Abstract
Vibrio parahaemolyticus is part of the natural microflora of estuarine and coastal marine waters and can be also present in seafood, especially shellfish and bivalve molluscs. In this study we compared the reference cultural method ISO 6887-3 with two molecular methods, multiplex PCR and real-time PCR, for the detection of two distinct genetic markers (tlh species-specific gene and tdh virulence gene) of V. parahaemolyticus in bivalve mollusc. The analyses were performed on clams inoculated with V. parahaemolyticus ATCC 43996 at T0 and after a 3 and 6 h of pre-enrichment in alkaline saline peptone water. Counts on agar plates were largely inaccurate, probably due to other Vibrio species grown on the TCBS selective agar. Multiplex PCR assays, performed using primers pairs for tdh and tlh genes, showed a detection limit of 104 CFU/g of shell stock within 6 h of pre-enrichment, respecting however the action level indicated by the National Seafood Sanitation Program guideline. Detection by tdh gene in real-time PCR reached the definitely highest sensitivity in shorter times, 101 CFU/g after 3 h of pre-enrichment, while the sensitivity for the tlh gene was not promising, detecting between 105 and 106 CFU/g after 6 h of pre-enrichment. Our findings provide a rapid routine method of detection of V. parahaemolyticus based on tdh gene by real-time PCR for commercial seafood analysis to identify the risk of gastrointestinal diseases.
Electronic supplementary material
The online version of this article (10.1007/s13197-017-2986-9) contains supplementary material, which is available to authorized users.
Keywords: V. parahaemolyticus, Seafood, Detection method multiplex PCR, Real-time PCR
Introduction
Many marine molluscs are harvested around the world for their meat and are considered as important resources that contribute considerable economic value to the world’s fisheries (Leiva and Castilla 2002). In the year 2013, the commercial harvest of at least 9.8 million tons of molluscs was reported as part of the world fisheries catch (FAO 2015) and the demand for global fisheries product, including molluscs, increases every year as the human population grows (Naylor et al. 2000; Diana 2009) leading to exploitation of new stocks (Dey 2015).
This phenomenon requires an improved stringency on the microbiological control, in particular to pathogenic vibrio species. Among these, Vibrio parahaemolyticus, widely distributed in estuarial and coastal marine waters, is often isolated from seawater, sediment and a variety of seafood such as oysters, clams, scallops, octopus, shrimps, crabs, lobsters, crawfish, and various kinds of fish (Raghunath et al. 2008; Shen et al. 2009). It is capable of causing sea-food borne gastroenteritis in humans with diarrhoea, vomiting, abdominal cramps, and fever (Lin et al. 2005). The pathogenicity of V. parahaemolyticus has been associated with thermostable direct haemolysin (TDH) and/or TDH-related haemolysin (TRH) coded by the tdh and/or trh genes, respectively (Honda and Iida 1993; Honda et al. 1987a, b).
According to current European Legislation (EC Reg. n. 2073/2005 and subsequent amendments), the evaluation of shellfish safety is based entirely on Escherichia coli as an indicator of fecal contamination, that is not related to the presence of seawater autochthonous bacteria, such as V. parahaemolyticus (Koh et al. 1994). Several reports indicate that pathogenic V. parahaemolyticus (carrying either tdh or trh genes) are poorly present in environmental and seafood samples (DePaola et al. 2003; Deter et al. 2010; Robert-Pillot et al. 2004) representing, therefore, a minority of the isolates especially in comparison to the prevalence of these genes in clinical isolates (Cook et al. 2002; DePaola et al. 2003; Hayat-Mahmud et al. 2006). As consequence, in the EC Reg. n. 2073/2005 no specific criteria for the detection of pathogenic V. parahaemolyticus in fish and seafood are mentioned, but the need to develop reliable methods for its research is underlined.
Conventional analytical methods to detect the population of virulent Vibrio are particularly time consuming: the method described by the ISO reference foresees an enrichment of the homogenized sample up to 18 h. Another disadvantage is the false-positive or false negative-results, due to the biochemical identification step (Croci et al. 2007; O’Hara et al. 2003). Real-time PCR is a rapid, sensitive and reliable method for detection of pathogens in food samples; detection and/or identification methods based on it have been extensively applied for total and pathogenic Vibrio spp., combining several primers for targeting different genes (Bej et al. 1999) or species (Park et al. 2013). Specific marker genes used to positively identify V. parahaemolyticus by PCR are the toxR, a regulatory gene firstly discovered in V. cholerae (Lin et al. 1993) with a V. parahaemolyticus-specific sequence (Kim et al. 1999); the chromosomal DNA fragment of V. parahaemolyticus named pR72H (Lee et al. 2002); the thermolabile haemolysin gene (tlh) (Bej et al. 1999) encoding a phospholipase A2 (Zhang and Austin 2005), considered a species-specific marker for V. parahaemolyticus and frequently employed to identify this species (DePaola et al. 2003; Nordstrom et al. 2007; Jones et al. 2012); the gyrB gene that encodes the B subunit protein of DNA gyrase (Venkateswaran et al. 1998). However, these genes provide no information regarding pathogenic potential. In fact, among the V. parahaemolyticus species, it is assumed that only the strains carrying the genes tdh and/or trh are implicated in gastroenteritis cases and thus considered as enteropathogenic (Nishibuchi and Kaper 1995; Zhang and Austin 2005). As consequence, as reviewed by Bisha et al. (2012), a number of PCR assays have been employed to affect detection of the tdh or trh genes. Garrido et al. (2012) tested a multiplex real-time PCR with different pathogenic strains of V. parahaemolyticus containing tdh and/or trh pathogenicity genes and results were compared with the cultured-based ISO method.
The aim of this study was to compare the reference cultural method ISO 6887-3 (Anonymous 2003) with two molecular methods, multiplex PCR and real-time PCR, for the detection of pathogenic V. parahaemolyticus in clams. Multiplex PCR and real-time PCR were performed to detect two distinct genetic markers (tlh species-specific gene and tdh virulence gene) of V. parahaemolyticus. In particular, the molecular methods were evaluated in order to increase the rapidity and sensitivity of V. parahaemolyticus detection in the routine laboratory analyses.
Materials and methods
Samples collection
Samples of clams (2 kg) were collected by MARE. A s.r.l. (Cattolica, Italy) during spring and summer 2009. Twenty-five grams of the samples were analysed by plating on TCBS cholera medium agar (Oxoid, Milan, Italy) following the protocol described by ISO/TS 21872-1:2007, in order to determine the absence both of V. parahaemolyticus (smooth and green colonies) and Vibrio cholerae (smooth and yellow colonies). If some samples resulted positively to these Vibrio species the presumptive confirmation was applied by Gram test, oxidase test, motility and halotolerance tests, while the biochemical confirmation was performed by API test (kit API 20NE) (BioMérieux, Marcy l’Etoile, France). The samples tested for the absence of V. parahaemolyticus and V. cholerae were stored at − 20 °C for the artificial inoculation.
Bacterial strain and growth conditions
Vibrio parahaemolyticus ATCC 43996, belonging to the American Type Culture Collection (ATCC, Manassas, VA, USA), was used for all the experiments and as control strain for tdh and tlh genes, after being tested for the presence of the two genes in PCR. V. parahaemolyticus ATCC 43996 was routinely grown on TCBS cholera medium agar (Oxoid, Milan, Italy) at 37 °C, while stock culture was keep at − 80 °C in Nutrient broth (Oxoid) with 15% of glycerol.
Inoculation of clams
According to the ISO 6887-3 (Anonymous 2003), an overnight bacterial culture of V. parahaemolyticus ATCC 43996 was used for artificial inoculation of the clams, as reported in flow diagram (Fig. 1). A loop of V. parahaemolyticus ATCC 43996 was inoculated in Tryptone Soya Broth (TSB, Oxoid) with 1% NaCl and incubated overnight at 37 °C. The culture was centrifuged, re-suspended in sterile physiological saline solution, and the number of bacterial cells was measured spectrophotometrically as optical density at 610 nm, corresponding to about 2 × 107 CFU/ml. Successively, tenfold serial dilutions of bacterial suspension were prepared in physiological solution for a final volume of 3 ml each. Clams were thawed and 25 g of sample (about 10 clams), including intervalvular water as described by the ISO/TS 21872-1 procedures, were homogenized in 225 ml of alkaline saline peptone water (ASPW, constituted by 20 g/L NaCl and 20 g/L peptone, adjusted at pH 8.6 with 5 N NaOH) in a Stomacher 400 circulator (Seaward, PBI). Twenty-seven ml of homogenized clams were inoculated with 3 ml of the bacterial suspension (2 × 107 CFU/ml) and its tenfold serial dilutions according to the ISO 6887-3, obtaining six differently contaminated samples ranging from 106 to 100 CFU/g for a total volume of 30 ml. Each sample was then divided into 3 aliquots, 10 ml each, to be analysed at three different time points: immediately after inoculation (T0), after 3 h (T3) and after 6 h (T6) of pre-enrichment in ASPW at 37 °C. After incubation, each sample was centrifuged at 800 rpm for 5 min for clarification; 500 µl of supernatant were taken for DNA extraction and 2.5 ml aliquots were frozen for further analyses. A positive control was obtained inoculating the homogenized clams with 107 CFU/g of V. parahaemolyticus ATCC 43996.
Fig. 1.
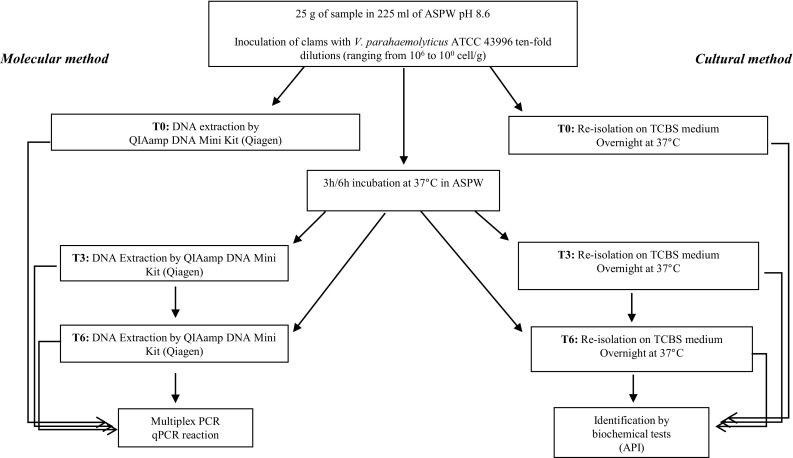
Flow diagram of artificial inoculation protocol in homogenized clams, performed following the ISO 6887-3 instructions (based on Rosec et al. 2012), and subsequent molecular, biochemical and cultural analyses. DNA was extracted from T0, T3 and T6 samples by QIAamp DNA Mini Kit (Qiagen); for V. parahaemolyticus ATCC 43996 the extraction was carried out by InstaGene Matrix (Bio-Rad). ASPW alkaline saline peptone water, TCBS thiosulfate citrate bile salts sucrose
Culture method
Cultural microbiological analyses were assessed on inoculated clams according to ISO 6887-3 method. Briefly, from each sample (T0, T3 and T6), artificially inoculated as described above, 100 µl were serially diluted in sterile physiological saline solution, plated onto TCBS agar (Oxoid) and enumerated after incubation of 24 h at 37 °C. Finally, typical V. parahaemolyticus blue-green colonies were identified by gram-staining and oxidase test and confirmed on the basis of their biochemical activity as described before.
DNA extraction methods
In order to obtain a positive control for PCR methods, DNA of pure cultures of V. parahaemolyticus ATCC 43996 were extracted by a commercially available extraction kit (InstaGene Matrix, Bio-Rad, Milan, Italy). The DNA of the artificially contaminated samples, after 0, 3 and 6 h of pre-enrichment in ASPW, were purified on QIAamp DNA Mini Kit columns (Qiagen, Milan, Italy) following the instructor’s procedure. The DNA was quantified spectrophometrically (OD260 nm) by Biophotometer (Eppendorf, Milan, Italy) and the samples were stored at − 20 °C.
Multiplex PCR
The primers targeting on tdh gene (rtdh 5′-TGG AAT AGA ACC TTC ATC TTC ACC-3′, and ltdh 5′-GTA AAG GTC TCT GAC TTT TGG AC-3′) and tlh gene (rtl, 5′-GCT ACT TTC TAG CAT TTT CTC TGC-3′, and ltl 5′-AAA GCG GAT TAT GCA GAA GCA CTG-3′) (Bej et al. 1999) were furnished by MWG (Milan, Italy). Conventional PCR was performed using tdh and tlh separately, in order to verify the presence of both genes in V. parahaemolyticus ATCC 43996, using the following conditions: 5 min at 95 °C, followed by 30 cycles of 60 s at 94 °C, 60 s at 54 °C and 60 s at 72 °C and final amplification 10 min at 72 °C (Baffone et al. 2006).
Multiplex PCR assay was performed in a final volume 25 μL using the following reaction mixture components (final concentrations): 1 × Green GoTaq® Reaction Buffer, 4 mM MgCl2 (Promega, USA), 0.2 mM of each of the deoxynucleoside triphosphates (Promega, USA), 1 µM each of the tdh and tlh forward and reverse primers (MWG, Milan, Italy) and 1.25 U of GoTaq® DNA polymerase (Promega, USA). DNA template was tested also diluted 1:10 and 1:100 in DNAse-free water. Amplification was performed with the T3000 Thermal Cycler (Biometra, Germany), following the thermal conditions above described. Positive and no template controls were included in each run. PCR products were separated by electrophoresis on 1.5% (w/v) agarose gel (Fermentas, Thermo Scientific, Italy) stained with ethidium bromide (0.5 μg/ml) (Sigma, Milan, Italy).
Detection of pathogenic V. parahaemolyticus by quantitative real-time PCR
In order to optimize real-time PCR procedure, we tested 58° and 60 °C as annealing temperatures, and three different final concentrations of MgCl2, equal to 2.5, 3.5 and 5 mM.
Quantitative real-time PCR was performed in a Rotor-Gene™ 6000 (Corbett, NSW); the PCR reaction mixture contained 1X SYBR® Premix Ex Taq II, 0.4 µM of each primer (MWG, Milan, Italy), MgCl2 2.5 mM, 5 µl of DNA template in a 25 μL final volume. The thermal protocol was 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. Positive and non-template controls were included in each run; each sample was run three times and values reported are the mean of the three repetitions.
Standard curves were obtained by tenfold serial dilutions of standard DNA of V. parahaemolyticus ATCC 43996 (American Type Culture Collection), and used to determine the correspondence of a known concentration with a Ct (Cycle threshold); on the basis of the calibration, the ng/µl of the DNA present in each samples were calculated. All the samples were finally processed for obtaining melting curve analyses by the software RotorGene 6000 version 1787 (Corbett). The amplification efficiency of the reactions was calculated using the slope (s) of the standard curve, and the following equation: E = 10(−1/slope) − 1.
The conversion of the genome equivalents obtained by real-time PCR in the contaminated clams in the amount of V. parahaemolyticus DNA was performed considering that the average genome size for V. parahaemolyticus was estimated 5.2 Mb (annotation from Genome, http://www.ncbi.nlm.nih.gov/genome/, NCBI) resulting in approximately 5.61 fg of DNA for each cell.
Statistical analysis
Statistical analysis was performed by Prism version 5.0 (GraphPad Software, Inc., La Jolla, USA). The assumptions for parametric test were checked prior to carrying out the analysis; when the assumptions for parametric test were not respected (such as for plate count enumeration), Kruskal–Wallis non-parametric test with Dunn’s multiple comparison test was applied. P values < 0.05 were considered to be statistically significant.
Results
Cultural enumeration of V. parahaemolyticus in artificially inoculated clams
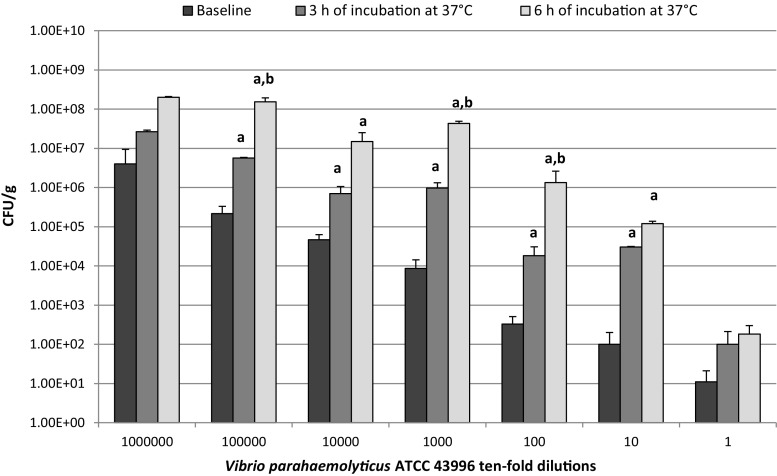
Clams artificially inoculated as described by the ISO-6887-3 were analysed by cultural method on TCBS agar after 0, 3 and 6 h of pre-enrichment in ASPW at 37 °C (Fig. 1). As shown in Fig. 2, after 3 h of incubation an increase in CFU/g values, with several fluctuations, was observed in all the tenfold serial dilution samples analysed compared to the relative initial cell load (T0) (P < 0.05). The increase was more remarkable after 6 h of incubation (T6) with values ranging from 1.3 × 106 to 2 × 108 CFU/g in 102 and 106 tenfold serial dilution samples respectively (P < 0.05).
Fig. 2.
Enumeration by standard plate count agar at baseline and after 3 and 6 h of incubation at 37 °C of V. parahemolyticus ATCC 43996 tenfold dilutions artificially inoculated in clams
Detection limit of multiplex end-point PCR
Multiplex end-point PCR was initially performed on DNA extracted from V. parahaemolyticus ATCC 43996 strain, giving the expected bands of 296 bp for tdh gene and 450 bp for tlh gene. Then, the same protocol was performed on the DNA obtained from the artificially inoculated clams samples after 0, 3 and 6 h of pre-enrichment incubation. The multiplex PCR on T0 samples gave a detectable signal until 105 CFU/ml of tenfold dilution sample (Fig. 3a, lane 6), while after 3 h of pre-enrichment a signal was visible at 104 CFU/ml (Fig. 3b, lane 5); the last detectable signal after 6 h of pre-enrichment showed a detection limit of 103 CFU/ml of shell stock (Fig. 3c, lane 4).
Fig. 3.
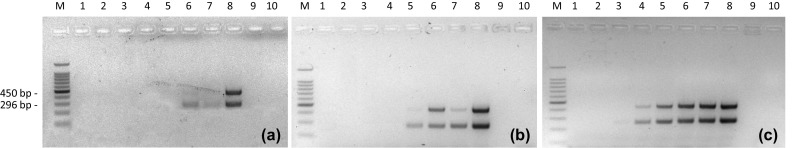
Multiplex PCR performed on tdh (296 bp) and tlh (450 bp) genes on DNA extracted from clams artificially inoculated with increasing concentrations of V. parahaemolyticus ATCC 43996 tenfold serial dilutions (M: 100 bp ladder, Catalogue number G2101, Promega; lane 1: 100 CFU/ml of tenfold dilution; lane 2: 101 CFU/ml; lane 3: 102 CFU/ml; lane 4: 103 CFU/ml; lane 5: 104 CFU/ml; lane 6: 105 CFU/ml; lane 7: 106 CFU/ml; lane 8: positive control; lane 9: negative control; line 10: non-template control) after 0 h (a), 3 h (b) and 6 h (c) of pre-enrichment at 37 °C
Detection sensitivity of real-time PCR
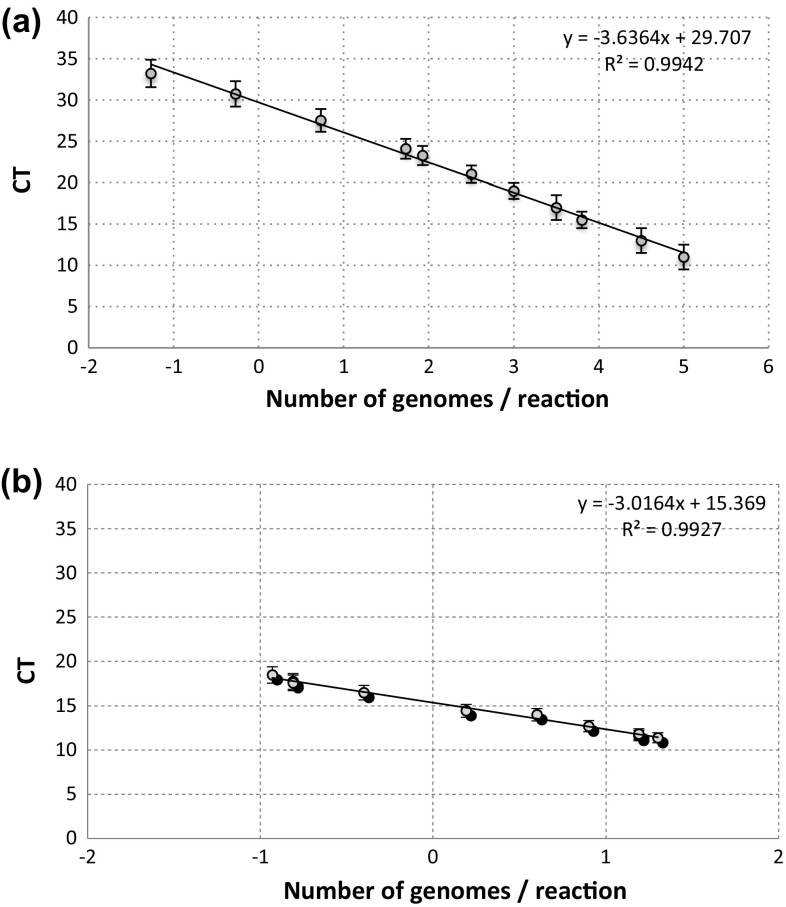
In order to assess the sensitivity of our method of extraction, we used two sets of primers, tdh and tlh to enumerate CFU of V. parahaemolyticus ATCC 43996 in artificially contaminated samples. In Fig. 4a, b, the curves for the tdh and tlh primers are reported; the limits of sensitivity of V. parahaemolyticus real-time PCR were determined from end point titration of DNA extracted from pure culture. The best results of amplification for both primers were obtained using 60 °C as annealing temperature, and 2.5 mM of MgCl2 (data not shown), giving for tdh gene a slope of the curve equal to − 3.63 (E = 1.14), while for tlh gene the slope was − 2.97 (E = 1.17). For both primers used, a linear relationship was observed between the Ct values and the standard quantity of cells inoculated.
Fig. 4.
Standard curves for the amount of purified genomic DNA of V. parahaemolyticus ATCC 43996, amplified with tdh (a) and tlh (b) primer pairs, versus CT value. The error bars indicate the standard deviations obtained in three independent experiments. CT, threshold cycle (i.e., the cycle at which a statistically significant increase in fluorescent signal is first detected)
The melting temperatures of the amplified products from tdh and tlh of V. parahaemolyticus were 80.5 and 89.5 °C respectively (Fig. 1S, supplementary data). Fluorescent signals for the negative control strains were under the threshold level. These real-time PCR results were used to obtain the calibration curves that allowed the V. parahaemolyticus quantification on the basis of tdh gene amplification. To evaluate the effect of the matrix on the extraction protocol and PCR reaction, the Ct values obtained from the extraction of artificially inoculated clams and of pure culture at the same concentration of V. parahaemolyticus were compared. The identical curves obtained with the artificially contaminated clams and pure culture showed no interference of matrix on the PCR amplification (data not shown).
In order to determine the detection limit of real-time PCR, DNA isolated from tenfold serial dilutions of inoculated clams (from 100 to 106 CFU/ml), immediately after the infection and after 3 and 6 h of the pre-enrichment were analysed (Fig. 5a–c respectively). The results of qPCR for the tdh gene showed positive signals for all the tenfold dilutions tested, and are reported in Table 1. The data evidenced a clear and robust quantification of V. parahaemolyticus in the inoculated clams containing tenfold serial dilutions of 105 and 106 CFU/ml, immediately after the inoculum (T0) (respectively quantified as 1.65E−03 and 8.02E−03 ng/µl). After 3 h of pre-enrichment, a good signal was obtained also in correspondence of the lowest tenfold dilution (100 CFU/ml, quantified by real-time PCR as 1.39E−05 ng). These results were consolidated also after 6 h of the pre-enrichment in all the tenfold serial dilution tested, where the lowest tenfold dilution was quantified as 1.18E−04 ng/µl. The results concerning the gene tlh gene evidenced the presence of V. parahaemolyticus only after 6 h of pre-enrichment with tenfold dilutions of 104 and 105 CFU/ml (data not shown).
Fig. 5.
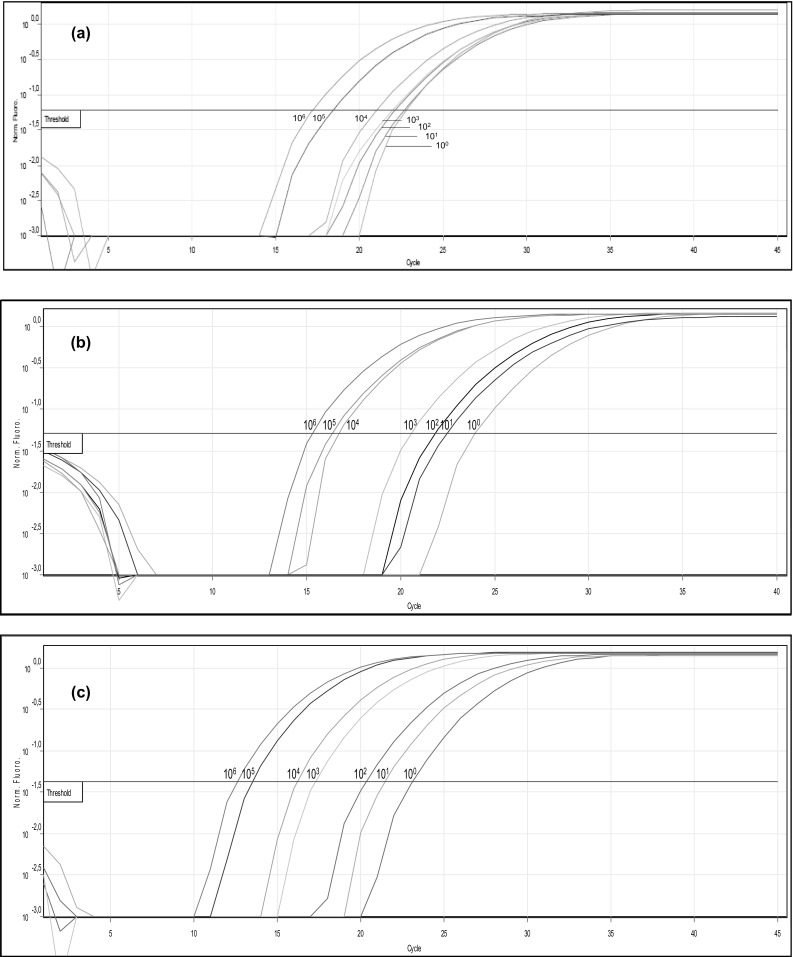
Fluorescent amplification curves for the tenfold dilution series of genomic DNA purified from clams artificially inoculated with V. parahaemolyticus ATCC 43996 and amplified by tdh primers at different T0 (a), T3 (b) and T6 (c) pre-enrichment incubation time
Table 1.
DNA quantification in real-time PCR by primers targeting tdh gene of Vibrio parahaemolyticus tenfold serial dilutions artificially inoculated in clams, assessed at baseline and after 3 and 6 h (T0, T3, T6) of pre-enrichment
| Vibrio parahaemolyticus inocula (CFU/ml) | Time of analysis | ||
|---|---|---|---|
| T0 | T3 | T6 | |
| 100 | 6.47E−06 | 1.39E−05 | 1.18E−04 |
| 101 | 8.05E−06 | 8.34E−05 | 6.51E−04 |
| 102 | 2.32E−05 | 1.48E−04 | 2.22E−03 |
| 103 | 5.61E−05 | 7.68E−04 | 9.46E−03 |
| 104 | 2.22E−04 | 6.09E−03 | 2.22E−03 |
| 105 | 1.65E−03 | 1.95E−02 | 6.07E−02 |
| 106 | 8.02E−03 | 1.01E−01 | 1.29E−01 |
The values are expressed in ng/µl
Discussion
V. parahaemolyticus is a common contaminant of sea water, sediments and seafood; though, not all the strains are recognized as pathogenic: thermostable direct haemolysin (TDH) and/or TDH-related haemolysin production have been associated with the pathogenicity of V. parahaemolyticus (Honda et al. 1987a, b; Honda and Iida 1993). Cultural standard methods don’t allow differentiation between pathogenic and non-pathogenic strains, bringing to unjustified waste of seafood batches, besides presenting the well-known limitations (time loss, variability given by operator, identification limited to species level).
Our results of V. parahaemolyticus enumeration on TCBS from artificially inoculated clams, showed an overestimation of CFU/ml due to the observed fluctuating data of the different inoculated samples. The presumptive V. parahaemolyticus colonies were screened for colour, and only the green ones were considered, but we can’t exclude the presence of other autochthonous bacteria including other Vibrio spp. In fact differentiation of Vibrio species on this medium is essentially due on sucrose fermentation, thus resulting in possible misleading among sucrose fermenting (V. cholerae with some strains of V. fluvialis or V. alginolyticus) or sucrose non-fermenting species (V. parahaemolyticus with V. mimicus or V. vulnificus) (Eddabra et al. 2011). Oppositely to our results, Kim and Lee (2014) who compared counts on TCBS agar plates with the sum of three Vibrio spp. counts obtained with different sets of primers by real-time PCR, observed a lower count on TCBS agar compared with real-time. This discrepancy could however be explained as TCBS, although selective for Vibrio spp., doesn’t allow growth of injured bacterial cell because of selective agents (Oliver et al. 1991). The same paper also reports that total bacterial counts performed on a rich medium as TSA gave comparable results with real-time PCR targeting the 16S rRNA gene, considering real-time PCR a remarkably more practical and sensitive method.Another disadvantage is represented by the following biochemical identification step (Croci et al. 2007; O’Hara et al. 2003) that can lead to false-positive or false negative-results and prolonged the identification time.
For the above mentioned limitations of counts on agar plate, several molecular approaches have been developed to detect this pathogen in seafood and marine samples, also using multiple sets of primers to detect different pathogenic species together (Kim and Lee 2014; Garrido-Maestu et al. 2014; Messelhäusser et al. 2010; Park et al. 2013). Nevertheless, molecular methods, not still considered by regulatory agencies, should require a fast and simple approach, in case of variable competences of the operative staff; moreover, considering the short shelf life of such food products (Garrido-Maestu et al. 2014) it would be advantageous completing analyses within a workday. The currently proposed PCR-based protocols, before DNA extraction from food matrix, require as enrichment process ranging between 18 and 24 h (Garrido-Maestu et al. 2014; Jones et al. 2012; Messelhäusser et al. 2010); in Tall et al. (2012) 4 subsequent steps of 24 h at 37 °C are required.
To our knowledge, not many papers focused on the optimization of the most rapid and feasible method for routine analyses on seafood. In our work, the results obtained by cultural method were compared to two different molecular methods, a multiplex end-point PCR and a real-time PCR, in samples incubated up to 6 h, in order to set the protocol in shorter times. The tlh gene is species-specific for V. parahaemolyticus (Bej et al. 1999), and the tdh gene, necessary to quantify pathogenic V. parahaemolyticus cells, was largely used by several authors (Garrido et al. 2012; He et al. 2014; Rosec et al. 2012; Zhu et al. 2012). In particular, Bej et al. (1999) provided a simple method for species-specific identification and pathogenic genes detection of V. parahaemolyticus in clams within 8 h of pre-enrichment within the level of 104 cfu per 1 g of shell stock, suggested by the National Seafood Sanitation Program guideline (NSSP 1997). In our investigation, the use of a multiplex PCR detected the tlh and tdh genes after 6 h of pre-enrichment, with a detection limit (103 CFU/ml) under the range recommended by the NSSP guidelines (1997). Therefore, the obtained reduction of pre-enrichment time to 6 h can allow the determination of contamination level in short times, if compared to the traditional cultural-based techniques in use. Some authors report that minimal infective dose of V. parahaemolyticus for a human infection is 105 CFU/g (Barker 1974); therefore, it is not reasonable to reject contaminated food detecting under this level (Messelhäusser et al. 2010). Our results could confirm the suitability of the multiplex PCR in laboratory for routine analyses on seafood, also when performed by untrained staff.
The real-time PCR, eliminating post-amplification analysis, further shortens times of analyses, providing quantitative data and improving the detection limit of about 100 times (Campbell and Wright 2003). Real-time PCR allowed at the same time to reduce the detection time of tdh gene up to 3 h of pre-enrichment and to increase the sensitivity up to 100 CFU/g of V. parahaemolyticus in clams, detected as 1.39E−05 ng/µl. Taking into account that, as recently reported by Suffredini et al. (2014), the average contamination levels in Italy for total V. parahaemolyticus were 84 and 73 CFU/g respectively for Sardinia and Veneto regions, our findings confirmed that this assay, developed for the gene of pathogenicity tdh, can be identified as a sensitive method for the detection of pathogenic V. parahaemolyticus in complex food matrices. Concerning the real-time PCR performed for tlh primer, the results were not promising; in fact, this primer pair cannot detect the presence of V. parahaemolyticus before the 6 h of pre-enrichment and the presence of tlh gene was determined only in the higher serial tenfold dilution (106 CFU/ml, data not shown).
Nordstrom et al. (2007) optimized a multiplex real-time PCR assay able to detected in food, clinical and environmental samples less than 10 CFU/reaction of pathogenic V. parahaemolyticus (tdh- and/or trh-positive) in presence of 104 CFU/reaction of total V. parahaemolyticus bacteria after overnight incubation. Park et al. (2013) optimized a multiplex real-time PCR for the simultaneous detection of V. cholerae, V. vulnificus and V. parahaemolyticus on environmental samples (seawater and plankton), obtaining a detection limit of 104 CFU/reaction for all the three species after an enrichment time of 6–8 h. Messelhäusser et al. (2010) reported for their method a sensitivity of 14 and 20 CFU/mL after a 24 h enrichment at 37 °C in multiplex real-time PCR, and an efficiency of standard curve between 97 and 98.3%, although using different primers.
Rizvi and Bej (2010) reported the positive result for an initial inoculum of 101 CFU of V. parahaemolyticus in oysters obtained at Ct value equal to 27.42, after 5 h of incubation; we could obtain a positive result for the 100 inoculum at Ct 20, after 6 h of incubation. A very low limit of detection, 4.8 fg or 1 CFU per reaction, was reached in real-time for purified V. parahaemolyticus genomic DNA and from pure culture respectively, after 10 h of incubation by Liu et al. (2012), but in naturally contaminated seafood samples only an indication of positive or negative result was reported.
Conclusion
The detection of Vibrio in clams is necessary to prevent infection that can lead to gastrointestinal disorders to humans, especially associated to V. parahaemolyticus and can be performed using biomolecular techniques in support of cultural method. A quick identification of this pathogen as contaminant of seafood, when present over the limits given by the regulatory agencies, becomes essential in case of VBNC cells, that were reported as able of resuscitation when stressful conditions are removed (Bates and Oliver 2004), or in case of an upshift of temperature to 25 °C (Wong et al. 2004). For this, we optimized a DNA extraction and employed multiplex end-point PCR and real-time PCR methods for the detection of Vibrio parahaemolyticus from a complex food matrix. Among the two molecular methods, multiplex-PCR showed high sensitivity allowing the detection of genes for identification and pathogenicity of V. parahaemolyticus in concentrations under the range recommended by the regulatory guidelines for this pathogen within 6 h of pre-enrichment. On the other hand, real-time PCR remarkably increased the sensitivity of the method and abolished post-amplification analyses, however not allowing the detection of the species-specific gene tlh.
In conclusion, we can recommend a careful evaluation of well ascertained molecular techniques, in order to optimize protocols for the best time requirements and sensitivity, that should satisfy food safety agencies. The high sensitivity, specificity and reproducibility of molecular techniques suggest that they could realistically replace cultural methods for routine analysis of commercial seafood samples to identify the risk of gastrointestinal diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The Authors would like to thank MARE. A s.r.l. (Cattolica, FC, Italy) to have provided the clam samples.
Footnotes
Sara Federici and Diana I. Serrazanetti have contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1007/s13197-017-2986-9) contains supplementary material, which is available to authorized users.
References
- Anonymous (2003) ISO 6887–3: 2003 Microbiology of food and animal feeding stuffs—preparation of test samples, initial suspension and decimal dilutions for microbiological examination—Part 3: Specific rules for the preparation of fish and fishery products. International Organization for Standardization (ISO) 1, Geneva, Switzerland
- Baffone W, Tarsi R, Pane L, Campana R, Repetto B, Mariottini GL, Pruzzo C. Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea (Italy) and study of their pathogenicity-associated properties. Environ Microbiol. 2006;8:1299–1305. doi: 10.1111/j.1462-2920.2006.01011.x. [DOI] [PubMed] [Google Scholar]
- Barker WHJ. Vibrio parahaemolyticus outbreaks in the United States. Lancet I. 1974;303:551–554. doi: 10.1016/S0140-6736(74)92727-5. [DOI] [PubMed] [Google Scholar]
- Bates TC, Oliver JD. The viable but nonculturable state of Kanagawa positive and negative strains of Vibrio parahaemolyticus. J Microbiol. 2004;42:74–79. [PubMed] [Google Scholar]
- Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tlh, tdh, and trh. J Microbiol Met. 1999;36:215–225. doi: 10.1016/S0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Bisha B, Simonson J, Janes M, Bauman K, Goodrige LD. A review of the current status of cultural and rapid detection of Vibrio parahaemolyticus. Int J Food Sci Technol. 2012;47:885–899. doi: 10.1111/j.1365-2621.2012.02950.x. [DOI] [Google Scholar]
- Campbell MS, Wright AC. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl Environ Microb. 2003;69:7137–7144. doi: 10.1128/AEM.69.12.7137-7144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DW, Bowers JC, DePaola A. Density of total and pathogenic (tdh +) Vibrio parahaemolyticus in Atlhantic and Gulf coast molluscan shellfish at harvest. J Food Protect. 2002;65:1873–1880. doi: 10.4315/0362-028X-65.12.1873. [DOI] [PubMed] [Google Scholar]
- Croci L, Suffredini E, Cozzi L, Toti L, Ottaviani D, Pruzzo C, Serratore P, Fischetti R, Goffredo E, Loffredo G, Mioni R. Comparison of different biochemical and molecular methods for the identification of Vibrio parahaemolyticus. J Appl Microbiol. 2007;102:229–237. doi: 10.1111/j.1365-2672.2006.03046.x. [DOI] [PubMed] [Google Scholar]
- DePaola A, Nordstrom J, Bowers C, Wells J, Cook D. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol. 2003;69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deter J, Lozach S, Veron A, Chollet J, Derrien A, Hervio-Heath D. Ecology of pathogenic and non-pathogenic Vibrio parahaemolyticus on the French Atlantic coast. Effects of temperature, salinity, turbidity and chlorophylla. Environ Microbiol. 2010;12:929–937. doi: 10.1111/j.1462-2920.2009.02136.x. [DOI] [PubMed] [Google Scholar]
- Dey MM. World and U.S. demand and supply relationships for seafood: implications for aquaculture producers. Aquac Mag. 2015;41(2):44–45. [Google Scholar]
- Diana JS. Aquaculture production and biodiversity conservation. J Biosci. 2009;59:27–38. doi: 10.1525/bio.2009.59.1.7. [DOI] [Google Scholar]
- Eddabra R, Piemont Y, Scheftel JM. Evaluation of a new chromogenic medium, chromID Vibrio, for the isolation and presumptive identification of Vibrio cholerae and Vibrio parahaemolyticus from human clinical specimens. Eur J Clin Microbiol. 2011;30:733–737. doi: 10.1007/s10096-010-1145-2. [DOI] [PubMed] [Google Scholar]
- FAO . Fishery statistical collections: global capture production. Rome: Food and Agriculture Organization of the United Nations; 2015. [Google Scholar]
- Garrido A, Chapela MJ, Ferreira M, Atanassova M, Fajardo P, Lago J, Vieites JM, Lago J, Vieites JM, Cabado AG. Development of a multiplex real-time PCR method for pathogenic Vibrio parahaemolyticus detection (tdh + and trh +) Food Control. 2012;24:128–135. doi: 10.1016/j.foodcont.2011.09.015. [DOI] [Google Scholar]
- Garrido-Maestu A, Chapela MJ, Peñaranda E, Vieites JM, Cabado AG. In-house validation of novel multiplex real-time PCR gene combination for the simultaneous detection of the main human pathogenic vibrios (Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus) Food Control. 2014;37:371–379. doi: 10.1016/j.foodcont.2013.09.026. [DOI] [Google Scholar]
- Hayat-Mahmud Z, Kassu A, Mohammad A, Yamato M, Bhuiyan NA, BalakrishNair G, Ota F. Isolation and molecular characterization of toxigenic Vibrio parahaemolyticus from the Kii Channel, Japan. Microbiol Res. 2006;161:25–37. doi: 10.1016/j.micres.2005.04.005. [DOI] [PubMed] [Google Scholar]
- He P, Chen Z, Luo J, Wang H, Yan Y, Chen L, Gao W. Multiplex real-time PCR assay for detection of pathogenic Vibrio parahaemolyticus strains. Mol Cell Probe. 2014;28:246–250. doi: 10.1016/j.mcp.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of thermostable direct haemolysin and related haemolysins. Rev Med Microbiol. 1993;4:106–113. doi: 10.1097/00013542-199304000-00006. [DOI] [Google Scholar]
- Honda S, Goto I, Minematsu I, Ikeda I, Asano N, Ishibashi M, Kinoshita Y, Nishibuchi M, Honda T, Miwatani T. Vibrio parahaemolyticus infectious disease caused by Kanagawa phenomenon-negative O3:K6 originated from Maldives. Jpn J Infect Dis. 1987;61:1070–1078. doi: 10.11150/kansenshogakuzasshi1970.61.1070. [DOI] [PubMed] [Google Scholar]
- Honda S, Goto I, Minematsu I, Ikeda I, Asano N, Ishibashi M, Kinoshita Y, Nishibuchi M, Honda T, Miwatani T. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet. 1987;29:331–332. doi: 10.1016/S0140-6736(87)92062-9. [DOI] [PubMed] [Google Scholar]
- Jones JL, Ludeke CM, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol. 2012;51:2343–2352. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Lee JL. Multipurpose assessment for the quantification of Vibrio spp. and total bacteria in fish and seawater using multiplex real-time polymerase chain reaction. J Sci Food Agric. 2014;94:2807–2817. doi: 10.1002/jsfa.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EG, Huyn JH, LaRock PA. Pertinence of indicator organisms and sampling variables to Vibrio concentrations. Appl Environ Microbiol. 1994;60:3897–3900. doi: 10.1128/aem.60.10.3897-3900.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SKY, Wang HZ, Law SHW, Wu RSS, Kong RYC. Analysis of the 16S–23S rDNA intergenic spacers (IGSs) of marine vibrios for species-specific signature DNA sequences. Mar Pollut Bull. 2002;44:412–420. doi: 10.1016/S0025-326X(01)00256-9. [DOI] [PubMed] [Google Scholar]
- Leiva GE, Castilla JC. A review of the world marine gastropod fishery: evolution of catches, management and the Chilean experience. Rev Fish Biol Fish. 2002;11:283–300. doi: 10.1023/A:1021368216294. [DOI] [Google Scholar]
- Lin ZK, Kumagai K, Baba J, Mekalanos J, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1993;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Labbe RG, Shetty K. Inhibition of Vibrio parahaemolyticus in seafood systems using oregano and cranberry phytochemical synergies and lactic acid. Innov Food Sci Emerg. 2005;6:453–458. doi: 10.1016/j.ifset.2005.04.002. [DOI] [Google Scholar]
- Liu B, He X, Chen W, Yu S, Shi C, Zhou X, Chen J, Wang D, Shi X. Development of a real time PCR assay for rapid detection of Vibrio parahaemolyticus from seafood. Protein Cell. 2012;3:204–212. doi: 10.1007/s13238-012-2017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messelhäusser U, Colditz J, Thärigen D, Kleih W, Höller C, Busch U. Detection and differentiation of Vibrio spp. in seafood and fish samples with cultural and molecular methods. Int J Food Microbiol. 2010;142:360–364. doi: 10.1016/j.ijfoodmicro.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Naylor RL, Goldburg RJ, Primavera JH, Kautsky N, Beveridge MCM, Clay J, et al. Effect of aquaculture on world fish supplies. Nature. 2000;405:1017–1024. doi: 10.1038/35016500. [DOI] [PubMed] [Google Scholar]
- Nishibuchi M, Kaper JB. Thermostable direct haemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;63:2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom JL, Vickery MCL, Blackstone GM, Murray SL, DePaola A. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol. 2007;73:5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSSP: National Shellfish Sanitation Program (1997) Sanitation of the harvesting, processing and distribution of shellfish. In: Manual of Operations Part II, U.S. Department of Health and Human Services, Food and Drug Administration, Washington, DC
- O’Hara CM, Sowers EG, Bopp CA, Duda SB, Strockbine NA. Accuracy of six commercially available systems for identification of members of the family Vibrionaceae. J Clin Microbiol. 2003;41:5654–5659. doi: 10.1128/JCM.41.12.5654-5659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, Nilsson L, Kjelleberg S. Formation of nonculturable vulnificus cells and its relationship to the starvation state. Appl Environ Microbiol. 1991;57:2640–2644. doi: 10.1128/aem.57.9.2640-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Jeon S, Kim JY, Park M, Kim S. Multiplex real-time polymerase chain reaction assays for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Osong Public Health Res Perspect. 2013;4:133–139. doi: 10.1016/j.phrp.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunath P, Acharya S, Bhanumathi A, Karunasagar I. Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 2008;25:824–830. doi: 10.1016/j.fm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Rizvi AV, Bej AK. Multiplexed real-time PCR amplification of tlh, tdh and trh genes in Vibrio parahaemolyticus and its rapid detection in shellfish and Gulf of Mexico water. Antonie Van Leeuwenhoek. 2010;98:279–290. doi: 10.1007/s10482-010-9436-2. [DOI] [PubMed] [Google Scholar]
- Robert-Pillot A, Guenole A, Lesne J, Delesmont R, Fournier JM, Quilici ML. Occurrence of the tdh and trh genes in Vibrio parahaemolyticus isolates from waters and raw shellfish collected in two French coastal areas and from seafood imported into France. Int J Food Microbiol. 2004;91:319–325. doi: 10.1016/j.ijfoodmicro.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Rosec JP, Causse V, Cruz B, Rauzier J, Carnat L. The international standard ISO/TS 21872-1 to study the occurence of total and pathogenic Vibrio parahaemolyticus and Vibrio cholerae in seafood: its improvement by use of a chromogenic medium and PCR. Int J Food Microbiol. 2012;157:189–194. doi: 10.1016/j.ijfoodmicro.2012.04.026. [DOI] [PubMed] [Google Scholar]
- Shen XS, Cai YQ, Liu CC, Liu WW, Hui YH, Su YC. Effect of temperature on uptake and survival of Vibrio parahaemolyticus in oysters (Crassostrea plicatula) Int J Food Microbiol. 2009;136:129–132. doi: 10.1016/j.ijfoodmicro.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Suffredini E, Cozzi L, Ciccaglioni G, Croci L. Development of a colony hybridization method for the enumeration of total and potentially enteropathogenic Vibrio parahaemolyticus in shellfish. Int J Food Microbiol. 2014;186:22–31. doi: 10.1016/j.ijfoodmicro.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Tall A, Teillon A, Boisset C, Delesmont R, Touron-Bodilis A, Hervio-Heath D. Real-time PCR optimization to identify environmental Vibrio spp. strains. J Appl Microbiol. 2012;113:361–372. doi: 10.1111/j.1365-2672.2012.05350.x. [DOI] [PubMed] [Google Scholar]
- Venkateswaran K, Dohmoto N, Harayama S. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol. 1998;64:681–687. doi: 10.1128/aem.64.2.681-687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HC, Wang P, Chen SY, Chiu SW. Resuscitation of viable but non-culturable Vibrio parahaemolyticus in a minimum salt medium. FEMS Microbiol Lett. 2004;233:269–275. doi: 10.1111/j.1574-6968.2004.tb09491.x. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Austin B. Haemolysins in Vibrio species. J Appl Microbiol. 2005;98:1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Zhu RG, Li TP, Jia YF, Song LF. Quantitative study of viable Vibrio parahaemolyticus cells in raw seafood using propidium monoazide in combination with quantitative PCR. J Microbiol Meth. 2012;90:262–266. doi: 10.1016/j.mimet.2012.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.