Abstract
Shigellosis (bacillary dysentery) is an acute enteric infection caused by members of Shigella genus. It causes annual deaths of approximately five million children in developing countries. Among Shigella spp., S. flexneri causes more serious forms of dysentery than other Shigella species. Due to the appearance of multidrug-resistant strains of Shigella spp., it is necessary to find alternative antimicrobial agents. The aims of this study were the isolation of a novel species-specific phage against S. flexneri and to evaluate its potential and efficacy for biocontrolling of S. flexneri in foods. Shigella flexneri PTCC 1234 was used as the host strain for bacteriophage isolation from waste water. A lytic phage of the Siphoviridae family was isolated and designated as vB_SflS-ISF001. The phage activity remained at high levels after 1 h of incubation at − 20 to 50 °C and was fairly stable for 1 h at pH values ranging from 7 to 9. The latent period and burst size were approximately 20 min and 53 ± 4 phages per host cell, respectively. Raw and cooked chicken breast were inoculated with a predetermined amount of S. flexneri and subjected to biocontrol test. The results showed that using vB_SflS-ISF001 phage led to more than two logs reduction in the count of viable S. flexneri. It was demonstrated that using vB_SflS-ISF001 phage is of high potential for developing an alternative strategy against S. flexneri contamination in foodstuffs.
Keywords: Shigellosis; Shigella flexneri; Siphoviridae; Biocontrol, Bacteriophage
Introduction
Shigella flexneri is a Gram-negative, non-motile and rod-shaped bacterium which belongs to Shigella genus of Enterobacteriaceae. Shigella flexneri and other members of Shigella genus (S. dysenteriae, S. boydii and S. sonnei) are causative agents of shigellosis (bacillary dysentery) (Zaidi and Estrada-García 2014). Shigellosis is responsible for annual deaths of approximately five million children in developing countries (Jafari et al. 2009). Shigella flexneri is endemic in many third world countries and causes more serious forms of dysentery than other Shigella species (Tajbakhsh et al. 2012). Shigella flexneri is more commonly known as responsible for shigellosis outbreaks in developing countries and also causes approximately two million infections per year worldwide (Jun et al. 2016a).
Shigella spp. are reported by the world health organization (WHO) as the third most frequently isolated bacteria from foodstuffs (WHO 2005). For example, porridge (Zhang et al. 2013), sugar snaps (Löfdahl et al. 2009) and fresh sugar pea (Muller et al. 2009) were found to be contaminated with Shigella spp. and reported as the suspected sources of the bacteria in Guangxi, Sweden and Denmark shigellosis outbreaks, respectively. Moreover, in 2014, chicken, beef and dairy products were reported as the sources for Shigella spread (Ahmed and Shimamoto 2014).
In the last few years, antibiotic resistance in bacteria has increased (Jamal et al. 2015). Although the first single antibiotic resistant Shigella was isolated in Japan in 1940, nowadays there are many reports on the emergence of multi-drug resistant (MDR) strains (Gu et al. 2015). Due to high similarity of Shigella and other members of Enterobacteriaceae especially Escherichia coli, Shigella strains (medical and food isolates) have gradually become resistant to most of the common recommended antimicrobial agents (Prabhurajeshwar et al. 2015; Taneja et al. 2012). Thus, there is an urgent need to find, develop and evaluate alternative strategies to fight against Shigella spp., especially in food sources. Using bacteriophages is suggested as a potential strategy (Shen et al. 2012) and they have been used to control foodborne pathogens such as Listeria monocytogenes, E. coli O157: H7, Salmonella enteritidis, Salmonella typhimurium, Campylobacter among others (Kazi and Annapure 2015).
This study aimed to isolate, identify and characterize a novel S. flexneri species-specific lytic phage, and to evaluate its potential for biocontrolling of S. flexneri in foodstuff.
Materials and methods
Bacterial strains and growth condition
Shigella flexneri serotype 1a strain (Persian Type Culture Collection 1234) (PTCC 1234), S. flexneri serotype 2b (PTCC 1865), Shigella sonnei (ATCC 9290), Shigella sonnei (PTCC 1777), Shigella dysenteriae (PTCC 1188), Shigella boydii (ATCC 9207), Escherichia coli (ATCC 25922), Klebsiella pneumonia (ATCC 13883), Streptococcus pneumonia (ATCC 49619), Pseudomonas aeruginosa (ATCC 27853), Enterococcus faecalis (ATCC 29212), Salmonella typhimurium (ATCC 14028), Proteus mirabilis (ATCC 43071), Staphylococcus epidermidis (ATCC 12228), Staphylococcus aureus (ATCC 35933), Streptococcus pyogenes (ATCC 19615) were purchased from Iranian Research Organization for Science and Technology (IROST), Tehran, Iran. Of these, S. flexneri PTCC 1234 was used to isolate and propagate bacteriophage and also used in all other experiments. It was cultured on brain heart infusion (BHI) agar or in BHI broth (Merck, Germany) with constant shaking at 37 °C. Other strains were used for determination of the isolated phage host range.
Lytic phage isolation and purification
A number of sewage samples were collected from wastewater treatment plant in Isfahan, Iran. Then, these samples were used to isolate S. flexneri specific phage according to standard enrichment method with some modifications (Ghasemi et al. 2014). Briefly, 25 ml of the samples were mixed with 25 ml of an early-exponential culture of S. flexneri PTCC 1234 and incubated at 37 °C for 24 h with constant shaking at 100 rpm. Then, the cultures were centrifuged at 10,000×g for 10 min, and the supernatants were filtered through 0.22 μm pore size membrane (Sartorious, India) to remove bacterial debris. Then, 20 μl of the filtrated media were spotted on the surface of solidified top agar layer (BHI with 0.7% agar) which contained 107 CFU/ml host cells. Following plaque appearance, the filtrated media of the previous step were serially diluted (10−1–10−10) in sterilized sodium chloride-magnesium sulphate (SM) buffer (100 mmol l−1 NaCl, 8 mmol 1−1 MgSO4, 2% gelatin, 50 mmol l−1 Tris–HCl, pH 7.5) and then, 10 μl of each serial dilutions was mixed with 0.1 ml of early-exponential culture of the host cells and 5 ml of molten soft agar (BHI with 0.7% agar) at 50 °C. Finally, the mixture was decanted onto hard layer agar (BHI with 1.5% agar) to make double-layered agar plates. After 24 h, a single clear plaque was removed by a sterile Pasteur pipette and was transferred to a tube containing 500 μl of SM buffer, then subjected to homogenization and incubation (4 °C for 1 h). Afterward, 100 μl of this sample was added to 25 ml of early-log phase culture of S. flexneri PTCC 1234. After an overnight incubation at 37 °C, it was centrifuged (10,000×g at 4 °C for 10 min) and the supernatant was subjected to filtration (0.22 μm, Sartorious, India). Subsequently, the double-layer agar plate method was used to form single-plaque as described previously (Jeon et al. 2016). In order to ensure the purity of the phage this procedure was repeated three successive times. The phage was propagated according to Sambrook and Russell protocol (Sambrook and Russell 2001) and double-layer agar method was used to determine phage titers (Kropinski et al. 2009a).
Determination of host range
The host range was examined by the spot test method as described elsewhere (Jun et al. 2014). As described above, S. flexneri strains, other Shigella members strains and different gram positive and negative standard strains were used in this test.
Electron microscopy
The phage suspension (1011 PFU/ml) was concentrated by ultracentrifugation at 25,000×g for 120 min (Beckman Optima L-80 XP ultracentrifuge). The supernatant discarded and the pellet was gently resuspended in distilled water. Afterward, 10 μl of the concentrated phage was applied to a 400-mesh grid and then negatively stained with 2% uranyl acetate as previously described (Ghasemi et al. 2014). Electron micrographs were taken using a Philips HMG 400 (Netherlands) at an accelerating voltage of 100 kV.
Thermal/pH/saline stability
The stability tests were performed as previously described with some modifications (Ghasemi et al. 2014). Briefly, to evaluate the stability of the phage at different temperatures, phage suspensions (diluted in SM buffer) were placed in microtubes and incubated at − 20, 4, 25, 40, 50, 60, 70 and 80 °C for 1 h. The stability of the phage at various pH values (3–12) was assessed by incubation at 25 °C for 1 h. The effect of hypersaline condition on phage stability was determined by incubation at several concentrations of sodium chloride (1, 5, 10, 15, 20, 25, 30 and 35%) at 25 °C for 1 h. Further, bacteriophage titer was estimated using double-layer agar method (Kropinski et al. 2009a). All assays were repeated at least three times.
Determination of phage adsorption rate
Two hundred milliliters of early-log phase culture of S. flexneri PTCC 1234 (0.5 McFarland) was prepared and divided equally into two sterile bottles. To determine the phage adsorption rate, phage suspension was added to one bottle with a multiplicity of infection (MOI) of 0.1. The other one was inoculated with SM buffer (control bottle). Samples (1 ml) were taken every minute (up to 20 min) and phage-adsorbed cells were removed by centrifugation at 13,000×g for 30 s. Finally, the titer of the free phages in the supernatant was determined by the double-layer method (Kropinski et al. 2009a). This assay was performed in triplicate.
One-step growth curve
Burst size and latent period were assessed by the one-step growth analysis as previously described (Haq et al. 2012). Briefly, S. flexneri PTCC 1234 cells were harvested by centrifuging following their culture reached to appropriate optical density (OD) (OD600 nm = 0.1). Then, the pellet obtained was resuspended in 0.5 ml SM buffer and mixed with 200 μl of diluted phage (MOI = 0.01). Afterward, the mixture was incubated for 1 min and then centrifuged to remove unabsorbed phages. The pellet was resuspended in 50 ml of fresh broth medium and incubated at 37 °C. Samples (500 μl) were taken every 5 min and double-layer agar method was used to determine phage titers (Kropinski et al. 2009a). All the procedures were repeated in three independent replicates.
Bacterial lysis assay
Bacteriolytic activity was evaluated under standard laboratory conditions (SLC) according to the method described previously with some modifications (Haq et al. 2012). The early-log phase of S. flexneri PTCC 1234 culture at OD 600 nm = 0.1 aliquoted in sterile labeled bottles equally. The bottles were inoculated with different volumes of phage suspension to give MOIs of approximately 10 and 100. The bottles were incubated at 37 °C with a constant shaking of 110 rpm. The OD 600 absorbance was measured by Eppendorf BioPhotometer in 2 h intervals for 16 h. Bacteria-free and phage-free cultures were used as controls. All experiments were performed in triplicate.
Phage DNA isolation and restriction endonuclease analysis
A very high titer of the isolated phage suspension was prepared as described above. DNase I, RNase A (SinaClon, Iran) and proteinase K (Sigma, China) were used to degrade officious bacterial DNA, RNA, and proteins in the phage suspension, respectively. Then, the suspension was subjected to whole genome extraction according to the method explained previously (Ghasemi et al. 2014). Phage’s nucleic acid was treated with the restriction enzymes including EcoRI, EcoRV, HindIII and BamHI (Thermo Fisher Scientific, US). The mixture of DNA and each endonuclease was prepared based on manufacturer’s standard protocol and incubated at 37 °C for 1 h. The digested DNAs were detected by electrophoresis in 0.6% (w/v) agarose gel and visualized under UV light after staining with ethidium bromide (1 mg/ml). The sizes of different cleaved nucleic acids were estimated using SequentiX Gel Analyzer software.
Food sample preparation
Chicken is the main meat in meat-based food in the location where these experiments were done (Iran). As a result, the possibility of anthropogenic or non-anthropogenic contamination of this source with Shigella spp. is higher than other foodstuffs. Therefore, it was used as the food mixture in the current study. Raw and cooked chicken breasts were collected at local groceries and were preliminarily checked for contamination with Shigella spp. according to WHO recommended protocol (protocol number: 2010GFNLAB001) (Mikoleit 2010); then, were minced aseptically and placed in Petri dishes (5 grams) in a biosafety cabinet. Finally, the samples were kept at low temperature (4 °C) until further use.
Food experiments
The early-exponential culture of S. flexneri PTCC 1234 was used as a bacterial host in all food experiments. The phage (108 PFU/g) was applied to Petri dishes containing raw or cooked chicken breast after (in treatment experiment) or before (in prevention experiment) inoculation with bacterial cells (104 CFU/g) at 4 °C to represent refrigerator temperatures.
The treatment experiment was designed to evaluate the reduction potential of the phage in the food matrix. The host bacterium (104 CFU/g) was pipetted onto the surface of three Petri dishes containing cooked chicken breast and other three Petri dishes containing raw chicken breast before incubating at 4 °C. Phage suspension (108 PFU/g) was added carefully to one of the Petri dishes after 1 h (B/P1), then, the same was performed after 24 h (B/P24) and 48 h (B/P48) of incubation for the rest of Petri dishes.
The prevention experiment was designed to evaluate preservation and inhibitory effects of the isolated phage, mimicking food conditions. For this, first, phage suspensions (108 PFU/g) were added completely onto the surface of three Petri dishes containing cooked chicken breast and also three Petri dishes containing raw chicken breast. The host bacterial cells (104 CFU/g) were added to one of the plates after 1 h incubation at 4 °C (P/B1). Afterward, the same was done for the two remained Petri dishes after 24 h (P/B24) and 48 h (P/B48) of incubation at 4 °C, respectively. Moreover, three controls including (1) negative control containing neither bacterium nor phage (C), (2) a positive control containing S. flexneri only (CN) and (3) a positive control containing the isolated phage (CP), were included in food experiment. After 2, 24, 48, 72, 96, 120 and 144 h samples were tested for viable bacterial counts and phage titers using pour plate and double-layer agar methods, respectively. For this, 0.5 g of each sample were transferred to sterile tubes and homogenized with 2 ml of 1 × phosphate buffered saline (PBS) and 2.5 ml of SM buffer at room temperature. The liquid portion was transferred to two sterile tubes. One of them was centrifuged at 100×g for 5 min, and 10 μl of the supernatant was used to count S. flexneri on xylose lysine deoxycholate agar (XLD agar) with pour plate method (Harley and Prescott 2002). The other one was centrifuged at 1100×g for 10 min, then filtered (0.45 μm, Sartorious, India) and used to count phage numbers by double-layer agar method (Kropinski et al. 2009a). All food samples were prepared and counts carried out in triplicate.
Statistical analysis
Statistical analysis carried out by applying One-way ANOVA method on the obtained data using GraphPad Prism software, ver. 5.0 (Graph Pad Software, USA).
Results
Phage isolation and electron microscopy analysis
Seven phages were isolated from wastewater samples of wastewater treatment plant samples in Isfahan, Iran; which among them one phage with a clear plaque (0.5–2 mm in diameter) was selected for further studies (Fig. 1a) and was designated as vB_SflS-ISF001 (Kropinski et al. 2009b). Electron microscopy revealed that vB_SflS-ISF001 had an icosahedral head (64 ± 0.4 nm) which was connected directly to a non-contractile tail. The tail length and width were 112 ± 0.6 and 15.5 ± 3 nm, respectively (Fig. 1b, c). Based on its morphology, vB_SflS-ISF001 was categorized as a member of Siphoviridae family of the Caudovirales order (Ackermann 2007).
Fig. 1.
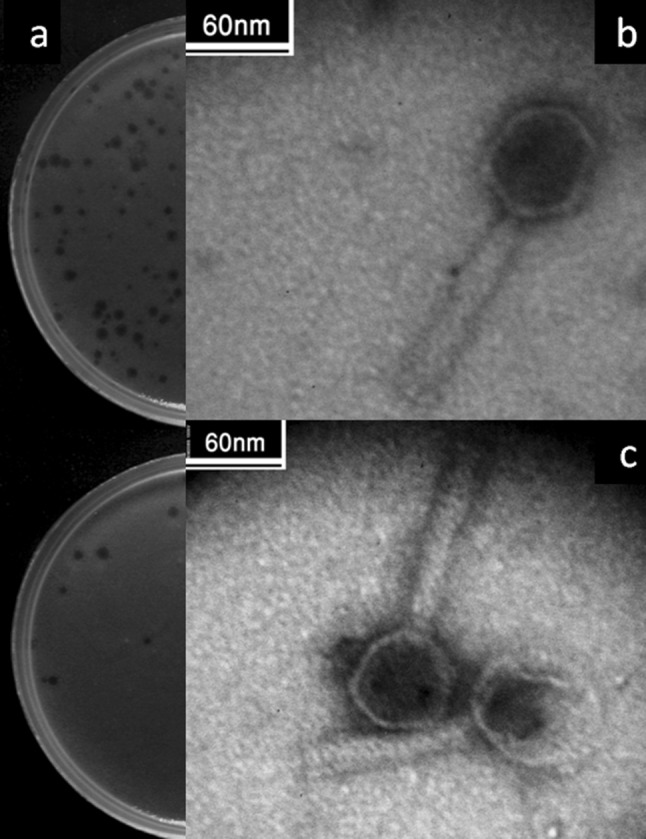
Morphology of vB_SflS-ISF001 bacteriophage. a Phage plaques. b, c Electron micrographs of vB_SflS-ISF001
Bacteriophage host range
To evaluate the vB_SflS-ISF001 host range, its infectivity was tested on all four species of Shigella using spot test method. Shigella flexneri (n = 2), S. sonnei (n = 2), S. boydii (n = 1), S. dysenteriae (n = 1), Escherichia coli, Klebsiella pneumonia, Streptococcus pneumonia, Pseudomonas aeruginosa, Enterococcus faecalis, Salmonella typhimurium, Proteus mirabilis, Staphylococcus epidermidis, Staphylococcus aureus and Streptococcus pyogenes were used to determine the host range of phage vB_SflS-ISF001. Only S. flexneri PTCC 1234 and S. flexneri PTCC 1865 were susceptible (clear plaque formation). Also, S. dysenteriae PTCC 1188 was partially inhibited (turbid plaque formation).
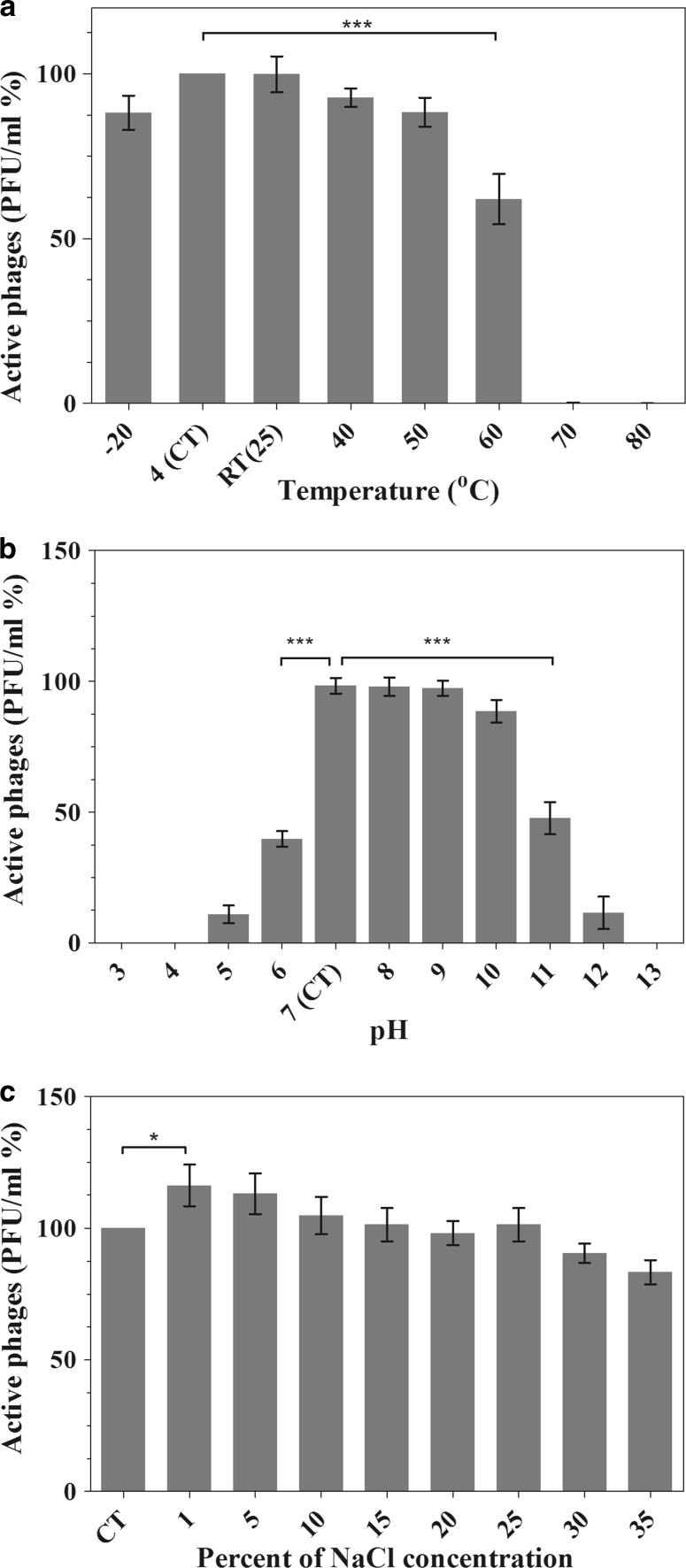
Thermal/pH/saline stability of vB_SflS-ISF001
Phage stability was determined by monitoring the changes in vB_SflS-ISF001 titer at different pH, saline and temperature conditions. Although decreases in the titers were observed, the phage activity remained at high levels after 1 h of incubation at − 20 to 50 °C, whereas a significant decrease in the titers was observed after incubation at 60 °C (P < 0.001) (Fig. 2a). vB_SflS-ISF001 was fairly stable for 1 h at pH values ranging from 7 to 9, while a significant decrease in the activity was observed at pH values of 6, 11 and 12 (P < 0.001). No active phage was detected at pH values of 4 and 13 after 1 h incubation (Fig. 2b). As shown in Fig. 3c, while no significant reduction was observed in the phage infectivity during incubation at different saline concentrations, there was an approximately 16% increment in its activity after incubation in 1% salt solution for 1 h (P < 0.05).
Fig. 2.
The effects of different temperatures (a), pH values (b), and saline concentrations (c) on vB_SflS-ISF001 activity. P values (*P < 0.05, **P < 0.01 and ***P < 0.001). CT: Compare to control in each experiment. RT: Room temperature
Fig. 3.
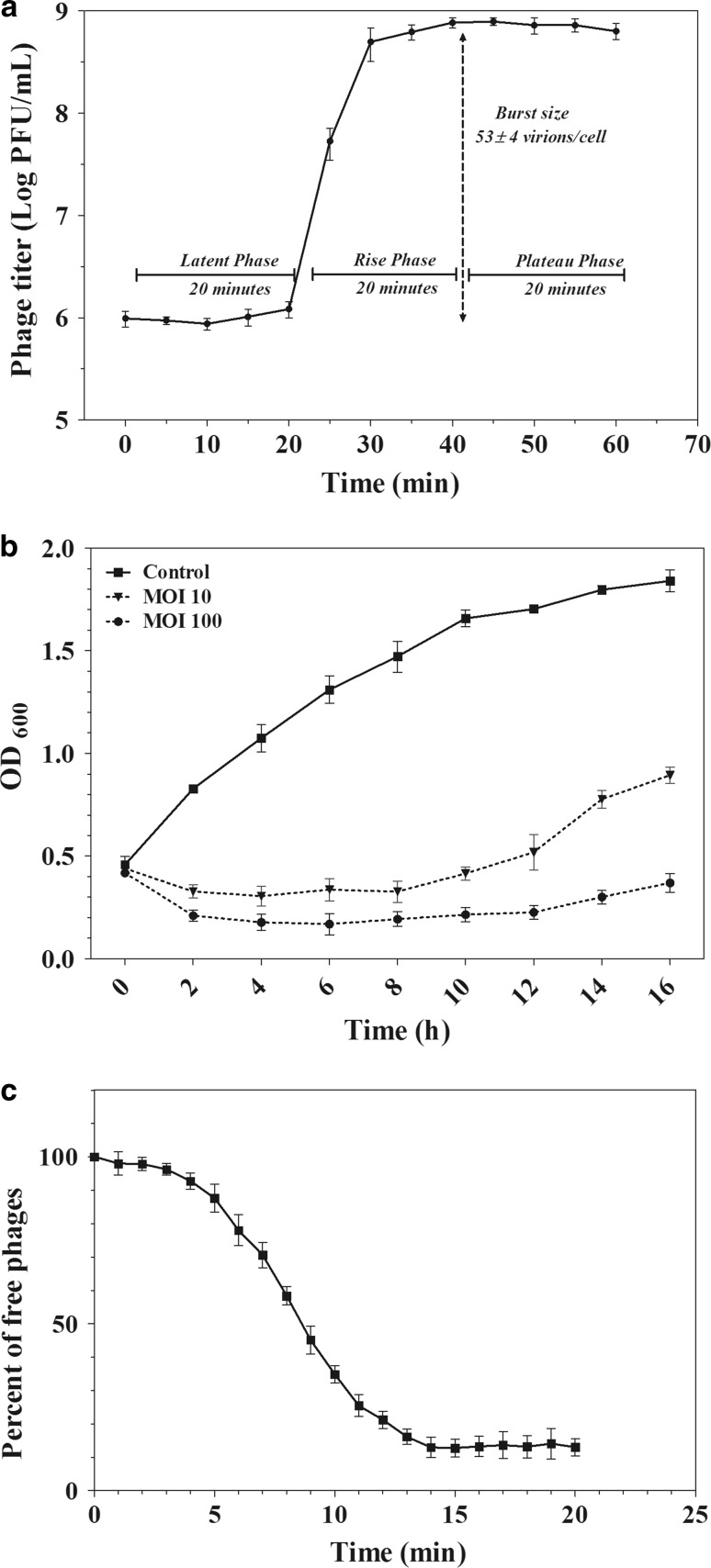
One step growth curve, reduction assay test and adsorption rate of vB_SflS-ISF001. a Latent period and burst size. b Effects of mid (MOI = 10) and high (MOI = 100) titers on the growth rate of S. flexneri. c phage adsorption rate. Values are the mean ± SD (standard deviation) from triplicate assays
Latent period, burst size, adsorption rate and antibacterial activity of vB_SflS-ISF001
The single-step growth experiment showed that the latent period and burst size were approximately 20 min and 53 ± 4 phages per host cell, respectively (Fig. 3a). As shown in Fig. 3b, the optical density of the bacterial culture was decreased after about 2 h, remained steady for 10 h and then increased slightly. Additionally, The adsorption rate of vB_SflS-ISF001 phage was 87.1% after 14 min at 25 °C (Fig. 3c).
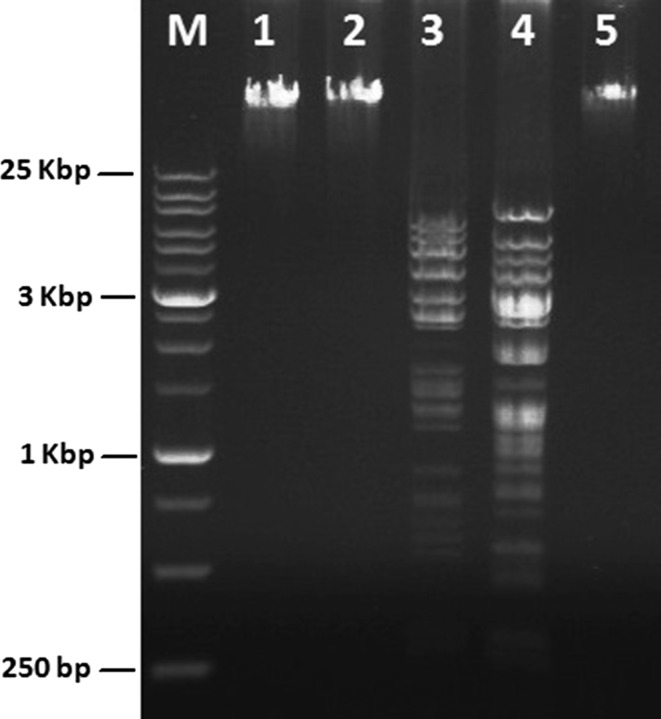
Phage genome restriction pattern
Digestion patterns of vB_SflS-ISF001 genomic DNA demonstrated that it was only digested with EcoRV and HindIII (Fig. 4). According to SequentiX Gel Analyzer software data, the genome size of phage vB_SflS-ISF001 was estimated to be approximately 44,474–46,280 bp.
Fig. 4.
Phage vB_SflS-ISF001 genomic DNA size. Lane M: X-large DNA marker (SinaClon, Iran); lane 1: Untreated genomic DNA; lane 2: EcoRI; lane 3: EcoRV; lane 4: HindIII; lane 5: BamHI
Efficacy of phage vB_SflS-ISF001 in biocontrolling of contaminated food
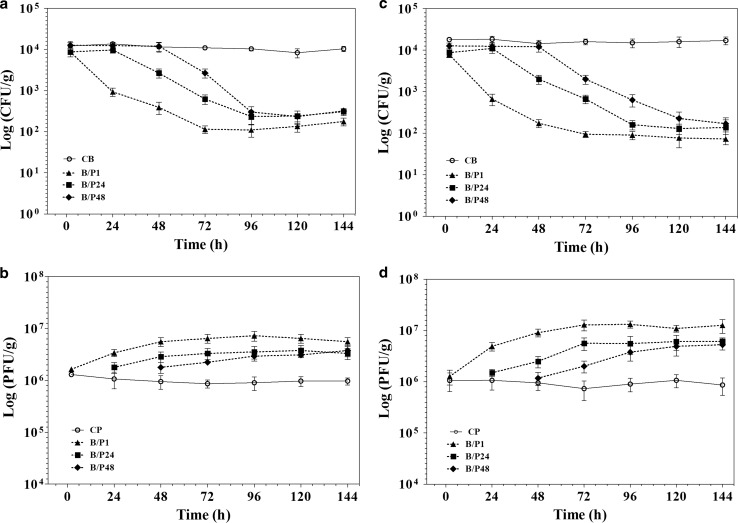
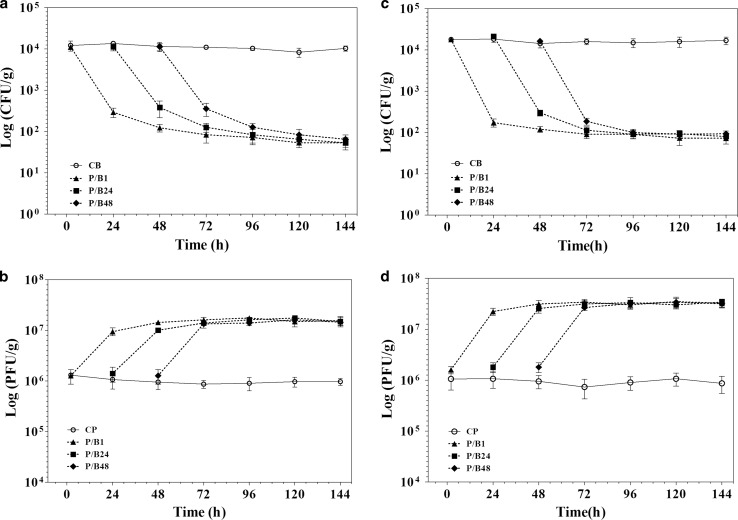
The shown results in Figs. 5 and 6 demonstrate that applying vB_SflS-ISF001 reduced the contamination rate by S. flexneri (in raw and cooked chicken breast) in both treatment and prevention experiments. In the treatment experiment, the efficacy of phage vB_SflS-ISF001 in the reduction of the number of viable target bacterial cells (S. flexneri PTCC 1234) in cooked chicken breast was better than the raw chicken breast. In fact, after phage addition, viable counts decreased greatly during first 24 h and then decreased slightly until 72 h (Fig. 5a, c). Phage propagation was clearly detected as the titers were increased after phage inoculation (Fig. 5b, d). The cell number of S. flexneri PTCC 1234 was reduced by approximately 2 logs in prevention experiment within the first 24 h after host inoculation in both cooked and raw chicken breast. After 48 and 72 h, the numbers of viable S. flexneri cells were reduced further and were even lower than 100 CFU/g in all groups (Fig. 6a, c). Moreover, phage titer (PFU/g) was increased sharply during 24 h after addition of the bacterium (Fig. 6b, d). No significant changes were observed in the titer of S. flexneri PTCC 1234 cells and phage vB_SflS-ISF001 in bacterial (CB) and phage (CP) control groups, respectively.
Fig. 5.
Treatment effects of the phage on the viable cell numbers of S. flexneri in raw and cooked chicken breast (treatment experiments). a Viable S. flexneri cells in the raw chicken breast. b Active phages in the raw chicken breast. c Viable S. flexneri cells in cooked chicken breast. d Active phages in cooked chicken breast. Data represent the mean ± SD. B/P1, 24 and 48: Bacterial inoculated samples/phage was added after 1, 24 and 48 h. CB: Positive control containing S. flexneri only, CP: Positive control containing isolated phage vB_SflS-ISF001 only
Fig. 6.
Preservation-inhibition effects of the phage on the growth of S. flexneri in raw and cooked chicken breast (prevention experiment). a Viable S. flexneri cells in the raw chicken breast. b Active phages in raw chicken breast. c Viable S. flexneri cells in cooked chicken breast. d Active phages in cooked chicken breast. Data represent the mean ± SD. P/B1, 24 and 48: Phage inoculated samples/bacterium was added after 1, 24 and 48 h. CB: Positive control containing S. flexneri only, CP: Positive control containing isolated phage vB_SflS-ISF001 only
Discussion
The Shigella genus is one of the most important groups of foodborne pathogens which has caused different outbreaks in many countries such as China, Sweden, and Denmark (Zhang et al. 2013). This genus is capable of contaminating various types of foods as well as drinking water and can spread quickly (Jun et al. 2014). Uncontrollable and overuse of different classes of antibiotics in agriculture, animal husbandry and clinical settings and the high potential of antibiotic resistance genes capable of spreading among different bacteria especially Enterobacteriaceae are the main reasons to investigate, develop and use a cost-effective alternative method (Jamal et al. 2015; Jun et al. 2014; Zhang et al. 2013).
Natural potential of bacteriophages against bacteria introduces them as possible alternative agents for antibiotics as well as a biocontrol tool in the food industry (Pérez Pulido et al. 2015). A recent study demonstrated that phage application led to 95–99% reduction in Campylobacter jejuni on chicken skin (Zhang et al. 2013). Furthermore, Salmonella was eliminated by specific phages (Bao et al. 2015; Guenther et al. 2012; Sklar and Loerger 2001). Moreover, a phage cocktail was able to reduce Listeria monocytogenes contamination on different food samples (Perera et al. 2015). However, studies on biocontrol of Shigella by phages are rare (Zhang et al. 2013; Ahmadi et al. 2015). Therefore, the aim of this study was to isolate and characterize a novel lytic phage for S. flexneri and also evaluate its potential and efficacy for controlling S. flexneri contamination in the matrix of raw or cooked chicken breast.
Regarding the fact that S. flexneri is mostly a water-borne pathogen, urban wastewater samples were used for isolation of virulent phages. Phage vB_SflS-ISF001 showed a very high host specificity and was able to lyse all S. flexneri strains while also was able to partially inhibit S. dysenteriae. Although it is generally agreed that Shigella members and E. coli are sufficiently similar and recently suggested that both could be taxonomically categorized in the same species (Jun et al. 2014), vB_SflS-ISF001 did not show any lytic activity against E. coli strain. This phage showed a highly specific host range that makes it an appropriate agent for phage therapy and biocontrol without damaging normal flora and useful bacteria.
Shigella specific phages of Myoviridae and Podoviridae family have already been reported (Faruque et al. 2003; Jakhetia and Verma 2015; Jamal et al. 2015; Jun et al. 2014, 2016a; Zhang et al. 2013). Additionally, some phages which belong to Siphoviridae such as pSf-1 and pSf-2 have been well characterized earlier and are able to produce clean plaques in S. boydii, S. flexneri and S. sonnei cultures (Jun et al. 2013, 2016b). Although vB_SflS-ISF001, pSf-1 and pSf-2 belong to the same family but differ in morphological characteristics and genome size. The pSf-1 tail length (103 ± 6 nm) and width (13 ± 3 nm) were smaller and the head diameter (73 ± 3 nm) was bigger than vB_SflS-ISF001 (Jun et al. 2013). In contrast, the tail length (136 ± 3 nm) and head diameter (57 ± 4 nm) of pSf-2 were respectively bigger and smaller than vB_SflS-ISF001 (Jun et al. 2016b). Moreover, pSf2 and vB_SflS-ISF001 had the same tail width (15 ± 3 nm).
The genome size of vB_SflS-ISF001 is smaller than pSf-1 (51,821 bp) (Jun et al. 2013), pSf-2 (50,109 bp) (Jun et al. 2016b) and Shfl1 (50,661 bp) (GenBank accession number: NC_015456). These results indicated that vB_SflS-ISF001 has the smallest genome size among S. flexneri specific Siphoviridae phages which had already been isolated.
Diverse phage tolerance against different pH values, temperatures, and saline concentrations has already been reported. This was examined for the phage vB_SflS-ISF001 which was stable in a significantly wide range of temperatures from − 20 to 60 °C for 1 h. It was completely inactivated after incubation at 70 °C for 1 h. Compare to pSf-1 which is active in temperatures ranging from 4 to 50 °C, the thermal stability of vB_SflS-ISF001 was higher (Jun et al. 2013). Also, the phage was able to tolerate salt concentrations ranging from 1 to 35%. Compare to the control group, low concentrations of NaCl had a slightly positive impact on the bacteriolytic activity of the phage. The optimal activity of vB_SflS-ISF001 was recorded at pH values ranging from 5 to 12 while pSf-1 was detectable only in pH values ranging from 5 to 9 (Jun et al. 2013). In other words, vB_SflS-ISF001 showed higher activity in basic conditions compared to pSf-1. Stability results indicated that vB_SflS-ISF001 has the potential to be used in diverse environments and different foodstuffs.
One step growth curve describes different stages involved in the multiplication of phages. Large burst size could enhance the efficacy of phage therapy as well as phage-based biocontrol, as burst size is obviously related to the lytic activity and phage reproduction (Shen et al. 2012; Silva et al. 2014). Different burst sizes have been reported for pSf-1 and pSf-2 (S. flexneri Siphoviridae phages) which have higher (86.86 virions/cell) and lower (16 virions/cell) burst sizes, respectively (Jun et al. 2013, 2016b). The phage vB_SflS-ISF001 has a relatively large burst size (53 ± 4 virions/cell) and short latent period (20 min); this indicates that the phage vB_SflS-ISF001 has a proper lytic activity to be used in biocontrol applications. Using S. flexneri PTCC 1234, the bacteriolytic activity of phage vB_SflS-ISF001 was evaluated under SLC. The results showed that the host cells growth was strongly inhibited after phage addition in different MOIs. Bacterial lysis assay also indicated that bacteriolytic activity of phage vB_SflS-ISF001 is MOI-dependent and has the highest bacteriolytic activity at MOI = 100.
This study investigated the bacterial lysis assay under SLC, yet further study is required to approve the phage potential in food biocontrol. The administrated biocontrol experiments on raw and cooked chicken breast indicate that phage vB_SflS-ISF001 has the potential to be used as a biocontrolling agent for S. flexneri contaminations. Phage application reduced the number of viable S. flexneri count at least 2 logs in both treatment and prevention experiments. This is the first study that investigated biocontrolling of S. flexneri using a Siphoviridae phage in foodstuff. As a result, no comparison was possible.
As it was shown, desirable phage stability was observed in both raw and cooked chicken breast. The newborn phage particles were released by infected cells in the first hours and increased the initial phage count for at least 1.5 logs which point toward a desirable advantage for biocontrol application. Although the phage count remained at high levels, but a significant decrease in S. flexneri number was recorded only up to the first 72 h. This finding could be as a result of inefficient diffusion or inhomogeneous distribution of the phage particles in food samples as well as phage trapping among food components (Guenther et al. 2012).
Conclusion
In this study, a novel S. flexneri specific phage (vB_SflS-ISF001) which lyses S. flexneri PTCC1234 and PTCC 1865 was well characterized. Its treatment and prevention potential against food contamination was evaluated. This is a fundamental research on bacteriophage application in controlling and inhibition of growth and survival of Shigella contamination in foodstuff. In conclusion, based on the host range, stability characterization, lysis activity, burst size and food experiments results, vB_SflS-ISF001 showed a great potential to be developed as an alternative strategy against S. flexneri contamination in foodstuff. Moreover, isolation of new virulent phages to make phage cocktail is necessary and also ensures complete elimination of the pathogen in different foods.
Acknowledgements
This research was funded by an operating grant of the Dean of Research and Graduate Studies at the University of Isfahan.
References
- Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152(2):227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- Ahmadi H, Anany H, Walkling-Ribeiro M, Griffiths M. Biocontrol of Shigella flexneri in ground beef and Vibrio cholerae in seafood with bacteriophage-assisted high hydrostatic pressure (HHP) treatment. Food Bioprocess Technol. 2015;8(5):1160–1167. doi: 10.1007/s11947-015-1471-6. [DOI] [Google Scholar]
- Ahmed AM, Shimamoto T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157: H7 and Shigella spp. from meat and dairy products in Egypt. Int J Food Microbiol. 2014;168:57–62. doi: 10.1016/j.ijfoodmicro.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Bao H, Zhang P, Zhang H, Zhou Y, Zhang L, Wang R. Bio-control of Salmonella enteritidis in foods using bacteriophages. Viruses. 2015;7(8):4836–4853. doi: 10.3390/v7082847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Chowdhury N, Khan R, Hasan MR, Nahar J, Islam MJ, Yamasaki S, Ghosh A, Nair GB, Sack DA. Shigella dysenteriae type 1-specific bacteriophage from environmental waters in Bangladesh. Appl Environ Microbiol. 2003;69(12):7028–7031. doi: 10.1128/AEM.69.12.7028-7031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi SM, Bouzari M, Emtiazi G. Preliminary characterization of Lactococcus garvieae bacteriophage isolated from wastewater as a potential agent for biological control of lactococcosis in aquaculture. Aquac Int. 2014;22(4):1469–1480. doi: 10.1007/s10499-014-9760-z. [DOI] [Google Scholar]
- Gu B, Zhou M, Ke X, Pan S, Cao Y, Huang Y, Zhuang L, Liu G, Tong M. Comparison of resistance to third-generation cephalosporins in Shigella between Europe-America and Asia-Africa from 1998 to 2012. Epidemiol Infect. 2015;143(13):2687–2699. doi: 10.1017/S0950268814003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S, Herzig O, Fieseler L, Klumpp J, Loessner MJ. Biocontrol of Salmonella typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int J Food Microbiol. 2012;154(1):66–72. doi: 10.1016/j.ijfoodmicro.2011.12.023. [DOI] [PubMed] [Google Scholar]
- Haq IU, Chaudhry WN, Andleeb S, Qadri I. Isolation and partial characterization of a virulent bacteriophage IHQ1 specific for Aeromonas punctata from stream water. Microb Ecol. 2012;63(4):954–963. doi: 10.1007/s00248-011-9944-2. [DOI] [PubMed] [Google Scholar]
- Harley JP, Prescott LM. Laboratory exercises in microbiology. 5. New York: McGraw Hill; 2002. pp. 105–108. [Google Scholar]
- Jafari F, Hamidian M, Rezadehbashi M, Doyle M, Salmanzadeh-ahrabi S, Derakhshan F, Reza Zali M. Prevalence and antimicrobial resistance of diarrheagenic Escherichia coli and Shigella species associated with acute diarrhea in Tehran, Iran. Can J Infect Dis Med Microbiol. 2009;20(3):e56–e62. doi: 10.1155/2009/341275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakhetia R, Verma NK. Identification and molecular characterisation of a novel Mu-Like bacteriophage, SfMu, of Shigella flexneri. PLoS ONE. 2015;10(4):e0124053. doi: 10.1371/journal.pone.0124053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal M, Chaudhry WN, Hussain T, Das CR, Andleeb S. Characterization of new Myoviridae bacteriophage WZ1 against multi-drug resistant (MDR) Shigella dysenteriae. J Basic Microbiol. 2015;55(4):420–431. doi: 10.1002/jobm.201400688. [DOI] [PubMed] [Google Scholar]
- Jeon J, D’Souza R, Pinto N, Ryu CM, Park J, Yong D, Lee K. Characterization and complete genome sequence analysis of two Myoviral bacteriophages infecting clinical carbapenem-resistant Acinetobacter baumannii isolates. J Appl Microbiol. 2016;121(1):68–77. doi: 10.1111/jam.13134. [DOI] [PubMed] [Google Scholar]
- Jun JW, Kim JH, Shin SP, Han JE, Chai JY, Park SC. Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res Microbiol. 2013;164(10):979–986. doi: 10.1016/j.resmic.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Jun JW, Yun SK, Kim HJ, Chai JY, Park SC. Characterization and complete genome sequence of a novel N4-like bacteriophage, pSb-1 infecting Shigella boydii. Res Microbiol. 2014;165(8):671–678. doi: 10.1016/j.resmic.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Jun JW, Giri SS, Kim HJ, Yun SK, Chi C, Chai JY, Lee BC, Park SC. Bacteriophage application to control the contaminated water with Shigella. Sci Rep. 2016;6:22636. doi: 10.1038/srep22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JW, Kim HJ, Yun SK, Chai JY, Lee BC, Park SC. Isolation and comparative genomic analysis of T1-like Shigella bacteriophage pSf-2. Curr Microbiol. 2016;72(3):235–241. doi: 10.1007/s00284-015-0935-2. [DOI] [PubMed] [Google Scholar]
- Kazi M, Annapure US. Bacteriophage biocontrol of foodborne pathogens. J Food Sci Technol. 2015;53(3):1–8. doi: 10.1007/s13197-015-1996-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay. Bacteriophages Methods Protoc. 2009;1:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- Kropinski AM, Prangishvili D, Lavigne R. Position paper: the creation of a rational scheme for the nomenclature of viruses of bacteria and archaea. Environ Microbiol. 2009;11(11):2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- Löfdahl M, Ivarsson S, Andersson S, Långmark J, Plym-Forshell L. An outbreak of Shigella dysenteriae in Sweden, May–June 2009, with sugar snaps as the suspected source. Eurosurveillance. 2009;14(28):29–35. doi: 10.2807/ese.14.28.19268-en. [DOI] [PubMed] [Google Scholar]
- Mikoleit M. Laboratory protocol: biochemical identification of Salmonella and Shigella using an abbreviated panel of tests. Geneva: WHO Global Foodborne Infections Network; 2010. [Google Scholar]
- Muller L, Jensen T, Petersen R, Mølbak K, Ethelberg S. Imported fresh sugar peas as suspected source of an outbreak of Shigella sonnei in Denmark, April–May 2009. Eurosurveillance. 2009;14(24):218–226. doi: 10.2807/ese.14.24.19241-en. [DOI] [PubMed] [Google Scholar]
- Perera MN, Abuladze T, Li M, Woolston J, Sulakvelidze A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015;52:42–48. doi: 10.1016/j.fm.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Pérez Pulido R, Grande Burgos MJ, Gálvez A, Lucas López R. Application of bacteriophages in post-harvest control of human pathogenic and food spoiling bacteria. Crit Rev Biotechnol. 2015;36(5):851–861. doi: 10.3109/07388551.2015.1049935. [DOI] [PubMed] [Google Scholar]
- Prabhurajeshwar, Oli Ak, Ashajyothi C, Chandrakanth RK. Prevalence and antibiotic susceptibility pattern of fluoroquinolone resistant Shigella species isolated from infants stool in Gulbarga district, Karnataka, India. Asian Pac J Trop Dis. 2015;5:S116–S120. doi: 10.1016/S2222-1808(15)60871-4. [DOI] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shen G-H, Wang J-L, Wen F-S, Chang K-M, Kuo C-F, Lin C-H, Luo H-R, Hung C-H. Isolation and characterization of φkm18p, a novel lytic phage with therapeutic potential against extensively drug resistant Acinetobacter baumannii. PLoS ONE. 2012;7(10):e46537. doi: 10.1371/journal.pone.0046537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva YJ, Costa L, Pereira C, Mateus C, Cunha Â, Calado R, Gomes NC, Pardo MA, Hernandez I, Almeida A. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS ONE. 2014;9(12):e114197. doi: 10.1371/journal.pone.0114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar IB, Loerger RD. Attempts to utilize bacteriophage to combat Salmonella Enterica serovar entemtidis infection in chickens. J Food Saf. 2001;21(1):15–29. doi: 10.1111/j.1745-4565.2001.tb00305.x. [DOI] [Google Scholar]
- Tajbakhsh M, Migura LG, Rahbar M, Svendsen CA, Mohammadzadeh M, Zali MR, Aarestrup FM, Hendriksen RS. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? J Antimicrob Chemother. 2012;67(5):1128–1133. doi: 10.1093/jac/dks023. [DOI] [PubMed] [Google Scholar]
- Taneja N, Mewara A, Kumar A, Verma G, Sharma M. Cephalosporin-resistant Shigella flexneri over 9 years (2001–09) in India. J Antimicrob Chemother. 2012;67(6):1347–1353. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- WHO Shigellosis: disease burden, epidemiology and case management. Wkly Epidemiol Rec. 2005;80:94–99. [PubMed] [Google Scholar]
- Zaidi MB, Estrada-García T. Shigella: a highly virulent and elusive pathogen. Curr Trop Med Rep. 2014;1(2):81–87. doi: 10.1007/s40475-014-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang R, Bao H. Phage inactivation of foodborne Shigella on ready-to-eat spiced chicken. Poult Sci. 2013;92(1):211–217. doi: 10.3382/ps.2011-02037. [DOI] [PubMed] [Google Scholar]