Summary
Lateral Organ Boundaries Domain (LBD) proteins are plant‐specific transcription factors playing crucial roles in growth and development. However, the function of LBD proteins in Eucalyptus grandis remains largely unexplored. In this study, LBD genes in E. grandis were identified and characterized using bioinformatics approaches. Gene expression patterns in various tissues and the transcriptional responses of EgLBDs to exogenous hormones were determined by qRT‐PCR. Functions of the selected EgLBDs were studied by ectopically overexpressing in a hybrid poplar (Populus alba × Populus glandulosa). Expression levels of genes in the transgenic plants were investigated by RNA‐seq. Our results showed that there were forty‐six EgLBD members in the E. grandis genome and three EgLBDs displayed xylem‐ (EgLBD29) or phloem‐preferential expression (EgLBD22 and EgLBD37). Confocal microscopy indicated that EgLBD22, EgLBD 29 and EgLBD37 were localized to the nucleus. Furthermore, we found that EgLBD22, EgLBD29 and EgLBD37 were responsive to the treatments of indol‐3‐acetic acid and gibberellic acid. More importantly, we demonstrated EgLBDs exerted different influences on secondary growth. Namely, 35S::EgLBD37 led to significantly increased secondary xylem, 35S::EgLBD29 led to greatly increased phloem fibre production, and 35S::EgLBD22 showed no obvious effects. We revealed that key genes related to gibberellin, ethylene and auxin signalling pathway as well as cell expansion were significantly up‐ or down‐regulated in transgenic plants. Our new findings suggest that LBD genes in E. grandis play important roles in secondary growth. This provides new mechanisms to increase wood or fibre production.
Keywords: Eucalyptus grandis, lateral organ boundaries domain, gene expression, secondary growth, xylem development, fibre production
Introduction
The Lateral Organ Boundaries Domain (LBD) proteins, also known as ASYMMETRIC LEAVES2‐LIKE (ASL) proteins, are a family of plant‐specific transcription factors with a highly conserved Lateral Organ Boundaries (LOB) Domain (Iwakawa et al., 2002). The first LBD gene was identified in Arabidopsis thaliana based on the distinctive gene expression pattern of an enhancer trap insertion (Shuai et al., 2002). The LBD gene family of A. thaliana can be divided into two classes according to the structure of the LOB domain. Class I have a completely conserved CX2CX6CX3C zinc finger‐like motif, GAS (Gly‐Ala‐Ser) block and an LX6LX3LX6L leucine zipper‐like coiled‐coil motif, whereas class II only contain a conserved zinc finger‐like motif (Iwakawa et al., 2002; Shuai et al., 2002). The zinc finger‐like motif is thought to be required for DNA binding, and the leucine zipper‐like motif presumably participates in protein dimerization (Majer and Hochholdinger, 2011; Matsumura et al., 2009).
The LBD gene families of Arabidopsis, rice, poplar, tomato, apple, Medicago, maize, grape, mulberry and barley has been studied with members ranging from 24 to 58 (Cao et al., 2016; Jia et al., 2014; Luo et al., 2016; Shuai et al., 2002; Wang, 2016; Wang et al., 2013a,b; Yang et al., 2006; Zhang et al., 2014; Zhu et al., 2007). The LBD genes are found only in plants, implying that this gene family may regulate plant‐specific growth and development processes (Shuai et al., 2002). Previous studies of the LBD gene family in various plant species have shown that they play an important role in many developmental processes, including leaf, lateral root, inflorescence, embryo and flower development (Borghi et al., 2007; Bortiri et al., 2006; Liu et al., 2005; Xu et al., 2003, 2008). In addition, LBD genes were also involved in plant secondary metabolism, assimilation of nitrogen nutrition as well as hormone‐mediated plant lateral organ development (Albinsky et al., 2010; Bell et al., 2012; Rubin et al., 2009).
Secondary growth results from cell division in the vascular cambium or lateral meristem, causing the stems and roots to thicken, and in woody plants, this process produces secondary xylem inward and secondary phloem outward (Mellerowicz et al., 2001; Spicer and Groover, 2010). This event is the engine of wood production and thus of substantial economic interest. Several reports have demonstrated that LBD genes play a significant role in control of secondary growth. Soyano et al. (2008) revealed that overexpression of AtLBD30 (AtASL19) and AtLBD18 (AtASL20) induced transdifferentiation of cells from nonvascular tissues into tracheary element‐like cells, the basic units that constitute xylem vessels. By analysing the expression patterns of Populus tremula × Populus alba (Pta) LBD gene family, Yordanov et al. (2010) showed that PtaLBD1 and PtaLBD4 were specifically expressed in secondary phloem, while PtaLBD15 and PtaLBD18 were preferentially expressed in secondary xylem. Additionally, they found that overexpression of PtaLBD1 resulted in significantly enhanced secondary phloem production due to up‐regulation of the Populus putative ortholog of ALTERED PHLOEM DEVELOPMENT (APL), a MYB transcription factor‐encoding gene that played a role in specifying phloem identity in Arabidopsis (Bonke et al., 2003). However, no attention has been paid to the question whether different LBD genes play different roles in secondary growth and what are the mechanisms.
Eucalyptus grandis is an important woody plant within the Eucalyptus genus that encompasses some of the fastest growing plantation forest species. Its wood is widely used in lignocellulosic biofuel production, paper, pulp and raw cellulose products (Carroll and Somerville, 2009; Rockwood et al., 2008). A previous study has proved that members of PtaLBD genes play an essential role in control of secondary woody growth (Yordanov et al., 2010). However, the function of the LBD genes in E. grandis remains largely unexplored. Therefore, identification and characterization of LBD genes in E. grandis is vital in understanding their roles in regulating secondary growth and thus is crucial to improving its wood quality.
In this article, we used various in silico approaches to identify and characterize E. grandis LBDs. We identified 46 EgLBD genes in E. grandis and performed phylogenetic analysis of all EgLBD proteins in this species. We then analysed of their gene structures, conserved domains and subcellular localizations. Moreover, we used qRT‐PCR to study the gene expression patterns in various tissues and the transcriptional responses of EgLBD genes to hormone treatments. Finally, we characterized the functions of selected LBD genes by ectopic overexpression in a hybrid poplar (Populus alba × Populus glandulosa). We demonstrate that three LBD members (EgLBD22, EgLBD29 and EgLBD37) had different effects on secondary growth and several key genes related to gibberellin, ethylene, auxin signalling pathway, as well as cell expansion, were significantly up‐regulated or down‐regulated in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. Our results demonstrate that manipulating particular EgLBD genes can result in large increases in secondary growth, wood formation or fibre production. These findings provide new insights into the mechanisms by which LBD genes control secondary growth in trees.
Results
Identification, sequence features and phylogeny of the EgLBD gene family
To identify LBD genes in E. grandis, we performed a genomewide prediction of EgLBD genes by BLAST analysis of 43 AtLBDs, 57 PtLBDs and 35 OsLBDs against the E. grandis genome using the tBLASTn algorithm, SMART and Pfam tools. A total of 46 LBD genes from E. grandis were identified (Table S2) and numbered according to genome location. The open reading frames (ORFs) of EgLBD genes ranged from 213 bp (EgLBD15) to 936 bp (EgLBD21 and EgLBD38), the predicted molecular weight varied from 7.6 kDa (EgLBD15) to 33.3 kDa (EgLBD30) and the theoretical pI varied from 5.03 (EgLBD26) to 10.72 (EgLBD40) (Table S3).
Analysis of the protein domains within the 46 EgLBDs revealed that 38 EgLBDs belonged to class I, and 8 EgLBDs to class II (Figure S1). Class I contain a completely conserved CX2CX6CX3C zinc finger‐like motif, GAS (Gly‐Ala‐Ser) block and an LX6LX3LX6L leucine zipper‐like coiled‐coil motif, but class II only have a conserved zinc finger‐like motif. The GAS block is 46 amino acids in length, beginning with a FX2VH motif and ending with a DP(V/I)YG motif. The Pro residue in the DP(V/I)YG signature is present in all class I proteins in Arabidopsis, but this is not the case in E. grandis, as both EgLBD15 and EgLBD11 do not contain the Pro residue.
To investigate the evolutionary relationships of the E. grandis LBD proteins, sequences of the 46 EgLBDs, all known Arabidopsis LBD proteins and Populus trichocarpa LBD proteins including four Populus tremula × Populus alba LBD (PtaLBD) proteins implicated in secondary growth were used to construct a neighbour‐joining (NJ) phylogenetic tree. According to evolutionary relationships, 46 EgLBDs were classified into eight subgroups (subgroups a to f within class I and subgroups a and b within class II) (Figure 1). Among them, 14 EgLBDs belonged to class I a, 10 EgLBDs fell into class I b. Three (PtaLBD1, PtaLBD4, PtaLBD15) out of four LBD proteins (PtaLBD1, PtaLBD4, PtaLBD15 and PtaLBD18) associated with secondary growth that have been identified in Populus tremula × Populus alba (Yordanov and Busov, 2011) fell into these two subgroups, while PtaLBD18 belonged to a different subgroup (class I d). Interestingly, EgLBD22 and PtaLBD4, EgLBD37 and PtaLBD1, EgLBD29 and PtaLBD15, EgLBD13 and PtaLBD18 were clustered into the same clade (Figure 1). Therefore, we picked EgLBD22, EgLBD29 and EgLBD37 as close homologs of the poplar proteins for further functional analysis.
Figure 1.

The phylogenetic relationship of LBD proteins in Eucalyptus grandis, Arabidopsis thaliana, Populus trichocarpa and Populus tremula × Populus alba. Triangle (▲), circle (●), rhombus (♦), square(■) represent E. grandis, A. thaliana, P. trichocarpa and P. tremula × P. alba, respectively. The blue branches represent class I, and the red branches represent class II. Each LBD subclass is indicated by a green arc (class I a‐f and class II a‐b);, the homologs of PtaLBD1, PtaLBD4, PtaLBD15, PtaLBD18 are in yellow shadow, PtaLBD1, PtaLBD4, PtaLBD15, PtaLBD18, EgLBD13, EgLBD22, EgLBD29 and EgLBD37 are shown in sky blue font. The numbers on the branches mean the reliability per cent of bootstraps value based on 1000 replication, the scale bar represents 0.1 substitutions per amino acid.
Structural features of EgLBD proteins were investigated as a function of their phylogeny. By analysing the structure of EgLBD genes, we revealed that closely related members had a similar exon/intron structure and gene length. The number of exons ranged from one to three, with 31 genes having two exons, 12 genes having one exon and only three genes (EgLBD8, EgLBD29 and EgLBD42) having three exons (Figure 2a, b). To further understand the EgLBD functional regions, conserved motifs were predicted by MEME. Twenty individual motifs were identified (Figure 2c). Motif sequences were provided in Table S4. Our results showed that the length of motifs ranged from 30 to 70 amino acids and the number of motifs varied between 1 and 6 in each EgLBD protein. Conserved motif 1, motif 2 and motif 5 were most closely related to DUF260 (Domain of Unknown Function 260) (Table S4) based on CDART analysis. Notably, all EgLBDs contain motif 1 except EgLBD15, motif 2 was widespread in class I, motif 4 and 5 were widespread in class II, while other motifs were specific for a particular subgroup. These data suggest that the motifs in EgLBD proteins play critical roles in specific functions or have similar functionality.
Figure 2.

The phylogenetic relationship, gene structure and motif composition analysis of the LBD gene family in Eucalyptus grandis. (a) The amino acid sequences of the EgLBD proteins were aligned with Clustal X, and the phylogenetic tree was constructed using the neighbour‐joining method in MEGA5; the scale bar represents 0.1 substitutions per amino acid, and the branches of different classes were painted with different colours (the blue branches represents class I, the red branches represents class II). (b) Exon/intron structures of the EgLBD genes, the exons, introns and UTR are represented by the green boxes, black lines and blue boxes, respectively, and the scale bar represents 1 kb. (c) Conserved motif of EgLBD proteins, different motifs are represented by different coloured boxes with number 1–20, and the scale bar represents 50 aa (amino acid).
The 46 EgLBD genes were unevenly distributed on the eleven chromosomes (Figure S2). Chromosome 10 contains the largest number with eight EgLBD genes, followed by chromosome 5 and chromosome 7 (seven genes per chromosome). Chromosome 1 and chromosome 4 contained the least, with only one EgLBD gene on them. Furthermore, we observed that distribution of each type of LBD gene was uneven, as three paralogous gene pairs (EgLBD8/EgLBD9, EgLBD16/EgLBD19 and EgLBD30/EgLBD31) were located in the same chromosome, but the other ten paralogous gene pairs (EgLBD1/EgLBD42, EgLBD2/EgLBD41, EgLBD3/EgLBD26, EgLBD7/EgLBD29, EgLBD12/EgLBD13, EgL BD23/EgLBD32, EgLBD24/EgLBD21, EgLBD25/EgLBD37, EgLBD28/EgLBD36, EgLBD33/EgLBD39) were located on different chromosomes. In EgLBD family, we found that EgLBD15, EgLBD16 and EgLBD17 were tandem duplicated locus on chromosome 5. The percentage of tandemly duplicated genes within this gene family was very low and only reached 6.5% (3/46), compared to the average of 34% across the whole genome annotation (Myburg et al., 2014).
Tissue‐specific expression of EgLBD genes and response to GA and IAA treatment
The expression profile of a LBD gene can reflect its biological function. To find candidate EgLBD genes involved in secondary cell wall development, we employed qRT‐PCR to detect the expression patterns of EgLBD genes in E. grandis root, stem, leaf, xylem and phloem. As shown in Figure 3, EgLBD22 was preferentially expressed in the phloem and EgLBD29 gene expression levels were highest in the xylem. Like EgLBD22, the expression of EgLBD37 was also predominately expressed in phloem.
Figure 3.

Expression patterns of the 46 LBD genes in root, stem, leaf, xylem and phloem of Eucalyptus grandis by qRT‐PCR analysis. Bars indicate standard deviations (SD) (n = 3).
To assess the transcriptional responses of EgLBD genes to hormone treatments, the roots of 2‐month‐old E. grandis seedlings were treated with 150 μm of IAA and 150 μm of GA3, respectively. The expression levels of three EgLBD genes (EgLBD22, EgLBD29 and EgLBD37) were analysed by qRT‐PCR with stem‐derived RNA after the treatment of IAA and GA3 for 0 h, 1 h, 3 h, 6 h and 12 h (Figure 4). The results showed that EgLBD29 (2.5‐fold) and EgLBD37 (fourfold) were highly up‐regulated by IAA treatment with EgLBD22 displaying only a minor up‐regulation after only 1 h (1.4‐fold). All IAA gene expression responses peaked within 1‐3 h and then declined, with the expression levels of EgLBD22 declining to 0.4‐fold after 12 h. The GA3 response of EgLBD37 showed a 4.7‐fold increase after 3 h and then declined back to normal after 12 h. However, EgLBD22 and EgLBD29 expression dropped to 0.4 and 0.2, respectively, at 6 h and 1 h under GA3 treatment. EgLBD29 gene expression remained low throughout the 12‐h period, while the expression of EgLBD22 increased back to 0.8‐fold after 12 h.
Figure 4.

Expression analysis of EgLBD22, EgLBD29 and EgLBD37 genes under IAA and GA3 treatment by qRT‐PCR. Bars represent standard deviation for three replicates.
Subcellular localization and possible functional partners of EgLBD proteins
We predicted the possible localization of EgLBD proteins using the protein subcellular localization prediction tool Plant‐mPLoc ( http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). The results showed that all EgLBD proteins were predicted to be localized to the nucleus (Table S3). To validate the predicted localizations, EgLBD22, EgLBD29 and EgLBD37 were transiently expressed in tobacco. The three EgLBD proteins were successfully expressed as fluorescent protein fusions. As shown in Figure S3, the fluorescent signals from fusion proteins LBD22‐GFP, LBD29‐GFP and LBD37‐GFP were all only observed in the nuclei of transformed tobacco leaf cells, while the fluorescence of protein encoded 35S::GFP could be observed everywhere in the cell including the membrane, cytoplasm and nucleus. Therefore, the prediction for subcellular location of EgLBD22, EgLBD29 and EgLBD37 proteins was confirmed by our transient expression experiment. To find out the possible functional protein association networks, protein–protein interactions were predicted using STRING ( http://string-db.org/). The results of protein–protein interaction prediction for functional protein association networks revealed that EgLBD22, EgLBD29 and EgLBD37 had 4, 10, 2 possible functional partners, respectively, all of them consisting of different proteins. The partner with the highest score for EgLBD22, EgLBD29 and EgLBD37 was EXPA14 (EXPANSIN A14), F3F9.16 (general regulatory factor 2) and DOF (DSB formation protein), respectively (Table S5‐S7).
Effects of key EgLBDs on secondary growth when overexpressed
To characterize the functions of key EgLBDs, multiple transgenic lines of hybrid poplar (Populus alba × Populus glandulosa) clone 84k carrying the constructs 35S::LBD22, 35S::LBD29 and 35S::LBD37 were generated, validated for the presence and expression of the transgenes (Figures S4 and S5). The most striking phenotype of EgLBD37‐oe plants is that all the transgenic lines were much taller (up to 59%) than wild‐type 84 k trees (Figure 5) and showed a great increase in internode length (up to 30%) (Figure 6a, b), diameter of stem (up to 44%) (Figure 6c, d) and leaf size (up to 61%) (Figure 6e, f). In contrast, the most obvious phenotype of EgLBD29‐oe plants was that the height of all the transgenic lines was much smaller (less than 22%) than wild‐type 84k trees (Figure 5) and showed a dramatic reduction in internode length (less than 46%) (Figure 6a, b) and leaf size (less than 72%) (Figure 6e, f), while 35S::EgLBD22 did not exhibit any obvious phenotypes (Figure 5, Figure 6).
Figure 5.

Plant height of EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe and wild‐type hybrid poplar (Populus glandulossa × Populus alba) trees. (a) Average height data for ten‐week‐old wild‐type 84k (Populus glandulossa × Populus alba), EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. Bars indicate standard errors (SE), and asterisks indicate significant differences relative to control plants (WT‐84k) with * denoting P < 0.01. Values are reported as means ± SE (n = 5). (b) Ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.
Figure 6.

Internode length, stem diameter and leaf size of EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe and wild‐type hybrid poplar (Populus glandulossa × Populus alba) trees. (a) Total internode length from 3rd to 8th nodes for ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. (b) Representative images of the internode from 3rd to 8th nodes of ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. (c) Average stem diameter data for ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. (d) Representative stem cross section at the base of ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. (e) Average area of the 6th leaf from the top of ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. (f) The 6th leave from the top of ten‐week‐old wild‐type 84k, EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants. Bars indicate standard error (SE), and asterisks indicate significant differences relative to control plants (WT‐84k) with * denoting P < 0.01. Values are reported as means ± SE (n = 5).
To evaluate the impact of EgLBD22, EgLBD29 and EgLBD37 on secondary growth, we sectioned the stems of the 10th node that undergo secondary growth from the control plant WT‐84k and EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe transgenics. An obvious difference in stem anatomy could be observed under light microscopy between WT‐84k, EgLBD37‐oe and EgLBD29‐oe transgenic plants (Figure 7a–c). In EgLBD37‐oe plants, the overall width of cortex region and the lignified component of the secondary xylem were significantly increased (Figure 7b, f). There was also an increase in the thickness of the cortex region and the lignified component of the secondary xylem in EgLBD29‐oe (Figure 7c, g). The most profound change in stem anatomy for EgLBD29‐oe transgenics was that all the transgenic lines produced more phloem fibres (Figure 7c, g) when compared with WT‐84k (Figure 7a, e), EgLBD22‐oe (Figure 7d, h) and EgLBD37‐oe transgenics (Figure 7b, f). For EgLBD22‐oe transgenics, no significant phenotypic changes could be detected when it was compared with that of the control WT‐84k (Figures 5, 6, 7d, h).
Figure 7.

Stem anatomy of EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe and wild‐type hybrid poplar (Populus glandulossa × Populus alba) trees. Fixed stem sections were stained with safranin and fast green (Gefan biotechnology Co. Ltd., Shanghai). (a, e) Stem structure of ten‐week‐old wild‐type 84k plant. (b, f) Stem structure of ten‐week‐old EgLBD37‐oe plant. (c, g) Stem structure of ten‐week‐old EgLBD29‐oe plant. (d, h) Stem structure of ten‐week‐old EgLBD22‐oe plant. In EgLBD37‐oe plant, the overall width of cortex region and the lignified component was significantly increased (b, f). Increased phloem fibre production in the EgLBD29‐oe plant was observed (arrow in g) when compared with wild‐type 84k (arrow in e), EgLBD37‐oe (arrow in f) and EgLBD22‐oe plant (arrow in h).
Transcriptome changes in EgLBD22‐oe, EgLBD29‐oe, EgLBD37‐oe plants
To gain further insight into the biological functions of EgLBD22, EgLBD29 and EgLBD37, we carried out RNA‐seq analysis of LBD22‐oe, EgLBD29‐oe and EgLBD37‐oe hybrid poplar (Populus alba × Populus glandulosa) plants. Compared with WT‐84k plant, there were 299, 421 and 118 unique sequences showing at least onefold changes in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants, respectively, (Tables S8–S10). In EgLBD22‐oe plant, the expansin gene c60809_g2 decreased 1.3‐fold and an auxin efflux carrier component 6 gene decreased 1.5‐fold, but a class‐I KNOX gene (c70392_g3) was shown to be up‐regulated to 1.3‐fold (Table S8). In EgLBD29‐oe plant, the expression of ethylene‐responsive transcription factor ERF023‐like gene increased 3.8‐fold and an ethylene‐responsive element‐binding family protein‐encoding gene increased 1.8‐fold, while the expansin gene c60809_g2 and expansin gene c60754_g1 was reduced 1.1‐ and 1.2‐fold, respectively (Table S9). Consistent with our expectation, a MYB family transcription factor APL‐like gene (c62428_g1) was observed to be up‐regulated to 1.8‐fold in EgLBD29‐oe plant (Table S9). However, there were no alterations in the expression level of APL‐like gene in EgLBD22‐oe plant and EgLBD37‐oe plant when compared with that of the WT‐84k plant (Tables S8 and S10). Notably, Populus trichocarpa gibberellin‐regulated protein 5 was up‐regulated to 4.6‐fold and expansin‐B3 was up‐regulated to 1.5‐fold in EgLBD37‐oe plants (Table S10); however, no changes in the expression of class‐I KNOX genes were detected in EgLBD29‐oe plant and EgLBD37‐oe plant as compared to that of WT‐84k plant (Tables S9 and S10).
To verify the gene expression results obtained from RNA‐seq experiments, we chose eight representative genes (gibberellin‐regulated protein 5, expansin‐B3, ethylene‐responsive transcription factor ERF023‐like gene, ethylene‐responsive element‐binding family protein‐encoding gene c59103_g1, expansin gene c60809_g2, expansin gene c60754_g1, auxin efflux carrier component 6 gene and APL‐like gene c62428_g1) (Table S11) and monitored their expression levels using qRT‐PCR method. The results showed that these genes were significantly up‐regulated or down‐regulated in the EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants as compared to the control WT‐84k (Figure 8), which is in good agreement with the results obtained by our RNA‐seq analysis of EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.
Figure 8.
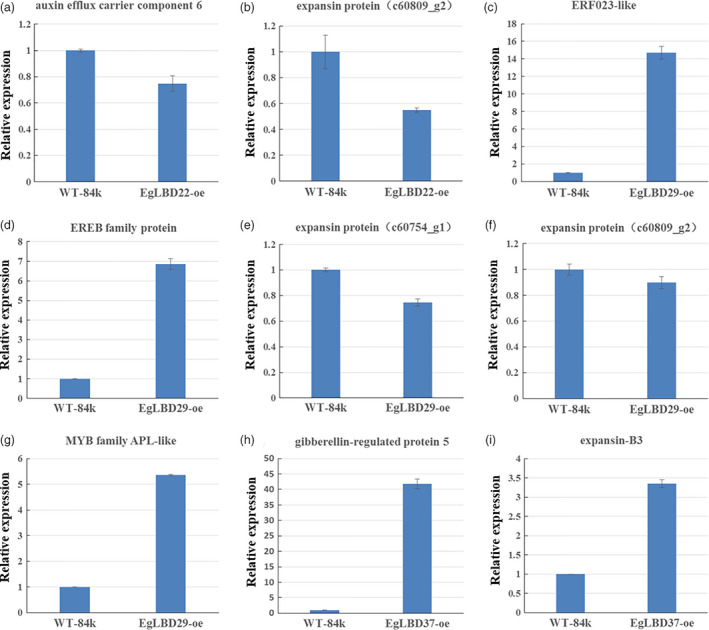
Expression analysis of selected genes in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants by qRT‐PCR. (a) Comparison of the expression level of auxin efflux carrier component 6 encoding gene between WT‐84k and EgLBD22‐oe plants. (b) Comparison of the expression level of expansin protein‐encoding gene (c60809_g2) between WT‐84k and EgLBD22‐oe plants. (c) Comparison of the expression level of ethylene‐responsive transcription factor ERF023‐like protein‐encoding gene between WT‐84k and EgLBD29‐oe plants. (d) Comparison of the expression level of ethylene‐responsive element‐binding (EREB) family protein‐encoding gene between WT‐84k and EgLBD29‐oe plants. (e) Comparison of the expression level of expansin protein‐encoding gene (c60754_g1) between WT‐84k and EgLBD29‐oe plants. (f) Comparison of the expression level of expansin protein‐encoding gene (c60809_g2) between WT‐84k and EgLBD29‐oe plants. (g) Comparison of the expression level of APL ‐like gene (c62428_g1) between WT‐84k and EgLBD29‐oe plants. (h) Comparison of the expression level of gibberellin‐regulated protein 5 encoding gene between WT‐84k and EgLBD37‐oe plants. (i) Comparison of the expression level of expansin‐B3 protein‐encoding gene between WT‐84k and EgLBD37‐oe plants. Bars represent standard deviation for three replicates. All expression estimates were normalized to the expression of an actin loading control gene.
Discussion
LBD genes exist ubiquitously in plants and are important regulators of plant‐specific processes (Majer and Hochholdinger, 2011; Xu et al., 2016). In this study, forty‐six EgLBDs were identified from the E. grandis genome sequences. The LBD gene family in E. grandis is similar to the estimates for other reported plant species, such as the forty‐three LBD genes in Arabidopsis and thirty‐five in rice (Shuai et al., 2002; Yang et al., 2006). Given that tandem duplications were reported to be 34% on average in the E. grandis genome (Myburg et al., 2014) and tandem duplications in the EgLBD family was only 6.5% (3/46) in this investigation, we conclude that the evolution of this family in E. grandis was not significantly affected by tandem duplications.
Structural analysis is a powerful method to mine valuable information concerning duplication events and phylogenetic relationships of genes within a gene family. In this study, we observed that LBD genes in E. grandis had simple gene structures and that most EgLBD members within the same subgroup had a similar exon/intron structure and gene length. This is similar to the LBD genes in Arabidopsis thaliana, rice and apple (Shuai et al., 2002; Wang et al., 2013b; Yang et al., 2006). Therefore, we infer that the structures of LBD genes are relatively conserved in different angiosperms. Through analysis of EgLBD proteins within each subgroup, we demonstrated that they generally possessed similar protein motifs. Among the motifs, the designation of motifs 1, 2 and 5 was DUF260 (domain of unknown function 260) (Table S4). EgLBD15 only had motif 2, while all the other class I EgLBD proteins had motif 1 and motif 2, and all the class II EgLBD proteins had motif 1 and motif 5. Because DUF260 motif contains the conserved DNA binding motif (CX2CX6CX3C zinc finger‐like motif), we believe that this protein motif is likely to play a role in transcriptional regulation of their target genes.
The mechanisms by which LBD genes control root and leaf development have been elucidated. Hay et al. (2006) demonstrated that auxin and ASYMMETRIC LEAVES1 (AS1) converged to repress expression of the KNOTTED1‐like homeobox (KNOX) gene BREVIPEDICELLUS (BP) and thus promote leaf development. Regulation of lateral root development by AtLBD16, AtLBD28, AtLBD29 and AtLBD33 was found to be associated with the auxin signal transduction pathway, and these genes were directly regulated by AtARF7 and AtARF19 through auxin responsive elements (Lee et al., 2009; Okushima et al., 2007). During lateral root initiation, AtLBD18 and AtLBD33 had been proven to activate plant cell division through transcriptional regulation of Arabidopsis E2Fa (Berckmans et al., 2011) or by directly binding to the promoter of EXPANSIN14 (Lee et al., 2013). Arabidopsis JAGGED LATERAL ORGANS (a member of the LBD gene family) acts with ASYMMETRIC LEAVES2 to coordinate KNOTTED1‐like homeobox (KNOX) and PIN‐FORMED1 (PIN1) expression during shoot and root development in Arabidopsis (Rast and Simon, 2012). However, there is still limited knowledge about the mechanisms by which LBD genes control secondary growth. Yordanov et al. (2010) reported that two key class I KNOTTED1‐like homeobox genes (ARBORKNOX1 and ARBORKNOX2) that promote meristem identity in the cambium were down‐regulated, while APL was up‐regulated in PtaLBD1‐oe plants showing enhanced woody growth and remarkable changes in bark texture. Based on these findings, they proposed a mechanistic model of LBD regulatory roles in secondary woody growth (Yordanov and Busov, 2011). In this model, PtaLBD1 and PtaLBD4 are expressed at the cambium/phloem boundary and regulate secondary phloem development by restraining the expression of meristem identity genes (ARBORKNOX1 and ARBORKNOX2) in the cambium zone and promote phloem development through activation of APL and likely other unknown set of genes. PtaLBD18 and PtaLBD15 are expressed at the cambium/xylem border and also restrict the expression of meristem identity genes to the cambium zone while at the same time promote xylem development through activation of other unknown genes. PtaLBDs play antagonistic roles with meristem maintenance genes to maintain meristem identity in cambium and/or promote xylem/phloem/ray cell tissue differentiation. Although this model seems plausible, the molecular mechanisms underlying secondary growth remain to be further elucidated.
Members of LBD genes in Arabidopsis, rice and other species have been found to respond to the treatments of cytokinin, gibberellic acid, auxin, brassinosteroid and abscisic acid (Bell et al., 2012; Berckmans et al., 2011; Naito et al., 2007; Wang et al., 2013b Zentella et al., 2007). Using qRT‐PCR method, we have demonstrated that expression of some EgLBD genes also responded to auxin and gibberellin (Figure 4). The expression levels of EgLBD22, EgLBD29 and EgLBD37 were all transiently up‐regulated following exogenous auxin treatment (Figure 4). Interestingly, EgLBD37 was up‐regulated, whereas EgLBD22 and EgLBD29 were down‐regulated under exogenous gibberellic acid treatment (Figure 4), suggesting that the EgLBD22/29 and EgLBD37 genes may differentially regulate GA‐mediated growth and developmental processes in E. grandis. Gibberellic acid‐stimulated Arabidopsis6 (AtGASA6) promoted cell elongation during the germination of Arabidopsis seeds (Zhong et al., 2015). In this study, we demonstrated that gibberellin‐regulated protein5 in hybrid poplar was up‐regulated to 4.6‐fold in EgLBD37‐oe plants (Table S10). Given that the link between the transcriptional response of EgLBD37 to GA treatment and dramatic increase in secondary growth and the connection between the expression level of gibberellin‐regulated protein5 and internode length of EgLBD37‐oe plants, it is reasonable for us to conclude that EgLBD37 enhances secondary growth and plant stem elongation through GA‐mediated signalling pathway. Moreover, we found that expansin‐B3 was up‐regulated in EgLBD37‐oe plants (Table S10), leading us to conclude that, like AtLBD18, EgLBD37 promotes cell expansion during secondary growth via activating the expression of expansin genes.
In our experiment, an ethylene‐responsive transcription factor ERF115 was predicted to be the functional partner of EgLBD29 (Table S6). Interestingly, we found that the expression of ethylene‐responsive transcription factor ERF023‐like gene increased 3.8‐fold and the expression of an ethylene‐responsive element‐binding family protein‐encoding gene (EREBP) increased 1.8‐fold in EgLBD29‐oe plants, which showed a significant broader phloem fibres region (Table S9, Figure 7g). Recently, a similar plant morphology to EgLBD29‐oe lines was observed in pLMX5::ERF139‐overexpressing hybrid aspen (Populus tremula × Populus tremuloides) which showed a dwarf phenotype with altered wood development (Vahala et al., 2013). It has already been well documented that ethylene can enhance plant fibre development (Shi et al., 2006). All of these evidences suggest that there is an ethylene‐mediated signalling pathway for the function of EgLBD29 in control of secondary growth, especially phloem fibre development. The expansin gene c60809_g2 and expansin gene c60754_g1 was reduced 1.1‐ and 1.2‐fold in EgLBD29‐oe plants (Table S9), while Expansin‐B3 was up‐regulated to 1.5‐fold in EgLBD37‐oe plants (Table S10). Because the phenotype of EgLBD29‐oe plants contrasted to that of EgLBD37‐oe plants in terms of height, leaf size and stem diameter (Figures 5 and 6), it is likely that EgLBD29 and EgLBD37 exert opposite influences on cell growth and cell wall expansion during secondary growth through their opposite roles in control of the expression of expansin genes.
We found that overexpression of EgLBD37 not only significantly increased diameter of stem (Figure 6c, d), but also dramatically increased plant height (Figure 5), internode length (Figure 6a, b) and leaf size (Figure 6e, f). Like EgLBD37, overexpression of PtaLBD1 increased stem diameter of transgenic plants via promoting secondary growth. However, overexpression of PtaLBD1 did not increase plant height, internode length and leaf size (Yordanov et al., 2010). Why did EgLBD37‐oe affect the cortex thickness and xylem formation but not phloem development? One plausible explanation is that EgLBD37 is not the functional ortholog of PtaLBD1 (for example, it could be EgLBD25 according to Figure 1). Another explanation could be that there is a functional divergence in terms of target genes between EgLBD37 and PtaLBD1. In addition, we found that EgLBD29‐oe plants have reduced plant height (Figure 5), shorter internode length (Figure 6a, b), thinner stem diameter (Figure 6c, d) and smaller leaf size (Figure 6e, f), which contrasts to the phenotype of EgLBD37‐oe plants, implying that EgLBD29 and EgLBD37 may have an antagonistic function. The potential application of EgLBD37 would be to increase timber production and EgLBD29 would be to improve pulp yield because of high‐quality fibres that may come from EgLBD29‐oe transgenic trees.
In conclusion, we have described the evolutionary relationship of the LBD gene family in E. grandis and functionally characterized three EgLBD genes, two of which showed significant effects on secondary growth. Our results will facilitate efforts to gain a deeper understanding of the structure‐function relationships of these genes and may enable novel breeding techniques to improve wood formation or fibre production in trees.
Experimental procedures
Plant materials and growth conditions
One‐year‐old E. grandis trees were planted in the greenhouse of Chinese Academy of Forestry under controlled conditions with a relative humidity of 50% at 25 °C. For detecting the expression level in different tissues, roots, stems, leaf, xylem and phloem of E. grandis were sampled in the spring of 2015. For phloem collection, a bark window approx. 30 × 50 mm was removed from the stem using a hammer and chisel. Xylem tissue was collected by scraping cells from the exposed wood with chisel. For determination of IAA (indol‐3‐acetic acid) and GA3 (gibberellic acid) response, roots of two‐month‐old seedlings (25–30 cm in height) were treated with 150 μm IAA (Sigma‐Aldrich China, Shanghai) or 150 μm GA3 (Biotopped, Beijing) solution for 0, 1, 3, 6 and 12 h, respectively. The first time point (0 h) served as a control. After IAA and GA3 solution treatment, stems were harvested from seedlings. Three biological replicates were performed for each sample. All the samples were immediately frozen in liquid nitrogen and stored at −80 °C freezer before total RNA isolation.
Identification of LBD genes in E. grandis
The E. grandis genome sequence version 2.0 was downloaded from the Phytozome database ( http://www.phytozome.net/) (Goodstein et al., 2012) and used to construct a local BLAST database. All known LBD gene sequences of Arabidopsis, poplar and rice were downloaded from the GenBank ( https://www.ncbi.nlm.nih.gov/genbank/) of NCBI (National Center for Biotechnology Information). These sequences were used as query to perform local BLAST searches against the E. grandis genome with e‐value cut‐off set to 10−5. All candidate LBD protein sequences were examined by the domain analysis programs SMART (Simple Modular Architecture Research Tool) ( http://smart.embl-heidelberg.de/) (Letunic et al., 2015) and Pfam ( http://pfam.xfam.org/) (Punta et al., 2012) with the default parameters. We then analysed the E. grandis candidate LBD protein sequence domain using a hidden Markov model (HMM) as described by Wu et al. (2002).
Bioinformatic analysis and phylogenetic analysis
ExPASy ( http://web.expasy.org/compute_pi/) was used to predict the pI and molecular weight (Artimo et al., 2012). All LBD proteins sequences from A. thaliana, E. grandis, Populus trichocarpa and four Populus tremula × Populus alba (Pta) LBD proteins were aligned using Clustal X (Thompson et al., 1997). Using MEGA5 software, the phylogenetic tree of full‐length sequences was constructed by the neighbour‐joining method with bootstrap to be 1000 (Tamura et al., 2011).
Gene structure, conserved motif analyses and chromosomal location
EgLBD genes exon/intron structure was identified with Gene Structure Display Server 2.0 (GSDS, http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). Conserved motifs of the proteins were analysed using the Multiple Em for Motif Elucidation (MEME) program ( http://meme-suite.org/index.html) (Bailey and Elkan, 1994). MEME was used by setting repetitions to any number, the number of motifs to 20 and optimum motif width to 30–70. Functional designation of the motifs was performed to get valid domain hits for architecture search with CDART (Conserved Domain Architecture Retrieval Tool) reported by Geer et al. (2002), which can be accessed at https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi. The chromosomal locations were retrieved from the genome data downloaded from the Phytozome database (Goodstein et al., 2012) and mapped to the chromosomes using MG2C (Map Gene2 Chromosome v2, http://mg2c.iask.in/mg2c_v2.0/).
RNA extraction, cDNA synthesis and gene expression profiling
Total RNA was isolated with RNA extraction kit (TIANGEN, Beijing, China) from E. grandis samples including root, stem, leaf, xylem and phloem. The purity and quality of RNA was checked by NanoDrop8000 (Thermo Fisher Scientific, Waltham, MA, USA) and analysed by gel electrophoresis. One microgram of total RNA was reverse‐transcribed using the Prime‐Script RT reagent kit (Takara, China). qRT‐PCR was conducted with SYBR Premix EX Taq II (Takara, China). PP2A‐3 (protein phosphatase 2A subunit A3) gene was used as internal reference, and each reaction was conducted in triplicate. The stem expression values were set to 1. Relative gene expression was calculated according to the delta–delta Ct method of the system. The primers used in qRT‐PCR analysis for tissue‐specific expression of EgLBD genes are listed in Table S1.
Subcellular localization and protein–protein interaction prediction
Protein subcellular localization was predicted using Plant‐mPLoc ( http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). To validate subcellular localization, the full‐length coding sequences (without the stop codon) of EgLBD22, EgLBD29 and EgLBD37 were amplified from RNA of E. grandis stem by RT‐PCR. The PCR product of EgLBD37 was digested with NcoI and SpeI and directionally ligated into vector pCAMBIA1302 to construct the EgLBD37‐GFP fusion gene driven by a CaMV35S promoter (Niwa, 2003). The PCR products of EgLBD22 and EgLBD29 were ligated to vector pCAMBIA1302, respectively, using Seamless Assembly Cloning Kit (CloneSmater, Beijing, China). The pCAMBIA1302‐GFP was used as the positive control. Transient expression in lower leaf epidermal cells of Nicotiana tabacum L. was performed as described by Zheng et al. (2005). The transient expression of the EgLBD22/29/37‐GFP fusion proteins was observed under Ultra‐VIEW VoX 3D Live Cell Imaging System Spinning Disk confocal laser scanning microscope (PerkinElmer, Waltham, MA, USA). All the primers used in subcellular localization are listed in Table S1. Protein–protein interaction prediction was performed using the online tool of STRING ( http://string-db.org/).
Plant transformation
Plantlets of hybrid poplar (Populus alba × Populus glandulosa) clone 84k were grown at 23–25 °C under a 16/8 h day/night cycle with a light intensity of 50 μm/m2/s provided by cool white fluorescent tubes. The full‐length coding sequences of EgLBD22, EgLBD29 and EgLBD37 were amplified from RNA of E. grandis stem by RT‐PCR. The PCR products of EgLBD22, EgLBD29 and EgLBD37 were digested with XbaI and XmaI and directionally ligated into vector pBI121. All the primers used to construct the overexpression vector are shown in Table S1. Leaf discs from 84k were infected with an overnight culture of Agrobacterium tumefaciens harbouring the 35S::EgLBD22, 35S::EgLBD29 or 35S::EgLBD37 construct at an OD600 of 0.5. Infected leaf discs were then cocultured in darkness with Agrobacterium in the shoot induction medium (Murashige‐Skoog basal medium containing 0.5 mg 6‐benzylaminopurine and 0.05 mg naphthalene acetic acid per litre) for 3 days at 23 ± 2 °C. The leaf discs were then transferred to the shoot induction medium containing 200 mg/L Timentin and 40 mg/L Kanamycin under a 16/8 h light/dark regime. After one month, individual regenerated shoots were cut off and transferred to root induction medium (half‐strength Murashige‐Skoog medium supplemented with 0.05 mg/L indole‐3‐butyric acid and 0.02 mg/L naphthalene acetic acid) containing 200 mg/L Timentin and 40 mg/L Kanamycin.
Phenotype and microscopy analysis
Phenotype analysis for plant height, stem diameter, leaf size and internode length was carried out with ten‐week‐old plants. Five independent lines were used for phenotype investigation for each construct. Significant differences relative to control wild‐type plants (WT‐84k) were determined by Student's t‐test. At the same time, stem segments (0.5 cm in length) of the 10th node from WT‐84 and transgenic plants were sampled, immediately fixed in FAA (formaldehyde 3.7%, ethyl alcohol 50.0% and acetic acid 5.0%) and embedded using Shandon Excelior and Histocentre 2 (Thermo Fisher Scientific, Waltham, MA, USA). Five‐μm‐thick sections were stained with safranin and fast green (Gefan biotechnology Co. Ltd., Shanghai). Images were taken using Leica DM 6000B fully automated upright microscope (Leica Microsystems GmbH, Wetzlar, Germany).
RNA‐seq analysis of transgenic plants
The total RNA was isolated from the shoots of one‐month‐old EgLBD22‐oe, EgLBD29‐oe or EgLBD37‐oe transgenic plants according to the method as described above. RNA integrity was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies Santa Clara, CA, USA ). A total amount of 1.5 μg RNA per sample was used for library preparations. Libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit (NEB Ipswich, MA, USA), and index codes were added to attribute sequences to each sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index‐coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3‐cBot‐HS (Illumina San Diego, CA, USA). After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform and paired‐end reads were generated.
Transcriptome assembly was accomplished based on the left.fq and right.fq using Trinity (Grabherr et al., 2011) with min_kmer_cov set to 2, and all other parameters set to default values. Gene function was annotated based on Nr (NCBI nonredundant protein sequences), Nt (NCBI nonredundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss‐Prot (a manually annotated and reviewed protein sequence database), KO (KEGG Ortholog database) and GO (Gene Ontology) databases. Clean reads were obtained by removing low‐quality reads and reads containing adapter, ploy‐N from raw data and were mapped back to the assembled transcriptome. Readcounts for each gene were obtained from the mapping results and then were adjusted by edgeR program package through one scaling normalized factor (Robinson et al., 2010). We estimated the expression levels of each unique sequence in the two samples using RSEM (Li and Dewey, 2011). We then executed statistical analysis using the mapped read numbers for each unique sequences calculated by RSEM. Differential expression analysis was performed using the DEGseq R package (Wang et al., 2010). P value was adjusted using q value (Storey and Tibshirani, 2003). q value <0.005 and |log2(fold change)| > 1 was set as the threshold. Gene ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R package based on Wallenius noncentral hypergeometric distribution (Young et al., 2010). Validation of the gene expression was carried out using qRT‐PCR. The specific primers for qRT‐PCR analysis are shown in Table S1.
Supporting information
Figure S1 Conserved domains of EgLBD protein family.
Figure S2 The chromosomal localization of the LBD gene family in Eucalyptus grandis.
Figure S3 Subcellular localization of EgLBD22, EgLBD29 and EgLBD37 proteins.
Figure S4 Gel electrophoresis analysis for the presence of the transgene in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.
Figure S5 Validation for the expression of the transgene in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants by qRT‐PCR.
Table S1 All the primers used in this study.
Table S2 The coding sequences of LBD genes in Eucalyptus grandis.
Table S3 The information of LBD gene family in Eucalyptus grandis.
Table S4 Conserved motifs predicted by MEME program in EgLBD proteins.
Table S5 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD22.
Table S6 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD29.
Table S7 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD37.
Table S8 The differentially expressed genes between EgLBD22‐oe and WT‐84k plants.
Table S9 The differentially expressed genes between EgLBD29‐oe and WT‐84k plants.
Table S10 The differentially expressed genes between EgLBD37‐oe and WT‐84k plants.
Table S11 The information of eight key differentially expressed genes in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.
Acknowledgements
This work was supported by the Basic Research Fund of RIF (RIF2014‐01), Natural Science Foundation of China (31670676), Mondi and Sappi through the Forest Molecular Genetics Programme, the Technology and Human Resources for Industry Programme (UID 80118) and the National Research Foundation (UID 18312, 71255 and 86936) of South Africa. The authors declare no conflict of interest.
References
- Albinsky, D. , Kusano, M. , Hiquchi, M. , Hayashi, N. , Kobayashi, M. , Fukushima, A. , Mori, M. et al. (2010) Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene‐changing nitrogen metabolism. Molecular Plant, 3, 125–142. [DOI] [PubMed] [Google Scholar]
- Artimo, P. , Jonnalagedda, M. , Arnold, K. , Baratin, D. , Csardi, G. and de Castro, E. (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Bell, E.M. , Lin, W.C. , Husbands, A.Y. , Yu, L. , Jaganatha, V. , Jablonska, B. , Mangeon, A. et al. (2012) Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl Acad. Sci. USA, 109, 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans, B. , Vassileva, V. , Schmid, S.P. , Maes, S. , Parizot, B. , Naramoto, S. , Magyar, Z. et al. (2011) Auxin‐dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell, 23, 3671–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke, M. , Thitamadee, S. , Mähönen, A.P. , Hauser, M.T. and Helariutta, Y. (2003) APL regulates vascular tissue identity in Arabidopsis . Nature, 426, 181–186. [DOI] [PubMed] [Google Scholar]
- Borghi, L. , Bureau, M. and Simon, R. (2007) Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell, 19, 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri, E. , Chuck, G. , Vollbrecht, E. , Rocheford, T. , Martienssen, R. and Hake, S. (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell, 18, 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Liu, C.Y. , Liu, C.X. , Zhao, Y.L. and Xu, R.R. (2016) Genome‐wide analysis of the lateral organ boundaries domain gene family in Vitis vinifera . J. Genet. 95, 1–12. [DOI] [PubMed] [Google Scholar]
- Carroll, A. and Somerville, C. (2009) Cellulosic biofuels. Annu. Rev. Plant Biol. 60, 165–182. [DOI] [PubMed] [Google Scholar]
- Geer, L.Y. , Domrachev, M. , Lipman, D.J. and Bryant, S.H. (2002) CDART: protein homology by domain architecture. Genome Res. 12, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein, D.M. , Shu, S. , Howson, R. , Neupane, R. , Hayes, R.D. and Fazo, J. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , Adiconis, X. et al. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A. , Barkoulas, M. and Tsiantis, M. (2006) ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis . Development, 133, 3955–3961. [DOI] [PubMed] [Google Scholar]
- Hu, B. , Jin, J. , Guo, A.Y. , Zhang, H. , Luo, J. and Gao, G. (2015) GSDS2.0: an upgraded gene feature visualization server. Bioinformatics, 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H. , Ueno, Y. , Semiarti, E. , Onouchi, H. , Kojima, S. , Tsukaya, H. , Hasebe, M. et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Jia, X.T. , Liu, W.X. , Xie, W.G. , Liu, Z.P. , Liu, Z.M. and Wang, Y.R. (2014) Genome‐wide analysis of the LBD transcription factor family in Medicago truncatula . Acta Botanica Boreali‐Occidentalia Sinica, 34, 2176–2187. [Google Scholar]
- Lee, H.W. , Kim, N.Y. , Lee, D.J. and Kim, J. (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis . Plant Physiol. 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W. , Kim, M.J. , Kim, N.Y. , Lee, S.H. and Kim, J. (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis . Plant J. 73, 212–224. [DOI] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinform. 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Wang, S. , Yu, X. , Yu, J. , He, X. , Zhang, S. , Shou, H. et al. (2005) ARL1, a LOB‐domain protein required for adventitious root formation in rice. Plant J. 43, 47–56. [DOI] [PubMed] [Google Scholar]
- Luo, Y.W. , Ma, B. , Zeng, Q.W. , Xiang, Z. and He, N.J. (2016) Identification and characterization of Lateral Organ Boundaries Domain genes in mulberry, Morus notabilis . Meta Gene, 8, 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer, C. and Hochholdinger, F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. [DOI] [PubMed] [Google Scholar]
- Matsumura, Y. , Iwakawa, H. , Machida, Y. and Machida, C. (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz, E.J. , Baucher, M. , Sundberg, B. and Boerjan, W. (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 47, 239–274. [PubMed] [Google Scholar]
- Myburg, A.A. , Grattapaglia, D. , Tuskan, G.A. , Hellsten, U. , Hayes, R.D. , Grimwood, J. , Jenkins, J. et al. (2014) The genome of Eucalyptus grandis . Nature, 510, 356–362. [DOI] [PubMed] [Google Scholar]
- Naito, T. , Yamashino, T. , Kiba, T. , Koizumi, N. , Kojima, M. , Sakakibara, H. and Mizuno, T. (2007) A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana . Biosci. Biotechnol. Biochem. 71, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Niwa, Y. (2003) A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol. 20, 1–11. [Google Scholar]
- Okushima, Y. , Fukaki, H. , Onoda, M. , Theologis, A. and Tasaka, M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . Plant Cell, 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta, M. , Coggill, P.C. , Eberhardt, R.Y. , Mistry, J. , Tate, J. , Boursnell, C. , Pang, N. et al. (2012) The Pfam protein families database. Nucleic Acids Res. 40, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast, M.I. and Simon, R. (2012) Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell, 24, 2917–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. and Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwood, D.L. , Rudie, A.W. , Ralph, S.A. , Zhu, J.Y. and Winandy, J.E. (2008) Energy product options for Eucalyptus species grown as short rotation woody crops. Int. J. Mol. Sci. 9, 1361–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. , Tohge, T. , Matsuda, F. , Saito, K. and Scheible, W.R. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis . Plant Cell, 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y.H. , Zhu, S.W. , Mao, X.Z. , Feng, J.X. , Qin, Y.M. , Zhang, L. , Cheng, J. et al. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell, 18, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, B. , Reynaga‐Pena, C.G. and Springer, P.S. (2002) The lateral organ boundaries gene defines a novel, plant‐specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano, T. , Thitamadee, S. , Machida, Y. and Chua, N.H. (2008) Asymmetric leaves2‐like19/lateral organ boundaries domain30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis . Plant Cell, 20, 3359–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer, R. and Groover, A. (2010) Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592. [DOI] [PubMed] [Google Scholar]
- Storey, J.D. and Tibshirani, R. (2003) Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA, 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. and Nei, M. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala, J. , Felten, J. , Love, J. , Gorzsás, A. , Gerber, L. , Lamminmäki, A. , Kangasjärvi, J. et al. (2013) A genome‐wide screen for ethylene‐induced Ethylene Response Factors (ERFs) in hybrid aspen stem identifies ERF genes that modify stem growth and wood properties. New Phytol. 200, 511–522. [DOI] [PubMed] [Google Scholar]
- Wang, J. (2016) A genome‐wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang University‐SCIENCE B (Biomedicine & Biotechnology), 17, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Feng, Z. , Wang, X. , Wang, X. and Zhang, X. (2010) DEGseq: an R package for identifying differentially expressed genes from RNA‐seq data. Bioinformatics, 2, 136–138. [DOI] [PubMed] [Google Scholar]
- Wang, X.F. , Liu, X. , Su, L. , Sun, Y.J. , Zhang, S.Z. , Hao, Y.J. and You, C.H.X. (2013a) Identification, evolution and expression analysis of the LBD gene family in tomato. Sci. Agric. Sin. 46, 2501–2513. [Google Scholar]
- Wang, X.F. , Zhang, S.Z. , Su, L. , Liu, X. and Hao, Y.J. (2013b) A genome‐wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE, 8, e57044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Song, C. , Wang, B. and Cheng, J. (2002) Hidden Markov model used in protein sequence analysis. J. Biomed. Eng. 19, 455–458. [PubMed] [Google Scholar]
- Xu, L. , Xu, Y. , Dong, A. , Sun, Y. , Pi, L. , Xu, Y. and Huang, H. (2003) Novel as1 and as2 defects in leaf adaxial‐abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development, 130, 4097–4107. [DOI] [PubMed] [Google Scholar]
- Xu, B. , Li, Z. , Zhu, Y. , Wang, H. , Ma, H. , Dong, A. and Huang, H. (2008) Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary‐specifying genes to promote sepal and petal development. Plant Physiol. 146, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Luo, F. and Hochholdinger, F. (2016) LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Yu, X. and Wu, P. (2006) Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis . Mol. Phylogenet. Evol. 39, 248–262. [DOI] [PubMed] [Google Scholar]
- Yordanov, Y.S. and Busov, V. (2011) Boundary genes in regulation and evolution of secondary growth. Plant Signal. Behavior, 6, 688–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanov, Y.S. , Regan, S. and Busov, V. (2010) Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus . Plant Cell, 22, 3662–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M.D. , Wakefield, M.J. , Smyth, G.K. and Oshlack, A. (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol. 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R. , Zhang, Z.L. , Park, M. , Thomas, S.G. , Endo, A. , Murase, K. , Fleet, C.M. et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis . Plant Cell, 19, 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.M. , Zhang, S.Z. and Zheng, C.C. (2014) Genome‐wide analysis of LATERAL ORGAN BOUNDARIES domain gene family in Zea mays . J. Genet. 93, 79–91. [DOI] [PubMed] [Google Scholar]
- Zheng, H. , Camacho, L. , Wee, E. , Batoko, H. , Legen, J. , Leaver, C.J. , Malho, R. et al. (2005) A Rab‐E GTPase mutant acts downstream of the Rab‐D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell, 17, 2020–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. , Xu, H. , Ye, S. , Wang, S. , Li, L. , Zhang, S. and Wang, X. (2015) Gibberellic acid‐stimulated Arabidopsis 6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 169, 2288–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q.H. , Guo, A.Y. , Gao, G. , Zhong, Y.F. and Xu, M. (2007) DPTF: a database of poplar transcription factors. Bioinformatics, 23, 1307–1308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Conserved domains of EgLBD protein family.
Figure S2 The chromosomal localization of the LBD gene family in Eucalyptus grandis.
Figure S3 Subcellular localization of EgLBD22, EgLBD29 and EgLBD37 proteins.
Figure S4 Gel electrophoresis analysis for the presence of the transgene in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.
Figure S5 Validation for the expression of the transgene in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants by qRT‐PCR.
Table S1 All the primers used in this study.
Table S2 The coding sequences of LBD genes in Eucalyptus grandis.
Table S3 The information of LBD gene family in Eucalyptus grandis.
Table S4 Conserved motifs predicted by MEME program in EgLBD proteins.
Table S5 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD22.
Table S6 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD29.
Table S7 Protein‐protein interaction prediction for possible functional protein association networks of EgLBD37.
Table S8 The differentially expressed genes between EgLBD22‐oe and WT‐84k plants.
Table S9 The differentially expressed genes between EgLBD29‐oe and WT‐84k plants.
Table S10 The differentially expressed genes between EgLBD37‐oe and WT‐84k plants.
Table S11 The information of eight key differentially expressed genes in EgLBD22‐oe, EgLBD29‐oe and EgLBD37‐oe plants.


