997–9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska, M.M. and Testerink, C. (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594. [DOI] [PubMed] [Google Scholar]
- Leidi, E.O. , Barragan, V. , Rubio, L. , El‐Hamdaoui, A. , Ruiz, M.T. , Cubero, B. , Fernandez, J.A. et al. (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61, 495–506. [DOI] [PubMed] [Google Scholar]
- Leshem, Y. , Melamed‐Book, N. , Cagnac, O. , Ronen, G. , Nishri, Y. , Solomon, M. , Cohen, G. et al. (2006) Suppression of Arabidopsis vesicle‐SNARE expression inhibited fusion of H2O2‐containing vesicles with tonoplast and increased salt tolerance. Proc. Natl Acad. Sci. USA, 103, 18008–18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.Y. , He, X.W. , Xu, L. , Zhou, J. , Wu, P. , Shou, H.X. and Zhang, F.C. (2008) Molecular and functional comparisons of the vacuolar Na+/H+ exchangers originated from glycophytic and halophytic species. J. Zhejiang Univ. Sci. B, 9, 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Tang, Z. , Hu, Y. , Yu, L. , Liu, Z. and Xu, G.H. (2014) Functional analyses of a putative plasma membrane Na+/H+ antiporter gene isolated from salt tolerant Helianthus tuberosus . Mol. Biol. Rep. 41, 5097–5108. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G. , Mitsukawa, N. , Oosumi, T. and Whittier, R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Long, X.H. , Mehta, S. and Liu, Z.P. (2008) Effect of NO3–N enrichment on seawater stress tolerance of Jerusalem artichoke (Helianthus tuberosus). Pedosphere, 18, 113–123. [Google Scholar]
- Lu, Y.X. , Li, C.J. and Zhang, F.S. (2005) Transpiration, potassium uptake and flow in tobacco as affected by nitrogen forms and nutrient levels. Ann. Bot. 95, 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin, T. , Bassil, E. , Zhang, S. and Blumwald, E. (2014) Vacuolar Na+/H+ antiporters are required for cellular K+ homeostasis, microtubule organization and directional root growth. Plants, 3, 409–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, T. , Kura‐Hotta, M. , Tsujimura, T. , Ohnishi, M. , Miura, M. , Okazaki, Y. , Mimura, M. et al. (2003) Rapid increase of vacuolar volume in response to salt stress. Planta, 216, 397–402. [DOI] [PubMed] [Google Scholar]
- Munns, R. and Gilliham, M. (2015) Salinity tolerance of crops—what is the cost? New Phytol. 208, 668–673. [DOI] [PubMed] [Google Scholar]
- Munns, R. and Tester, M. (2008) Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Peuke, A.D. (2010) Correlations in concentrations, xylem and phloem flows, and partitioning of elements and ions in intact plants. A summary and statistical re‐evaluation of modelling experiments in Ricinus communis . J. Exp. Bot. 61, 635–655. [DOI] [PubMed] [Google Scholar]
- Qi, Z. and Spalding, E.P. (2004) Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+‐H+ antiporter during salinity stress. Plant Physiol. 136, 2548–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Q.S. and Fratti, R.A. (2010) The Na+/H+ exchanger Nhx1p regulates the initiation of Saccharomyces cerevisiae vacuole fusion. J. Cell Sci. 123, 3266–3275. [DOI] [PubMed] [Google Scholar]
- Reguera, M. , Bassil, E. and Blumwald, E. (2014) Intracellular NHX‐type cation/H+ antiporters in plants. Mol. Plant. 7, 261–263. [DOI] [PubMed] [Google Scholar]
- Shabala, S. , Bose, J. , Fuglsang, A.T. and Pottosin, I. (2016) On a quest for stress tolerance genes: membrane transporters in sensing and adapting to hostile soils. J. Exp. Bot. 67, 1015–1031. [DOI] [PubMed] [Google Scholar]
- Tang, Z. , Fan, X.R. , Li, Q. , Feng, H.M. , Miller, A.J. , Shen, Q.R. and Xu, G.H. (2012) Knock down of a rice stelar nitrate transporter alters long distance translocation but not root influx. Plant Physiol. 160, 2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T. , Takesawa, T. , Kanzaki, H. and Nakamura, I. (2004) Genomic structure and differential expression of two tandem‐arranged GSTZ genes in rice. Gene, 335, 141–149. [DOI] [PubMed] [Google Scholar]
- Upadhyaya, N.M. , Surin, B. , Ramm, K. , Gaudron, J. , Schunmann, P.H.D. , Taylor, W. , Waterhouse, P.M. , et al. (2000) Agrobacterium‐mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust. J. Plant Physiol. 27, 201–210. [Google Scholar]
- Venema, K. , Belver, A. , Marin‐Manzano, M.C. , Rodríguez‐Rosales, M.P. and Donaire, J.P. (2003) A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 278, 22453–22459. [DOI] [PubMed] [Google Scholar]
- Walker, D.J. , Leigh, R.A. and Miller, A.J. (1996) Potassium homeostasis in vacuolate plant cells. Proc. Natl Acad. Sci. USA, 93, 10510–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Zhang, H. , Blumwald, E. and Xia, T. (2010) A novel plant vacuolar Na+/H+ antiporter gene evolved by DNA shuffling confers improved salt tolerance in yeast. J. Biol. Chem. 285, 22999–23006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G.H. , Fan, X.R. and Miller, A.J. (2012) Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T. , Apse, M.P. , Shi, H. and Blumwald, E. (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl Acad. Sci. USA, 100, 12510–12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T. , Aharon, G.S. , Sottosanto, J.B. and Blumwald, E. (2005) Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2 + ‐ and pH‐dependent manner. Proc. Natl Acad. Sci. USA, 102, 16107–16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, M. , Fan, X.R. , Feng, H.M. , Miller, A.J. , Shen, Q.R. and Xu, G.H. (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 34, 1360–1372. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Richter, G.L. , Wang, X. , Mlodzinska, E. , Carraro, N. , Ma, G. , Jenness, M. , et al. (2013) Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J. 74, 37–47. [DOI] [PubMed] [Google Scholar]
- Yang, T.Y. , Zhang, S. , Hu, Y.B. , Hu, F.C. , Hu, Q.D. , Chen, G. , Cai, J. et al. (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi, S. , Quintero, F.J. , Cubero, B. , Ruiz, M.T. , Bressan, R.A. , Hasegawa, P.M. and Pardo, J.M. (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30, 529–539. [DOI] [PubMed] [Google Scholar]
- Zhang, H.X. and Blumwald, E. (2001) Transgenic salt‐tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19, 765–768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Bioinformatic prediction of HtNHX1 and HtNHX2 structures.
Figure S2 Isolation of single copy inserted rice lines of HtNHX1 or HtNHX2 expression.
Figure S3 Effects of overexpression of HtNHX1 and HtNHX2 on grain yield, harvest index, and NPK concentrations in the rice grown in low K supplied soil.
Figure S4 Effects of HtNHX1 and HtNHX2 expression on ammonium N acquisition and distribution under low N and K supply condition.
Figure S5 HtNHX1, but not HtNHX2, functioned in enhancing tolerance to external hygromycin in both nhx1 mutated yeast cells (A) and Arabidopsis (B).
Table S1 The primers used in the experiments.
Table S2 The properties of the soil used in the experiments.


