Abstract
The PITX1 transcription factor is expressed during hindlimb development, where it plays a critical role in directing hindlimb growth and the specification of hindlimb morphology. While it is known that PITX1 regulates hindlimb formation, in part, through activation of the Tbx4 gene, other transcriptional targets remain to be elucidated. We have used a combination of ChIP-seq and RNA-seq to investigate enhancer regions and target genes that are directly regulated by PITX1 in embryonic mouse hindlimbs. In addition, we have analyzed PITX1 binding sites in hindlimbs of Anolis lizards to identify ancient PITX1 regulatory targets. We find that PITX1-bound regions in both mouse and Anolis hindlimbs are strongly associated with genes implicated in limb and skeletal system development. Gene expression analyses reveal a large number of misexpressed genes in the hindlimbs of Pitx1−/− mouse embryos. By intersecting misexpressed genes with genes that have neighboring mouse PITX1 binding sites, we identified 440 candidate targets of PITX1. Of these candidates, 68 exhibit ultra-conserved PITX1 binding events that are shared between mouse and Anolis hindlimbs. Among the ancient targets of PITX1 are important regulators of cartilage and skeletal muscle development, including Sox9 and Six1. Our data suggest that PITX1 promotes chondrogenesis and myogenesis in the hindlimb by direct regulation of several key members of the cartilage and muscle transcriptional networks.
Keywords: PITX1, Hindlimb, Anolis, Mouse, Chondrogenesis, Myogenesis
1. Introduction
The Pitx1 gene encodes a bicoid-class homeodomain transcription factor that plays a central role in growth and patterning of the vertebrate hindlimb (Lanctôt et al., 1999; Szeto et al., 1999). The complete ablation of Pitx1 function in mice results in reduced hindlimb size and the loss of hindlimb-specific features, as well as developmental defects of the mandible, teeth, palate, and pituitary gland. Haploinsufficiency for Pitx1 in mice and humans can result in clubfoot and other malformations of the leg, demonstrating that hindlimb development is sensitive to Pitx1 dosage (Alvarado et al., 2011; Gurnett et al., 2008). Since ectopic expression of Pitx1 in the developing forelimbs of chickens, mice, and humans results in the forelimb developing a more hindlimb-like morphology (Delaurier et al., 2006; Logan and Tabin, 1999; Spielmann et al., 2012), it is apparent that the role of Pitx1 in hindlimb formation extends to the control of limb-type identity. Furthermore, reductions in Pitx1 expression have been linked to the evolution of pelvic fin loss in natural populations of threespine sticklebacks and to the formation of wing-like feathers on the hindlimbs of certain breeds of domesticated pigeon (Chan et al., 2010; Domyan et al., 2016; Shapiro et al., 2004). Thus, Pitx1 is not only important for the formation of the hindlimb, but changes in Pitx1 hindlimb expression have contributed to the evolution of differences in hindlimb morphology.
In Pitx1 knockout mice, impaired hindlimb development is apparent by embryonic day 10.5 (E10.5), by which point there is a clear reduction in hindlimb bud size relative to wild-type embryos (Marcil et al., 2003). In Pitx1 null embryos, this reduction in hindlimb size is more severe in the right hindlimb than the left due to partial compensation by Pitx2, a paralog of Pitx1 which is expressed in the left lateral plate mesoderm during early embryogenesis. At later stages of hindlimb development, dramatic morphological alterations to the hindlimb skeleton are evident in Pitx1 null embryos. For instance, the ilium of the pelvis is either completely missing or present as a small rudiment, femur and tibia dimensions are greatly reduced, the patella is absent, and the relative proportions of the fibula and tibia are shifted to become more similar in size (Lanctôt et al., 1999; Szeto et al., 1999). Moreover, alterations to the hindlimb skeleton of Pitx1 knockout mice are not limited to changes in the position, shape, and size of the hindlimb bones, but also include a decrease in mineralization and reduced formation of extracellular matrix. Finally, the development of soft tissues is also impacted in the Pitx1 knockout embryos with patterning alterations that include shifts in the size and position or even the complete loss of certain hindlimb muscles and tendons (Delaurier et al., 2006).
Initial studies of Pitx1 mouse mutants determined that hindlimb expression of the Tbx4 gene is significantly reduced in the absence of Pitx1 function (Lanctôt et al., 1999; Szeto et al., 1999). Additional work showed that the ectopic expression of Pitx1 in the embryonic forelimb is sufficient to induce Tbx4 expression (Delaurier et al., 2006; Logan and Tabin, 1999). Our further investigations revealed that PITX1 binds to and regulates hindlimb enhancers of the Tbx4 gene, demonstrating that Tbx4 is a direct transcriptional target of PITX1 (Infante et al., 2013). Since Tbx4 encodes a T-box transcription factor that is critical for hindlimb bud outgrowth (Naiche and Papaioannou, 2003), it was proposed that reduced Tbx4 expression contributes to the hindlimb phenotypes found in Pitx1 knockout mice. Consistent with this hypothesis, subsequent work demonstrated that restoration of Tbx4 expression in Pitx1 null embryos is sufficient to rescue hindlimb growth defects and restore femur length, tibia length, and formation of the ilium (Duboc and Logan, 2011; Ouimette et al., 2010). However, restoring Tbx4 expression does not rescue other hindlimb patterning defects of the skeleton, muscles, and tendons. Therefore, the misregulation of additional PITX1 transcriptional targets likely contributes to Pitx1 mutant phenotypes. While we have shown that the PITX1 transcription factor binds to a large number of limb cis-regulatory elements and may promote hindlimb formation by regulating many different genes, Tbx4 remains the only well-validated direct, regulatory target of PITX1 (Infante et al., 2013).
In this study, we apply a comparative ChIP-seq approach to discover deeply conserved PITX1 binding events that occur in the embryonic hindlimbs of mice and Anolis lizards. We find that ancient PITX1 binding sites are enriched near many limb patterning genes and near components of the Hedgehog, BMP, and WNT signaling pathways. We also find that PITX1 binding events are enriched near genes that are misregulated in the hindlimbs of Pitx1 knockout mice. We compare the location of PITX1 binding events to the position of genes that exhibit PITX1-dependent expression to reveal putative direct transcriptional targets of PITX1. Our results suggest that PITX1 promotes hindlimb development through ancient regulatory interactions with several key members of the chondrocyte and muscle transcriptional networks.
2. Materials and methods
2.1. Animals
Pitx1 knockout mice were previously described (Szeto et al., 1999). The Pitx1 knockout allele was maintained on a 129/Sv background but outcrossed onto an outbred ICR background (Envigo) for the generation of embryos for RNA-seq and in situ hybridization experiments. Adult Anolis carolinensis were purchased from Candy’s Quality Reptiles (Reserve, LA, USA), housed at the University of Georgia, and bred to produce embryos. All procedures involving animals were performed in accordance with guidelines issued by the Institutional Animal Care and Use Committees (IACUC) at the University of Georgia under approved Animal Use Protocols (mouse protocol A2014 06-019; Anolis protocol A2015 02-020).
2.2. ChIP-seq
PITX1 ChIP-seq data for E11.5 mouse hindlimbs was previously described (GEO accession GSE41591; (Infante et al., 2013)). For PITX1 ChIP-seq on Anolis carolinensis hindlimbs, embryos were staged according to Sanger (Sanger et al., 2008) and hindlimbs from stages 6 to 7 were collected. Two independent Anolis ChIP-seq replicates and input control libraries were generated, using two separate pools of chromatin collected from embryonic lizard hindlimbs (50 pairs of hindlimbs per replicate for a total of 700 μg of chromatin in each ChIP). PureProteome Protein G Magnetic Beads (Millipore) were pre-incubated with PITX1 antibody (Santa Cruz Biotechnology, sc-18922) before incubating overnight with chromatin. All ChIP and input chromatin control libraries were produced using the NEBNext Ultra DNA Library Prep Kit for Illumina as previously reported (Infante et al., 2015). Libraries were submitted to the Georgia Genomics Facility and sequenced on the NextSeq 500 platform. Anolis PITX1 ChIP-seq data generated for this work have been deposited in the Gene Expression Omnibus (GSE104460) (Edgar et al., 2002).
2.3. ChIP-seq Data Analysis
Sequencing read quality was evaluated using FastQC (version 0.5.1, https://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). ChIP-seq reads were aligned to the mouse genome (mm9) or Anolis carolinensis genome (anoCar2) using bowtie v1.1.0 (Langmead et al., 2009) with the parameters described previously (Infante et al., 2013). PITX1 peaks were identified using MACS2 with default parameters except for the effective genome size, which was set for either mouse (mm9) or lizard (anoCar2). Peaks were associated with Gene Ontology (GO) terms using the Genomic Regions Enrichment of Annotation Tool (GREAT) (Mclean et al., 2010). The assignment of target genes was performed by associating PITX1 peaks with neighboring genes using GREAT. Anolis enhancer coordinates (anoCar2) were translated to the mouse genome (mm9) using the UCSC liftOver Tool. Only the Anolis enhancer regions that were successfully lifted-over to the mouse genome were used to compare PITX1 signal at orthologous enhancers between two species. The significance of PITX1 peak enrichment near putative targets was evaluated using Pearson’s Chi-Square Test. A permutation test was used to determine significant overlap between enhancer datasets. A distribution was created by randomly reshuffling genomic region coordinates 1000 times and performing overlaps using BEDTools v2.26.0 (Quinlan and Hall, 2010). A p-value was calculated by ranking the observed number of overlaps within the number of overlaps from the random distribution.
2.4. RNA-seq
In order to minimize variation in RNA-seq data collected from E9.5 embryos we collected the entire hindlimb field (ectoderm and underlying lateral plate mesoderm) together with the paraxial and axial mesoderm from both wild-type and Pitx1−/− embryos. For E11.5 and E12.5 RNA-seq, only the hindlimb buds on the right side were collected from wild-type and Pitx1 null embryos since expression of Pitx2 on the left side of embryos can partially compensate for the loss of Pitx1. Embryos were sexed via PCR and only XY embryos were used. All embryos used in RNA-seq were staged based on the forelimb morphology as described previously (Wanek et al., 1989). For each developmental stage and each genotype, RNA-seq libraries were made from three separate embryos. Total RNA was isolated using the mirVana RNA Isolation Kit (ThermoFisher Scientific). RNA quality was evaluated on an Agilent 2100 Bioanalyzer, and only samples with RIN values >8 were used. Libraries were constructed with TruSeq Stranded mRNA Sample Prep Kit for Illumina. For construction of E9.5 libraries 250 ng of total RNA was used. 500 ng of total RNA was used to construct E11.5 and E12.5 hindlimb libraries. Libraries were submitted to the Georgia Genomics Facility and sequenced on the Illumina NextSeq platform to produce approximately 50 million, single-end, 75bp reads per library. Mouse RNA-seq data generated for this work have been deposited in the Gene Expression Omnibus (GSE104460).
2.5. RNA-seq Data Analysis
Sequenced libraries were evaluated and trimmed with FastQC. R package (https://www.r-project.org/) was used to calculate the correlations among libraries. Sequenced reads were aligned to mm9 using Tophat2 on the Galaxy platform (Kim et al., 2013). Cuffdiff was used to generate qualified RPKM and to identify differentially expressed genes and transcripts (Trapnell et al., 2012). GO Analysis was performed with DAVID Bioinformatics Resources 6.7 (Huang et al., 2009). DAVID annotation categories that were applied to identify functional annotation clusters include “Functional Categories”, “Literature”, “Pathways” and “Tissue Expression”.
2.6. Comparison of PITX1 Binding to PITX1-depedent Transcription
To examine the correlation of mouse PITX1-enriched regions with the transcriptional regulation of neighboring genes, we compared the genomic distribution of PITX1 peaks with the location of the misregulated genes that we identified through RNA-seq analysis. We defined the regulatory domains of 976 misexpressed genes from E11.5 Pitx1−/− hindlimbs based upon the “Basal plus extension” approach using GREAT (Mclean et al., 2010). Each gene was assigned a basal regulatory domain with the minimum distance both upstream (5 kb) and downstream (1 kb) of the gene’s transcription start site (TSS). The whole gene regulatory domain was then extended in both directions to the nearest gene’s basal domain but no more than the maximum extension (1000 kb), and intersections between mouse PITX1 ChIP-Seq peaks and gene regulatory domains were performed. We used the same approach to search for overlapping regions between mouse/Anolis conserved PITX1-bound regions and the regulatory domains of misexpressed genes.
2.7. Whole-mount In Situ Hybridization and qRT-PCR
Whole-mount mRNA in situ hybridization was performed as described previously (Wilkinson, 1992). A minimum of three wild-type and three Pitx1−/− embryos were stained for each gene, and all replicates showed similar expression patterns. Embryos were staged based on the forelimb morphology. Riboprobes were generated by synthesizing a 500–640 bp template using gBlocks Gene Fragments (Integrated DNA Technologies), amplifying the template by PCR, and transcribing with T3 RNA polymerase (Jarvis and Condie, 2017). Quantitative real time-PCR was used to verify expression differences between wide-type and Pitx1−/− hindlimbs that were detected by RNA-seq. E11.5 and E12.5 right hindlimb total RNA was isolated with the mirVana RNA Isolation Kit (ThermoFisher Scientific) and cDNA was synthesized using the ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs). qPCR assays were performed on a LightCycler® 480 using SYBR Green I Master reagent (Roche). Gapdh was used as the internal control for normalization. The 2−ΔΔCt method was used to detect expression fold change for each target gene with three biological replicates. The primers used in qPCR were as follows: Alx3-F: 5′-CTATGACATCTCCGTACTGCC-3′; Alx3-R: 5′-TCTGGAGACATGAGACAGGG-3′; Pax9-F: 5′-GTGAATGGATTGGAGAAGGGAG-3′; Pax9-R: 5′-CATGTAGGGTGACACTTGGG-3′; Cxcl12-F: 5′-CGCTCTGCATCAGTGACG-3′; Cxcl12-R: 5′-GTTTGGAGTGTTGAGGATTTTCAG-3′; Sox9-F: 5′-GCCGACTCCCCACATTC-3′; Sox9-R: 5′-CGCTTCAGATCAACTTTGCC-3′; Col2a1-F: 5′-GGTTCACATACACTGCCCTG-3′; Col2a1-R: 5′-AAATTCCTGTTCAGCCCCTC-3′; Runx2-F: 5′-GTAGCCAGGTTCAACGATCTG-3′; Runx2-R: 5′-CCGTCCACTGTCACTTTAATAGC-3′; Six1-F: 5′-CAGGTCAGCAACTGGTTTAAG-3′; Six1-R: 5′-CAGAGGAGAGAGTTGATTCTGC-3′; Mef2c-F: 5′-CCAGATCTCCGCGTTCTTATC-3′; Mef2c-R: 5′-CCTCCCATTCCTTGTCCTG-3′; Myod1-F: 5′-CAGAATGGCTACGACACCG-3′; Myod1-R: 5′-ATGCGCTCCACTATGCTG-3′; Gapdh-F: 5′-AAGGTCGGTGTGAACGGATTTG-3′; Gapdh-R: 5′-GTCGTTGATGGCAACAATCTCC-3
3. Results
3.1. Conserved PITX1 binding events are enriched near limb genes
In previous work, we performed PITX1 ChIP-seq on mouse E11.5 hindlimbs (Infante et al., 2013). These experiments revealed the genome-wide distribution of mouse PITX1 binding sites during hindlimb development. While many of these PITX1-associated regions fall within well-conserved non-coding sequences, DNA sequence conservation is not sufficient to conclude that PITX1 is actually bound to orthologous cis-regulatory elements that are conserved in other species. In order to identify deeply conserved PITX1-binding events, we performed PITX1 ChIP-seq on embryonic hindlimb chromatin from the green anole lizard, Anolis carolinensis. The common ancestor of mammals and lizards lived more than 300 million years ago (Pyron, 2010). Therefore, PITX1-binding events that are shared between these species are likely to represent ancient binding interactions that are common to diverse species of limbed amniotes.
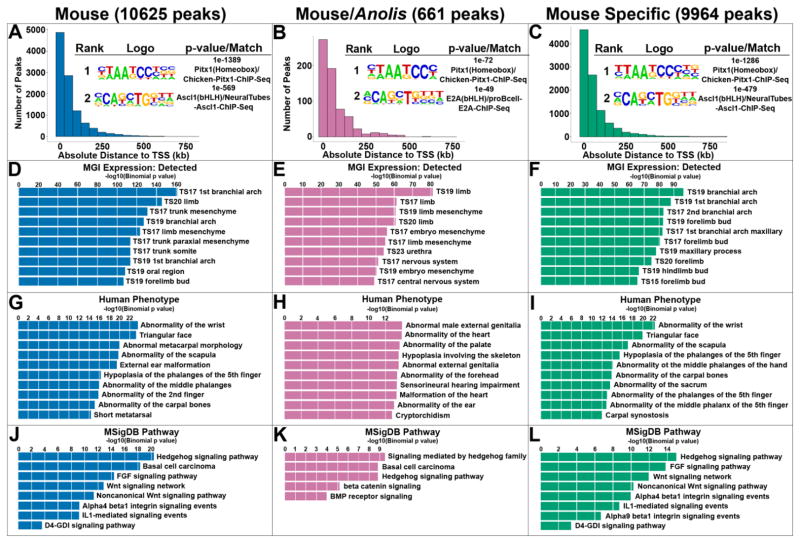
Anolis PITX1 ChIP-seq was carried out on chromatin isolated from the hindlimbs of stage 6 to 7 lizard embryos (Sanger et al., 2008). At these embryonic stages, Anolis hindlimbs are roughly comparable in development to E11.5 mouse hindlimbs. We determined that the PITX1-specific antibody that we previously used for mouse PITX1 ChIP-seq, also cross-reacts with Anolis PITX1 (Fig. S1). Therefore, we were able to use the same PITX1 antibody for mouse and Anolis ChIP-seq experiments. Analysis of our Anolis hindlimb PITX1 ChIP-seq replicates resulted in the identification of 11,090 peaks, a number similar to the 10,625 PITX1 ChIP-seq peaks that we identified in mouse hindlimbs (Table S2) (Infante et al., 2013). De novo motif searches performed within +/− 50 bps of PITX1 peak summits revealed that TAATCC, the core PITX1 binding motif, was significantly enriched in both Anolis (p-value = 1 x 10−986; 61% of target sequences) and mouse (p-value = 1 x 10−1044; 69% of target sequences) PITX1 peaks (Fig. 1A and Table S1). Moreover, we found that both mouse and Anolis PITX1 peaks are significantly enriched near genes associated with limb development (Fig. 1D, G, and J and Fig. S2).
Fig. 1.
Genome-wide enrichment of PITX1 binding activity in mouse and Anolis hindlimbs. A – C) Distribution of PITX1 peaks relative to transcription start sites (TSSs) and top two de novo motifs enriched within +/− 50 bps of A) mouse, B) mouse/Anolis conserved and C) mouse-specific PITX1 peak summits and their best matches to known motifs using HOMER. D – F) The top 10 MGI (Mouse Genome Informatics) Expression terms associated with D) mouse, E) mouse/Anolis conserved and F) mouse-specific PITX1 peaks. G – I) The top 10 Human Phenotype terms associated with G) mouse, H) mouse/Anolis conserved and I) mouse-specific PITX1 peaks. J – L) The top 10 MSigDB (Molecular Signature Database) Pathway terms associated with J) mouse, K) mouse/Anolis conserved and L) mouse-specific PITX1 peaks.
To identify deeply conserved enhancers that are bound by PITX1 in the developing hindlimbs of mouse and Anolis embryos, we began by determining the fraction of Anolis PITX1 peaks that have sequence orthologs in the mouse genome. Of 11,090 Anolis peaks, 2479 (22.4%) exhibit DNA sequence conservation between the Anolis and mouse genomes (Table S2). We intersected the mouse genome coordinates of these 2479 conserved sequences with the coordinates of the 10,625 mouse PITX1 ChIP-seq peaks to reveal 574 overlapping regions. These 574 regions represent the subset of putative cis-regulatory elements that exhibit sequence conservation between mouse and Anolis and that are bound by PITX1 in the developing hindlimbs of both species. The degree of overlap between mouse peaks and Anolis peaks is significantly higher than expected by chance (p<0.001). Prior work has demonstrated that binding site turnover is a common feature of enhancer evolution, and individual binding sites for a transcription factor can be lost and then compensated for by the gain of new binding sites for the same transcription factor (Hare et al., 2008). Therefore, we extended our search for putative cis-regulatory elements that are bound by PITX1 in mouse and Anolis by identifying regions where the sequence underlying an Anolis PITX1 peak is conserved in mouse and occurs within 500 base pairs of a neighboring mouse PITX1 peak. This led to the identification of an additional 87 putative cis-regulatory regions that are conserved between mouse and Anolis but where the location of the PITX1 binding event within the region differs between the mouse and Anolis orthologs. Together, our analyses revealed a total of 661 putative cis-regulatory elements that exhibit primary sequence conservation between mouse and Anolis and that are bound by PITX1 in both species (Table S2).
The majority of the mouse/Anolis PITX1-bound regions are located within 100 kb of transcription start sites with a distribution that is similar to the larger set of all mouse PITX1 peaks (Fig. 1A and 1B). As expected, the top ranked de novo motif found within the mouse/Anolis PITX1 regions matches the known PITX1 motif (Fig. 1B and Table S1). Notably, this motif is essentially indistinguishable from the top motif found in “mouse-specific” PITX1 peaks, the subset of regions that are bound by PITX1 in mice but that are not bound by PITX1 in Anolis or that do not have detectable sequence conservation in Anolis (Fig. 1C). To determine whether conserved PITX1 binding events are enriched near particular gene categories, we used GREAT (Mclean et al., 2010). This analysis demonstrated that mouse/Anolis PITX1 bound regions are significantly enriched near genes that are expressed in developing limbs, with 5 of the top 10 ranked gene expression GO terms associated with embryonic limb expression (Fig. 1E). Examination of signaling pathway GO terms showed that genes associated with the Hedgehog, WNT, and BMP signaling pathways are also enriched near conserved PITX1 bound regions (Fig. 1K). For PITX1 peaks that are mouse-specific, enrichments for similar sets of limb-related and signaling pathway GO terms were observed (Fig. 1F, I and L). Thus, mouse/Anolis conserved and mouse-specific PITX1 binding events display similar gene associations.
3.2. Genes related to patterning, chondrogenesis, and myogenesis are misregulated in Pitx1−/− hindlimbs
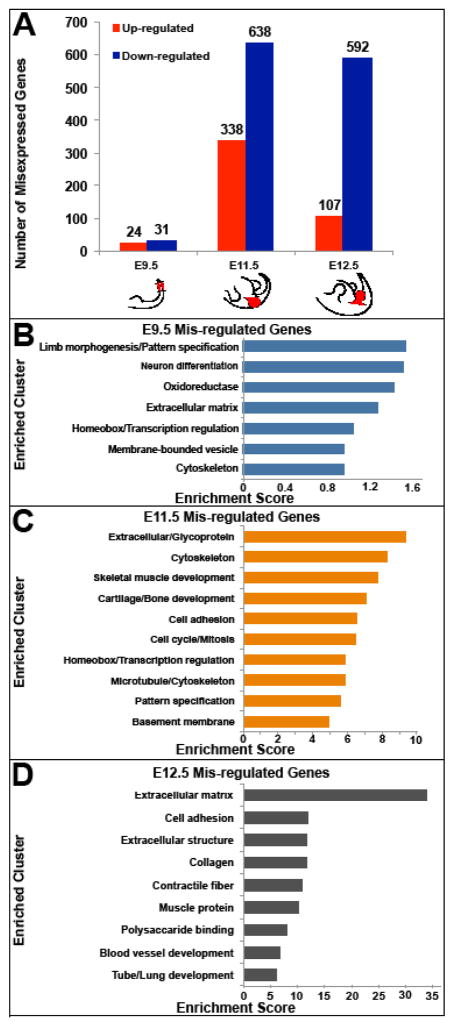
In order to identify genes with PITX1-dependent expression, we performed global gene expression comparisons between the developing hindlimbs of Pitx1−/− and wild-type mouse embryos. For our analyses, RNA-seq was carried out separately on E9.5 hindlimb fields, E11.5 hindlimbs, and E12.5 hindlimbs. Given the small size of the E9.5 hindlimb field, we collected the prospective hindlimb region (ectoderm and underlying lateral plate mesoderm) together the associated paraxial and axial mesoderm. This helped to decrease variation in the tissue samples that we collected. Because the E9.5 samples contained a substantial amount of tissue outside of the prospective hindlimb, we deeply sequenced the RNA-seq libraries that we generated to improve our ability to detect differentially expressed genes (~50 million reads were sequenced per library). Since Pitx2 expression on the left side of embryos can partially compensate for the loss of Pitx1 (Marcil et al., 2003), we exclusively used right hindlimb buds to prepare RNA-seq libraries at E11.5 and E12.5 as the hindlimbs can be precisely dissected at these stages. In the early hindlimb field, we found that a relatively modest number of genes (55 in total) are significantly misregulated in Pitx1 mutants (Fig. 2A and Table S3). In contrast, several hundred genes are misregulated at E11.5 and E12.5. At these later stages, we also observed that a greater portion of misregulated genes are down-regulated than up-regulated, consistent with PITX1 acting predominantly as a transcriptional activator.
Fig. 2.
Misregulated genes in the hindlimbs of Pitx1−/− mice at different stages of embryonic development. A) The number of misexpressed genes in Pitx1−/− compared to wild-type mouse hindlimbs at E9.5, E11.5 and E12.5. Red shading in cartoons represents the Pitx1 expression domain in the hindlimbs. Solid lines indicate where cuts were made for collection of tissue for RNA-Seq. B – D) The top scoring gene clusters associated with misexpressed genes in Pitx1−/− hindlimbs at B) E9.5, C) E11.5, and D) E12.5.
Previous studies have demonstrated that Tbx4 expression is reduced in the hindlimb buds of Pitx1 knockout embryos and that Tbx4 is a direct regulatory target of PITX1 (Infante et al., 2013; Lanctôt et al., 1999; Szeto et al., 1999). Consistent with these earlier findings, our RNA-seq analyses demonstrate that Tbx4 expression is significantly reduced in the absence of PITX1 (expressed at 34% and 56% of wild-type at E11.5 and E12.5, respectively; Table S3). Prior work also established that the expression of hindlimb-specific HoxC10 and HoxC11 genes can be induced in the forelimb through ectopic expression of Pitx1 (Delaurier et al., 2006; Logan and Tabin, 1999; Park et al., 2014). However, we found that the expression levels of HoxC10 and HoxC11 are not significantly reduced during hindlimb development of Pitx1−/− embryos, indicating that PITX1 function is not essential for expression of these genes. Finally, we examined the expression of Isl1, a gene that is essential for hindlimb bud formation but plays no role in forelimb development (Kawakami et al., 2011). In wild-type embryos, Isl1 is expressed transiently in the hindlimb field and early hindlimb bud but is down-regulated by E10.5. In E9.5 Pitx1−/− embryos, we find that Isl1 is expressed at significantly higher levels than wild-type (1.5x higher; Table S3). Therefore, of the known transcription factors with robust, hindlimb-biased expression, only Tbx4 exhibits reduced expression in the absence of PITX1.
We next used the DAVID functional annotation tool to determine whether specific pathways or gene categories are enriched among genes that are misexpressed in Pitx1 mutants (Huang et al., 2009). Among the categories of genes that are misregulated in Pitx1−/− hindlimb field at E9.5, the top ranked cluster relates to limb morphogenesis and pattern specification (Fig. 2B). By E11.5, the top ranked terms that are associated with misregulated genes include extracellular matrix, cartilage and bone development, and skeletal muscle development (Fig. 2C). Similar enrichments are observed at E12.5, with significant associations with genes related to extracellular matrix, cell adhesion, collagen, and muscle function (Fig. 2D). Thus, in Pitx1−/− embryos we find evidence of altered transcription of limb patterning genes in the early hindlimb field by E9.5, just prior to hindlimb bud outgrowth. By E11.5 large-scale alterations in gene expression are apparent with genes related to extracellular matrix, cartilage and skeletal muscle being enriched among misregulated genes.
3.3. PITX1 binding events are enriched near misregulated genes
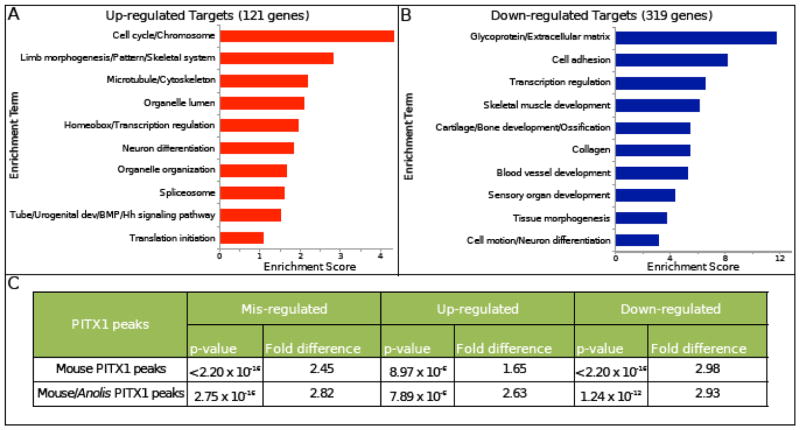
To examine the relationship between PITX1-bound regions and the transcriptional regulation of neighboring genes, we compared the genomic distribution of PITX1 peaks with the location of the misregulated genes identified through RNA-seq on Pitx1−/− hindlimbs. Since our PITX1 ChIP-seq data was collected from E11.5 hindlimb chromatin, we compared PITX1 binding sites with our list of E11.5 misregulated genes. We began by associating the 10,625 mouse PITX1 ChIP-seq peaks with gene locations using GREAT (Mclean et al., 2010). We found that PITX1 binding sites are significantly enriched near genes that are misexpressed in Pitx1−/− hindlimbs (2.45-fold enriched; p-value < 2.2 x 10−16) (Fig. 3C). In addition, the association between PITX1 binding events is stronger with down-regulated genes (2.98-fold enriched; p-value < 2.2 x 10−16) than up-regulated genes (1.65-fold enriched; p-value = 8.97 x 10−6). When we limited our analysis to the PITX1 binding events that are shared between mouse and Anolis, we observed that these sites are also enriched near genes that are misregulated in the absence of PITX1 (2.82-fold enriched; p = 2.8 x 10−16).
Fig. 3.
Putative direct targets of PITX1 are strongly associated with limb patterning, chondrogenesis and myogenesis in mouse hindlimbs. A–B) The top 10 enriched clusters associated with A) up- and B) down-regulated candidate targets of PITX1 in mouse hindlimbs. C) Mouse and mouse/Anolis conserved PITX1 binding sites are both significantly enriched near genes that are misexpressed in Pitx1−/− mouse hindlimbs.
In order to identify direct transcriptional targets of PITX1, we next compared the genomic positions of misexpressed genes to the location of PITX1 bound regions. Since precise maps of gene regulatory domains are not currently available for embryonic limbs, we associated PITX1 peaks with specific genes by using GREAT (Mclean et al., 2010). GREAT operates by assigning each gene a basal regulatory domain around the gene’s transcriptional start site and then extends the regulatory domain to the nearest upstream and downstream genes to a maximum of 1 megabase. Therefore, each PITX1 binding site has the potential to be assigned to more than one gene. Using this approach, we identified 440 genes as candidate, direct transcriptional targets of PITX1 (Table S4). Among the candidate targets of PITX1, the expression levels of 121 are increased and the levels of 319 are decreased in hindlimbs of Pitx1 knockout embryos. Of the 440 putative transcriptional targets of PITX1, 68 are associated with one or more neighboring mouse/Anolis conserved PITX1 binding event. The PITX1-dependent expression of these 68 genes coupled with the presence of conserved PITX1 binding events suggests that these genes are ancient transcriptional targets of PITX1 (Table S4).
3.4. PITX1 directly influences hindlimb development through essential regulators of cartilage and muscle development
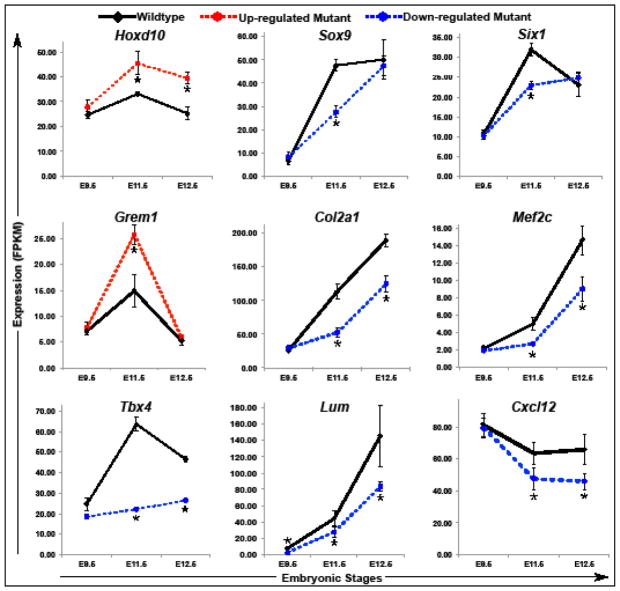
Our initial DAVID analysis of genes that are misregulated in Pitx1 null embryos included both direct transcriptional targets of PITX1 and genes that are misregulated as a secondary consequence of Pitx1 inactivation (see section 3.2). We revisited this analysis by focusing exclusively on our list of putative direct transcriptional targets of PITX1 and by performing separate DAVID analyses on up-regulated and down-regulated PITX1 targets. This revealed that up-regulated targets are enriched for terms related to cell cycle control, limb morphogenesis/patterning, homeobox genes, and components of the BMP and Hedgehog signaling pathways (Fig. 3A). The up-regulated genes underlying these enriched GO terms include a large number of factors with well-established roles in the initiation of limb bud formation and early patterning of the limb buds and include Alx3, Bmp2, Bmp4, Bmp7, Fgf10, Grem1, HoxA9, HoxD10, Irx3, Irx5, Meis2, and Tbx2 (Table S4). In contrast, the PITX1 target genes that are down-regulated in Pitx1 mutants are strongly enriched for terms related to extracellular matrix, cell adhesion, cartilage/bone development, and skeletal muscle development (Fig. 3B). The cartilage and bone related genes include FoxP4, Snai1, Sox6, Sox8, and Sox9 (Table S4), which are all transcription factors that regulate chondrogenesis (Bi et al., 1999; Chen and Gridley, 2013; Smits et al., 2001; Zhao et al., 2015). In addition, multiple collagens and other extracellular structural protein important for cartilage and bone formation have reduced expression in the absence of PITX1. Further examination of down-regulated PITX1 targets revealed multiple regulators of myogenesis including Cxcl12, Eya1, Mef2c, Nfatc2, and Six1 (Table S4; (Daou et al., 2013; Grifone et al., 2007; 2005; Molkentin et al., 1995; Vasyutina et al., 2005)). We also found that the Dcn, Lum, and Mkx genes, which are all important for tendon development (Danielson et al., 1997; Ito et al., 2010; Jepsen et al., 2002), were among the putative PITX1 transcriptional targets that we identified. In order to gain a better understanding of the Pitx1−/− expression defects we detected, we generated time-course charts from our RNA-seq data. This allowed us to examine the expression of misregulated genes at different developmental stages. We found that HoxD10 expression is significantly elevated in both E11.5 and E12.5 Pitx1−/− hindlimbs (Fig. 4). Similar patterns of elevated expression were observed for Alx3 and Meis2 (Table S4). These genes were up-regulated by 1.3x–2x in Pitx1−/− hindlimbs relative to wild-type at E11.5. Other early patterning genes, including Grem1, Bmp2, Bmp4, Bmp7, Fgf10, and Tbx2 (Fig. 4 and Table S4), exhibited transient up-regulation in Pitx1 mutants hindlimbs at E11.5 before returning to normal or near normal expression levels at E12.5. In contrast, chondrogenesis and myogenesis PITX1 target genes exhibited a delay in their up-regulation with decreased expression apparent at E11.5 (Fig. 4 and Table S4).
Fig. 4.
Expression levels of putative PITX1 transcriptional targets in wild-type and Pitx1−/− hindlimbs at different developmental stages. Time charts display the average RNA-Seq expression level from three independent biological replicates. Asterisks represent developmental stages with significantly different expression in Pitx1−/− compared to wild-type hindlimbs (FDR-adjusted p-value < 0.05). Error bars represent the standard error of the mean.
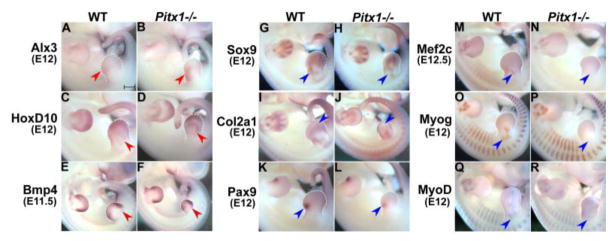
We characterized the expression patterns of several misregulated genes by whole-mount in situ hybridization (Fig. 5). In general, the overall pattern of up-regulated genes was very similar between the hindlimbs of Pitx1−/− and wild-type embryos, though we did find that Alx3 expression was expanded towards the center of Pitx1−/− hindlimb buds (Fig. 5A–B). In situ hybridization for genes associated with cartilage and muscle development produced more dramatic differences. For instance, Sox9 displayed altered expression in the proximal portion of the hindlimb bud at E12 (Fig. 5G–H), and Mef2c and other markers of myogenesis had qualitatively less staining by in situ hybridization at E11.5–E12.5 (Fig. 5M–R).
Fig. 5.
Whole-mount in situ hybridization of PITX1-dependent genes in wide-type and Pitx1−/− mouse embryos. A–F) The expression pattern of limb patterning genes (Alx3, HoxD10 and Bmp4). G–L) The expression pattern of genes related to cartilage development (Sox9, Col2a1 and Pax9). M–R) The expression pattern of genes related to muscle development (Mef2c, MyoD and Myogenin). Red arrowheads point to regions with increased expression while blue ones point to regions with decreased expression in Pitx1−/− mouse embryos. All images were taken at the same magnification. Scale bar = 500 μm.
3.5. PITX1 binding profiles at conserved transcriptional targets
To explore the ancient transcriptional targets of PITX1 in hindlimbs, we performed a final DAVID analysis on the 68 putative conserved targets of PITX1. This analysis shows that skeletal- and muscle-related GO terms (“Mesenchyme/Bone development”, “Skeletal system development” and “Muscle organ development”) are enriched among these 68 genes (Fig. S2A). Patterning-related terms (“Transcription factor activity” and “Pattern specification”) are also significantly enriched in the conserved target gene set (Fig. S2A). Thus, the functional terms associated with these 68 conserved PITX1 targets are similar to the terms enriched in the larger set of 440 putative PITX1 targets.
One of the predicted ancient regulatory targets of PITX1 is Tbx4. In mice, we previously demonstrated that PITX1 regulates Tbx4, at least in part, through binding of the HLEA cis-regulatory element (Infante et al., 2013; Menke et al., 2008). While HLEA is not conserved in Anolis, we find that there are two conserved PITX1 binding events downstream of Tbx4 (Fig. 6C). Therefore, in the mouse hindlimb there are both conserved and mouse-specific binding events occur at the Tbx4 locus. A further examination of the 68 predicted ancient targets of PITX1 showed that this group includes Sox9 and Six1, important regulators of cartilage and skeletal muscle development, respectively. In the case of Sox9, a significant PITX1 peak is identified 232 kb downstream of its transcription start site (Fig. 6A). The sequence of this binding region is conserved in the Anolis genome, and a PITX1 peak is also located at the orthologous sequence located near the Anolis sox9 gene. Similarly, there is a strongly enriched PITX1 peak located just upstream of Six1 in mouse (Fig. 6B). This region is conserved in the Anolis genome and is bound by PITX1 in embryonic Anolis hindlimbs. These results are consistent with the idea that PITX1 promotes chondrocyte and myoblast differentiation in the hindlimb by direct regulation of key factors involved in the chondrogenic and myogenic regulatory networks. Moreover, some of these factors are likely to be ancient regulatory targets of PITX1.
Fig. 6.
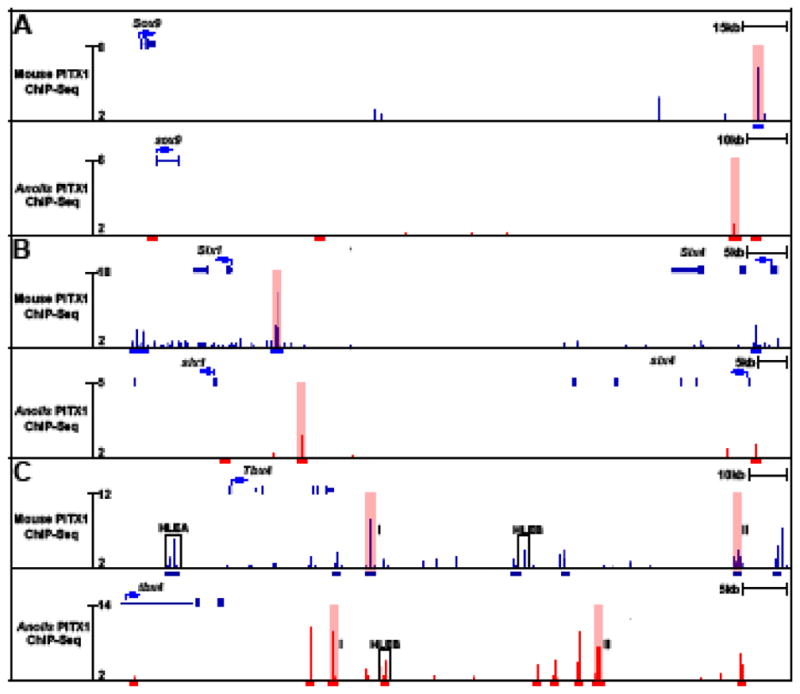
PITX1 ChIP-seq profiles in mouse and Anolis hindlimbs at putative PITX1 transcriptional targets. A) Sox9, B) Six1 and C) Tbx4. Blue bars denote mouse PITX1 peaks and red bars denote Anolis PITX1 peaks. Conserved binding events identified in both species are highlighted in pink. I and II are used to distinguish two different pairs of conserved peaks at the Tbx4 locus. The location of known Tbx4 hindlimb-specific enhancers, HLEA and HLEB, are outlined by black boxes.
4. Discussion
Nearly two decades ago functional studies of PITX1 demonstrated that this transcription factor performs pivotal roles in promoting hindlimb growth and specifying hindlimb morphology (Delaurier et al., 2006; Lanctôt et al., 1999; Logan and Tabin, 1999; Szeto et al., 1999). Yet the direct regulatory targets of PITX1 have remained largely unknown. Our previous work demonstrated that PITX1 is bound to thousands of different sites during mouse hindlimb development, including many known limb enhancers. However, the presence of a PITX1 binding interaction does not itself prove that the interaction directly influences gene expression or enhancer activity. Since it is not practical to perform direct functional tests on each of the thousands of different PITX1 binding sites that occur in developing mouse hindlimbs, we have used ChIP-seq combined with expression analyses to identify putative regulatory targets of PITX1.
Our RNA-seq analyses of early and later stages of hindlimb development revealed that many genes important for the initial stages of limb growth and patterning are up-regulated in the hindlimbs of Pitx1−/− embryos. The expression of several of these genes, including Bmp7, Fgf10, Meis2, and Tbx2, have previously been analyzed in Pitx1−/−;Pitx2−/+ embryos by whole-mount in situ hybridization and were found to have grossly normal patterns of expression (Marcil et al., 2003). However, our quantitative RNA-seq analyses show that the expression of these genes is elevated in the absence of PITX1. All four of these genes have neighboring PITX1 ChIP-seq peaks and are putative direct transcriptional targets of PITX1. Prior work has suggested that PITX1 can act as a transcriptional repressor in some contexts (Qi et al., 2011), but additional studies will be required to determine whether the binding of PITX1 is directly acting to reduce gene expression of certain limb-patterning genes.
Previous anatomical and histological analysis of Pitx1 knockout mice showed that the hindlimbs are reduced in overall size and the relative size and shape of hindlimb skeletal elements are altered. Lanctôt and colleagues also demonstrated that skeletal elements of Pitx1 knockout hindlimbs exhibit impaired ossification as evidenced by abnormal mineralization of the femur and tibia at E16.5 (Lanctôt et al., 1999). Therefore, the reduced and delayed expression of chondrogenesis genes that we have detected in Pitx1−/− hindlimbs aligns well with earlier histological studies. However, it has remained unclear whether the ossification defects of Pitx1 knockout embryos are directly due to a role of PITX1 in cartilage and bone differentiation or are a secondary consequence of growth and patterning defects that occur earlier in hindlimb development. Our mouse PITX1 ChIP-seq data strongly suggest that PITX1 directly influences the differentiation of skeletal tissues by regulating several factors that are involved in cartilage and bone development (Fig. 7A). For instance, we found that Sox9, an early chondrogenic marker necessary for mesenchymal condensation and early chondrocyte differentiation (Akiyama et al., 2002; Bi et al., 1999), has significantly reduced expression at E11.5 in the absence of PITX1. We also identified a strong PITX1 binding site downstream of Sox9’s transcription start site (Fig. 6A). This PITX1-bound element is a known limb enhancer (VISTA enhancer mm636; (Visel et al., 2007)) that is conserved among amniotes, and we found that PITX1 is bound to the orthologous Anolis sequence during embryonic development of lizard hindlimbs. Additionally, our ChIP-seq data revealed a series of PITX1 binding sites located within a large (>1MB; mm9 chr11:111,424,585–112,518,902) region upstream of Sox9. This region contains 12 mouse/Anolis conserved PITX1 binding events, and this genomic domain has previously been identified as an extended transcriptional control region for Sox9 (Gordon et al., 2009; Pfeifer et al., 1999; Wunderle et al., 1998). A number of distinct, tissue-specific enhancers have been found in this interval, including three limb enhancers (Gordon et al., 2014; Visel et al., 2007), and we have found that each of these limb enhancers overlaps with a conserved PITX1 binding event. We therefore hypothesize that that the Sox9 gene is an ancient, direct transcriptional target of PITX1.
Fig. 7.
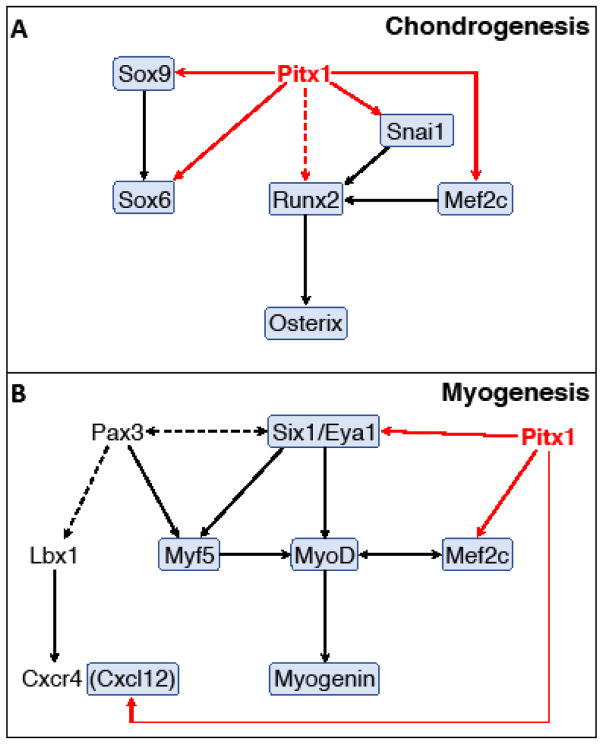
Proposed PITX1 regulatory interactions with components of the chondrogenesis and myogenesis regulatory networks. Solid arrows represent direct regulatory interactions and dashed arrows represent indirect or unproven regulatory interactions. Red arrows highlight PITX1 regulation of downstream targets identified from our data and black arrows represent interactions identified from the published literature. Genes in shaded boxes are down-regulated in Pitx1−/− hindlimbs.
In addition to Sox9, our ChIP-seq and RNA-seq data suggest that FoxP4, Mef2c, Snai1, and Sox6 are all direct targets of PITX1 during hindlimb development. Each of these genes has been implicated in chondrogenesis or osteogenesis (Arnold et al., 2007; Chen and Gridley, 2013; Smits et al., 2001; Zhao et al., 2015), and two of these loci (FoxP4 and Sox6) have conserved non-coding sequences that are bound by PITX1 in mouse and Anolis. Although we detected no conserved PITX1 binding events near Mef2c or Snai1, the mouse and Anolis orthologs of these genes both have species-specific PITX1 peaks (Fig. S2D and data not shown). Therefore, despite the apparent absence of conserved PITX1 bound sequences, Mef2c and Snai1 may also be ancient regulatory targets of PITX1. We also note that Runx2 and Osterix have reduced expression in Pitx1−/− hindlimbs by E12.5 (>2x reduced; Table S3). Both of these genes encode transcription factors that have well-established functions in promoting chondrocyte hypertrophy and osteoblast differentiation, and these genes are normally up-regulated shortly after Sox9 (Komori et al., 1997; Nakashima et al., 2002; Otto et al., 1997; Yoshida et al., 2004). Though we detected conserved PITX1 binding events adjacent to Runx2 and Osterix genes, our ChIP-seq data was collected at E11.5 rather than at E12.5, the stage at which we detected significant misregulation of the Runx2 and Osterix genes in Pitx1 knockout mice. Therefore, while we have not included these genes in our list of putative PITX1 targets, it remains possible that Runx2 and Osterix are directly regulated by PITX1.
Prior studies established that the Pitx2 gene is expressed in muscle progenitors throughout the developing embryo, including those that contribute to the limb musculature (L’Honor et al., 2007; Shih et al., 2007). Further investigations by L’Honoré demonstrated that PITX2 is required for the onset of MyoD expression in muscle precursors of the forelimbs and hindlimbs and showed that PITX2 directly binds to the MyoD core enhancer to activate MyoD expression (L’Honore et al., 2010). In contrast to Pitx2, the Pitx1 gene is expressed throughout the hindlimb mesenchyme and can be detected in both muscle progenitors and cells that give rise to the hindlimb skeleton (Marcil et al., 2003). Though Pitx1 is expressed in hindlimb muscle progenitors, a direct role for PITX1 in promoting myogenesis has not previously been demonstrated. Our expression analyses reveal that MyoD expression is delayed and decreased in Pitx1−/− hindlimbs in a manner similar to the MyoD expression defects observed in Pitx2 mutants (Fig. 5Q and R; Table S3). However, unlike PITX2 we find no evidence of PITX1 binding at the MyoD core enhancer or any other location within 600 kb upstream or downstream of the MyoD locus. Instead, we have identified direct myogenic targets of PITX1 that include Six1, Eya1 and Mef2c (Fig. 7B). The SIX1/EYA1 transcriptional complex functions as an upstream activator of myogenic determination genes, including MyoD, as evidenced by the loss of all hypaxial dermomyotome derived muscles in Six1/4 and Eya1/2 double mutants (Grifone et al., 2007; 2005). We found that Six1 is significantly down-regulated in E11.5 Pitx1 null hindlimbs (Fig. 4 and Fig. S2B), and conserved PITX1 peaks are located in its regulatory domain (Fig. 6B). Eya1 also shows significantly reduced expression in E11.5 Pitx1−/− hindlimbs (Table S3) and has species-specific PITX1 binding events in both mouse and Anolis hindlimbs (Table S2). In addition to the function of Mef2c in skeletal development (see above), this gene also promotes myogenic differentiation (Molkentin et al., 1995). Thus, we believe the reduced MyoD expression in Pitx1 knockout hindlimbs is a secondary consequence of the absence of PITX1.
Beyond myogenesis genes that encode transcription factors, we also identified Cxcl12 as a candidate PITX1 regulatory target. CXCL12 (also known as SDF1) is a secreted chemokine that acts as a ligand for the chemokine receptor CXCR4 (Vasyutina et al., 2005). CXCR4/CXCL12 signaling plays an important role in the migration of muscle progenitor cells into the limb buds with the Cxcr4 gene expressed in migrating muscle progenitors and Cxcl12 in the developing limbs. Taken together, we find strong evidence that PITX1 promotes myogenesis through direct regulation of multiple myogenesis genes that include transcriptional regulators and the Cxcl12 chemokine.
In conclusion, we have found significant delays in the expression of important cartilage, bone, and muscle genes in the absence of PITX1. While the relative importance of individual PITX1-binding sites remains to be tested, the presence of deeply conserved binding events that are shared between mammals and reptiles strongly suggests that PITX1 directly regulates the transcription of key components of the chondrogenesis and myogenesis regulatory networks.
Supplementary Material
Highlights.
ChIP-seq on mice and Anolis hindlimbs revealed ancient PITX1 binding events
PITX1 binding is enriched near genes misregulated in Pitx1−/− mouse hindlimbs
Loss of PITX1 causes reduced expression of key chondrogenesis and myogenesis genes
Deeply conserved PITX1 transcriptional targets include Sox9 and Six1
Acknowledgments
This work was supported by grants awarded to D.B.M. from the NSF (#1149453) and NIH (HD081034) and by two Junior Faculty Research Grants (#1847 and #2503) from the Office of the Vice President of Research at the University of Georgia. This study was supported in part by resources from the Georgia Advanced Computing Resource Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado DM, McCall K, Aferol H, Silva MJ, Garbow JR, Spees WM, Patel T, Siegel M, Dobbs MB, Gurnett CA. Pitx1 haploinsufficiency causes clubfoot in humans and a clubfoot-like phenotype in mice. Hum Mol Genet. 2011;20:3943–3952. doi: 10.1093/hmg/ddr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jónsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gridley T. Compensatory regulation of the Snai1 and Snai2 genes during chondrogenesis. J Bone Miner Res. 2013;28:1412–1421. doi: 10.1002/jbmr.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol. 1997;136:729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou N, Lecolle S, Lefebvre S, Gaspera BD, Charbonnier F, Chanoine C, Armand AS. A new role for the calcineurin/NFAT pathway in neonatal myosin heavy chain expression via the NFATc2/MyoD complex during mouse myogenesis. Development. 2013;140:4914–4925. doi: 10.1242/dev.097428. [DOI] [PubMed] [Google Scholar]
- Delaurier A, Schweitzer R, Logan MPO. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev Biol. 2006;299:22–34. doi: 10.1016/j.ydbio.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Domyan ET, Kronenberg Z, Infante CR, Vickrey AI, Stringham SA, Bruders R, Guernsey MW, Park S, Payne J, Beckstead RB, Kardon G, Menke DB, Yandell M, Shapiro MD. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. eLife. 2016;5:e12115. doi: 10.7554/eLife.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Logan MPO. Pitx1 is necessary for normal initiation of hindlimb outgrowth through regulation of Tbx4 expression and shapes hindlimb morphologies via targeted growth control. Development. 2011;138:5301–5309. doi: 10.1242/dev.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Attanasio C, Bhatia S, Benko S, Ansari M, Tan TY, Munnich A, Pennacchio LA, Abadie VR, Temple IK, Goldenberg A, van Heyningen V, Amiel J, FitzPatrick D, Kleinjan DA, Visel A, Lyonnet S. Identification of Novel Craniofacial Regulatory Domains Located far Upstream of SOX9and Disrupted in Pierre Robin Sequence. Hum Mutat. 2014;35:1011–1020. doi: 10.1002/humu.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, FitzPatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Gurnett CA, Alaee F, Kruse LM, Desruisseau DM, Hecht JT, Wise CA, Bowcock AM, Dobbs MB. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet. 2008;83:616–622. doi: 10.1016/j.ajhg.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped Enhancers Are Functionally Conserved in Drosophila Despite Lack of Sequence Conservation. PLoS Genet. 2008;4:e1000106–13. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Infante CR, Mihala AG, Park S, Wang JS, Johnson KK, Lauderdale JD, Menke DB. Shared Enhancer Activity in the Limbs and Phallus and Functional Divergence of a Limb-Genital cis-Regulatory Element in Snakes. Dev Cell. 2015;35:107–119. doi: 10.1016/j.devcel.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante CR, Park S, Mihala AG, Kingsley DM, Menke DB. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev Biol. 2013;374:234–244. doi: 10.1016/j.ydbio.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci USA. 2010;107:10538–10542. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis BR, Condie BG. Synthetic DNA templates for the production of in situ hybridization probes. bioRxiv. 2017:108613. doi: 10.1101/108613. [DOI] [Google Scholar]
- Jepsen KJ, Wu F, Peragallo JH, Paul J, Roberts L, Ezura Y, Oldberg A, Birk DE, Chakravarti S. A Syndrome of Joint Laxity and Impaired Tendon Integrity in Lumican- and Fibromodulin-deficient Mice. J Biol Chem. 2002;277:35532–35540. doi: 10.1074/jbc.M205398200. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Marti M, Kawakami H, Itou J, Quach T, Johnson A, Sahara S, O’Leary DDM, Nakagawa Y, Lewandoski M, Pfaff S, Evans SM, Belmonte JCI. Islet1-mediated activation of the β-catenin pathway is necessary for hindlimb initiation in mice. Development. 2011;138:4465–4473. doi: 10.1242/dev.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes inthe presence of insertions, deletions and genefusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- L’Honor A, Coulon V, Marcil A, Lebel ML, Lafrance-Vanasse J, Gage P, Camper S, Drouin J. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307:421–433. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- L’Honore A, Ouimette JF, Lavertu-Jolin M, Drouin J. Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development. 2010;137:3847–3856. doi: 10.1242/dev.053421. [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MPO, Tabin CJ. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx1 and Pitx2 are required for development of hindlimb buds. Development. 2003;130:45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Ouimette JF, Jolin ML, L’honoré A, Gifuni A, Drouin J. Divergent transcriptional activities determine limb identity. Nat Commun. 2010;1:1–9. doi: 10.1038/ncomms1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Infante CR, Rivera-Davila LC, Menke DB. Conserved regulation of hoxc11 by pitx1 in Anolis lizards. J Exp Zool B Mol Dev Evol. 2014;322:156–165. doi: 10.1002/jez.b.22554. [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, Korniszewski L, Back E, Scherer G. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet. 1999;65:111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA. A likelihood method for assessing molecular divergence time estimates and the placement of fossil calibrations. Syst Biol. 2010;59:185–194. doi: 10.1093/sysbio/syp090. [DOI] [PubMed] [Google Scholar]
- Qi DL, Ohhira T, Fujisaki C, Inoue T, Ohta T, Osaki M, Ohshiro E, Seko Tomomi, Aoki Shinsuke, Oshimura Mitsuo, Kugoh H. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol. 2011;31:1624–1636. doi: 10.1128/MCB.00470-10. doi.org/10.1128/MCB.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger TJ, Losos JB, Gibson-Brown JJ. A developmental staging series for the lizard genus Anolis: a new system for the integration of evolution, development, and ecology. J Morphol. 2008;269:129–137. doi: 10.1002/jmor.10563. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr Patterns. 2007;7:441–451. doi: 10.1016/j.modgep.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Spielmann M, Brancati F, Krawitz PM, Robinson PN, Ibrahim DM, Franke M, Hecht J, Lohan S, Dathe K, Nardone AM, Ferrari P, Landi A, Wittler L, Timmermann B, Chan D, Mennen U, Klopocki E, Mundlos S. Homeotic Arm-to-Leg Transformation Associated with Genomic Rearrangements at the PITX1 Locus. Am J Hum Genet. 2012;91:629–635. doi: 10.1016/j.ajhg.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisúa-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 2005;19:2187–2198. doi: 10.1101/gad.346205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanek N, Muneoka K, Holler-Dinsmore G, Burton R, Bryant SV. A staging system for mouse limb development. J Exp Zool. 1989;249:41–49. doi: 10.1002/jez.1402490109. doi.org/10.1002/jez.1402490109. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. In situ hybridization: a practical approach. Oxford University Press; 1992. [Google Scholar]
- Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA. 1998;95:10649–10654. doi: 10.1073/pnas.95.18.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue KI, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhou W, Yao Z, Wan Y, Cao J, Zhang L, Zhao J, Li H, Zhou R, Li B, Wei G, Zhang Z, French CA, Dekker JD, Yang Y, Fisher SE, Tucker HO, Guo X. Foxp1/2/4 regulate endochondral ossification as a suppresser complex. Dev Biol. 2015;398:242–254. doi: 10.1016/j.ydbio.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.