Abstract
Objectives
Telomere length (TL) is a biomarker of aging and age-related decline. Although venous blood is considered the “gold standard” for TL measurement, its collection is often not feasible or desired in non-clinical settings. Saliva and dried blood spots (DBS) have been used as alternatives when venipuncture cannot be performed. However, it is not known whether these sample types yield TL measurements comparable to those obtained from venous blood. We sought to determine whether different samples from the same individual yield comparable TL measurements.
Methods
We extracted DNA from matched buffy coat, saliva (Oragene and Oasis), and DBS (venous and capillary) samples from 40 women aged 18 to 77 years. We used the monochrome multiplex qPCR (MMQPCR) assay to measure TL in all sample types for each participant and applied quality control measures to retain only high-quality samples for analysis. We then compared TL from buffy coat and saliva to examine how these measurements differ and to test if TL is correlated across sample types.
Results
TL differed significantly across buffy coat, Oragene saliva, and Oasis saliva samples. TL from buffy coat and Oragene saliva was moderately correlated (ρ = 0.48, p = 0.002) and the most similar in size. Oasis saliva TL was not correlated with buffy coat or Oragene saliva TL, and was the shortest. DBS DNA yields were inadequate for TL measurement using the MMQPCR assay.
Conclusions
Using a matched dataset we demonstrate that sample type significantly influences the TL measurement obtained using the MMQPCR assay.
Keywords: genomic DNA, biomarker of aging, multiplex qPCR, saliva, dried blood spots
Introduction
Telomere length (TL) is a biomarker of age, age-related decline and disease (Baker and Sprott, 1988; Epel et al., 2004; Harley et al., 1990; Baird, 2006; Hornsby, 2007; see von Zglinicki and Martin-Ruiz, 2005 for review). TL has shown a predictable pattern of decline with age when measured from DNA extracted from whole blood collected by venipuncture (Aviv et al., 2008; Iwama et al., 1998; Rufer et al., 1999; Vaziri et al., 1994; see Müezzinler et al., 2013 for review). Although use of venous blood is common in clinical and biomedical settings, venipuncture is a relatively invasive, costly, and labor-intensive means of sample collection. This presents several obstacles for population- and field-based research studies, establishing a need for minimally invasive, cost-effective protocols to facilitate the collection of large sample sets across a range of non-clinical settings (McDade et al., 2007; Nussey et al., 2014). As population aging is a growing concern worldwide, there is an urgent need to understand aging and age-related health decline in understudied populations. The ability to measure TL in saliva and dried blood spots (DBS) is an attractive alternative to blood telomere length (BTL – often called leukocyte telomere length due to the predominance of DNA contributed by white blood cells) because these samples can be collected using minimally invasive techniques, pose less risk to participants, and tend to be more acceptable cross-culturally. Additionally, these sample types do not require immediate processing or cold storage and can be more easily transported.
Although many studies have measured TL in saliva (e.g., Chen et al., 2015; Gotlib et al., 2015; Lapham et al., 2015; Theall et al., 2013; Whisman et al., 2016) and DBS (Chae et al., 2014; Edmonds et al., 2015; Zanet et al., 2013), few studies have explicitly evaluated whether TL measured using minimally invasive sample types (specifically those that are suitable for field-based research) is correlated with TL measured from venous blood (see Mitchell et al., 2014, supplemental material), or examined the degree of similarity between TL measurements derived from different sample types. We measured TL in a matched dataset of saliva, capillary blood and venous blood from 40 women. We expected that biological sample types composed primarily of leukocytes would yield TL measurements correlated with TL measured from buffy coat (the concentrated layer of leukocytes that accumulates between the plasma and red blood cell [RBC] liquid layers following centrifugation of blood). We used the monochrome multiplex quantitative polymerase chain reaction (MMQPCR) assay to measure TL in buffy coat, saliva and DBS samples. Where possible, we compared TL measured from buffy coat to TL measurements from Oragene-collected saliva (passive drool method), Oasis-collected saliva (combined buccal abrasion/mouthwash method), venous dried blood spots (vDBS), and dried blood spots from finger stick (fDBS; herein referred to as capillary blood, but see Daae et al., 1988) to determine if TL from the same individual correlated across sample types and to evaluate the degree of similarity between these values.
Methods
Samples
Samples came from venous blood, capillary blood and saliva collected from a convenience sample of 200 residents of Eugene, OR aged 18 to 77 years. These samples, known as the Eugene200 Validation Sample Set, were collected for use in the validation of potential biomarker assays to incorporate into future data collection waves and analyses carried out by the WHO’s Study on global AGEing and adult health (SAGE) (Kowal et al., 2012).
We selected a subset of 40 females from among the 88 female samples for this study who represented the two extremes of the age range. Participants were divided into two cohorts. The younger cohort comprised 20 women aged 18–24 years. The older cohort comprised 20 women aged 40–77 years.
Participants completed a brief, self-report health questionnaire and provided four different sample types (Figure 1). We collected three 7 mL EDTA-coated glass vials of venous blood from each individual. One vial of venous blood was used to pipette five 50 μL drops of blood per card onto 15 standard Whatman 903 protein saver cards (GE Healthcare, Little Chalfont, Buckinghamshire, UK) for a total of 75 venous DBS per participant. The other two tubes were centrifuged to obtain plasma, and buffy coat was collected from one of these two tubes. Capillary blood was collected using a single-use finger stick lancet, and two ~50 μL drops of blood per card were stored on a total of five DBS cards for a maximum of 10 capillary DBS (fDBS or finger stick DBS) per participant. All blood samples were stored at −80°C prior to analysis. The fDBS collection was carried out according to the protocol outlined in McDade et al., (2007). For a detailed explanation of DBS and whole blood collection procedure, see Eick et al., (2016).
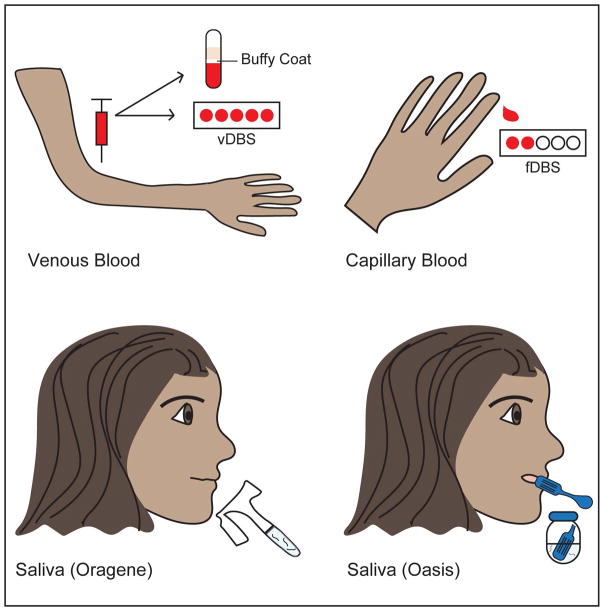
Figure 1. Sample types collected.
Four different sample types were collected using five different methods: buffy coat; venous blood stored on DBS cards (vDBS); capillary blood stored on DBS cards (fDBS); saliva collected using the Oragene kit; and, saliva collected using the Oasis kit. The Oragene collection protocol requires participants to spit into a tube until 2 mL of saliva has been collected (indicated by a black line). The Oasis collection kit calls for participants to gently abrade the inside of their cheek using the plastic cheek-scraping tool provided, rinse their mouth with solution provided in a collection cup, and then spit this solution into the collection cup. The end of the cheek-scraping tool used to abrade the cheek is also placed in the collection cup before sealing.
Participants provided saliva samples using two different modes of collection, the Oragene DNA Self-Collection kit (DNAGenotek by OraSure Technologies, Inc., Bethlehem, PA) and Oasis DNA.SAL kit (Oasis Diagnostics Corporation, Vancouver, WA). Both types of saliva sample were collected according to the respective manufacturer’s protocol and were stored at room temperature.
IRB approval for sample collection was obtained from the University of Oregon Committee for the Protection of Human Subjects (CPHS) and all participants provided informed consent.
DNA Extractions
DNA from frozen buffy coat samples was extracted using the QIAGEN PureGene Blood Core kit (QIAGEN Inc., Hilden, Germany). DNA from Oragene saliva was extracted using PrepIT L2P (DNA Genotek) following the manufacturer’s protocol. DNA from Oasis saliva was extracted using the MiniSAL DNA Isolation kit (Oasis Diagnostics) following the manufacturer’s protocol. DNA was extracted from DBS stored on Whatman 903 protein saver cards using the magnetic bead-based ChargeSwitch Extraction kit (Thermo Fisher Scientific, Inc., Waltham, MA) according to the manufacturer’s protocol for dried blood spots with the following modifications: two 6-mm DBS punches (sliced as thinly as possible with a razor blade) were incubated for ~8 hours in the buffer provided, and samples were vortexed after the first and second hours of incubation. DBS samples were eluted in 60 μL Elution Buffer to concentrate them. We tested several different protocols to determine which extraction method yielded the largest quantity of DNA from DBS samples (Table 1). All DNA extracts were stored at −20°C.
Table 1.
DNA extraction methods tested and used for all sample types.
| Sample Type | Major Predicted Cell Population | Extraction Method | Modifications to Protocol | Alternative Methods Tested |
|---|---|---|---|---|
| Buffy coat | Leukocytes | QIAGEN PureGene Blood Core kit | ||
| Oragene | Leukocytes | Oragene OG-500 PrepIT L2P | ||
| Oasis | Buccal Epithelial Cells | Oasis MiniSAL DNA Isolation kit | ||
| vDBS | Leukocytes | Invitrogen ChargeSwitch Extraction kit | Reduced elution volume from 150 μl to 60 μl | NucleoSpin Extraction kit, QiAMP DNA Investigator & QiAMP Mini kit |
| fDBS | Leukocytes | Invitrogen ChargeSwitch Extraction kit | Reduced elution volume from 150 μl to 60 μl | NucleoSpin Extraction kit, QiAMP DNA Investigator & QiAMP Mini kit |
DNA Quantification and A260/A280 ratio
Concentration of double-stranded DNA extracted from buffy coat and saliva samples was determined using the high sensitivity Qubit kit (accurate for initial sample concentrations from 10 pg/μL to 100 ng/μL; Thermo Fisher Scientific) and a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). All buffy coat samples and saliva samples were diluted to a working stock concentration of 2 ng/μL in PCR-grade water and single-use aliquots were kept frozen at −20°C. DBS extractions were not further diluted due to their low DNA concentrations. A NanoDrop spectrophotometer (Thermo Fisher Scientific) was used to assess the A260/A280 ratios of the buffy coat and saliva samples. Six buffy coat and 14 Oasis samples yielded A260/A280 values outside the typical range of sufficient sample purity. However, because provisional exclusion of these samples from the dataset did not significantly change associations between TL and age or between TL in different sample types, these samples were included in all subsequent analyses.
Multiplex Assay to Determine Relative TL (T/S Ratio)
We assessed relative TL using the monochrome multiplex TL assay (Cawthon 2009) on a BioRad CFX96 thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA) using the following cycling profile: denaturation and Taq activation for 15 min at 95°C; two cycles of 2 s at 98°C followed by 30 s at 49°C; 50 cycles of 2 s at 98°C; 30 s at 59°C; 15 s at 72°C with signal acquisition; 30 s at 84°C and 15 s at 85°C with signal acquisition; 60°C for 1 min, and then a melt curve from 60°C to 97°C at increments of 1°C for 5 s at each temperature.
PCR reactions were performed in a volume of 25 μL using the following reagents (final concentrations in the reaction are indicated in parentheses): AmpliTaq Gold DNA polymerase (0.625U; Applied Biosystems by Thermo Fisher Scientific), 1X PCR Gold buffer (50 mM KCl, 1.5 mM Tris-HCl; Applied Biosystems), MgCl2 (3 mM; Applied Biosystems), Betaine (1 M; Sigma-Aldrich, St. Louis, MO), DTT (1 mM; Sigma-Aldrich), Sybr Green I (0.75x; Applied Biosystems), dNTPs (0.2 mM each; Applied Biosystems), telomere primers (telg, ACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT and telc, TGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCTAACA; 500 nM; Eurofins Genomics, Louisville, KY), albumin primers (albu/albd; 300 nM; Eurofins Genomics), template DNA (10 ng for positive controls and saliva and buffy coat samples, 5 ng for DBS). Four concentrations of reference HeLa DNA (3-fold serial dilutions) (New England BioLabs, Ipswich, MA) were also included on each plate in triplicate (50 ng – 1.87 ng for buffy coat and saliva plates, 16.7 ng – 0.62 ng for DBS plates) to create a standard curve. Four positive control wells (10 ng/well HeLa genomic DNA) and two negative controls (no template) control wells were included on each plate. All experimental samples were assayed in triplicate. Assays were performed using 96-well PCR plates (8 rows × 12 columns; Applied Biosystems). The first and last rows and the first and last columns were not used to avoid “edge effects” (Eisenberg et al., 2015). Plates were sealed using clear qPCR strip-caps (Thermo Fisher Scientific).
Data Processing
For all runs, melt curves and amplification curves were examined to assess run and sample quality. Outlier wells were excluded, and we confirmed the absence of contamination in the negative controls.
Cawthon’s standard curve-based T/S ratio is a relative measure of TL. This is defined as the number of nanograms of the standard DNA that matches the experimental sample for copy number of the telomere template (T), divided by the number of nanograms of the standard DNA that matches the experimental sample for copy number of the single copy gene (S, albumin) (Cawthon, 2009). Baseline-subtracted curve fit predicted concentration data were exported to Microsoft Excel for both the telomere product and the albumin product. T/S ratios for each sample replicate were calculated, and subsequently the mean was calculated. For the standard curve telomere product (T), mean R2 was 0.99, with a mean efficiency of 90.48% across all plates. For the standard curve single copy gene product (S), mean R2 was 0.99 with a mean efficiency of 101.52%.
Samples that did not fall within the standard curve for either the telomere amplicon or the albumin amplicon were rerun on a plate with a 6-point standard curve (range 150 ng – 0.62 ng, 3-fold dilutions). In addition, samples with an intra-assay coefficient of variation (CV) based on three replicate wells of greater than 15% were rerun. For samples with an intra-assay CV of less than 15%, the mean T/S value of a sample was calculated, and the replicate that deviated most from the mean was marked as a potential outlier. The mean was then recalculated without the outlier, and this value was then divided by the value of the potential outlier. If the absolute log value of this ratio was greater than 0.4, the value was excluded (Needham et al., 2015).
Data Filtering and Quality Control
The MMQPCR assay for TL measurement has been shown to be sensitive to error (Aubert et al., 2012; Bustin et al., 2009; Eisenberg et al., 2015). We therefore implemented several quality control measures. The filtering criteria are given in Figure 2. Briefly, samples that did not amplify were removed. Next, samples that fell outside the range of at least one of the standard curves (for either T or S) were removed. Subsequently, percent standard error (SE) was calculated for each T/S ratio and used as the third criterion for sample exclusion. Unlike CV, SE is sensitive to sample size and is therefore a better measure of the validity of TL values yielded by the qPCR assay (Eisenberg et al., 2015). Samples were excluded if the SE exceeded 8.66%; this value is comparable to a CV of 15% for three replicates (Eisenberg et al., 2015). Our final dataset included 35 buffy coat samples, 39 Oragene samples, 37 Oasis samples, 4 venous DBS, and 6 capillary DBS samples. Due to the marked reduction in DBS sample size, no further statistical analyses were performed on these datasets. T/S ratios for all buffy coat, Oragene, and Oasis samples that were successfully amplified and included in statistical analyses are provided in Table S1 and descriptive statistics for each of these datasets can be found in Table S2.
Figure 2. Data filtering process.
Datasets for each sample type underwent filtering to exclude samples of insufficient quality according to the steps outlined above. This was done to account for the sensitive nature of the qPCR assay, to ensure that inaccurate telomere length measurements were not included in the subsequent statistical analyses.
Age Association
Leukocyte TL has consistently shown a negative association with age (see Müezzinler et al., 2013). Because of this, we tested for the presence of this association in each sample type as a measure of external validity before carrying out inter-sample correlation analyses (Eisenberg et al., 2015). Spearman’s rank correlation was used to determine if TL in buffy coat, Oragene, and Oasis samples correlated with chronological age. Buffy coat and Oragene saliva TL showed the expected negative correlation with age (ρ = −0.47, p = 0.002; 95% CI: −0.71, −0.15; ρ = −0.29, p = 0.04; 95% CI: −0.58, 0.05, for buffy coat and Oragene, respectively; Table S3). TL measurements from Oasis saliva failed to display the expected negative association with age (Table S3). After applying our quality control and filtering criteria the DBS datasets were drastically reduced. As a result, we did not test for correlations between age and TL in vDBS and fDBS samples.
In addition, to compare how well buffy coat, Oragene, and Oasis measurements of TL reflect chronological age, we fit linear regression models and compared their fit using the Akaike Information Criterion (AIC), where lower AIC scores indicate a better fit. For this analysis, the three datasets were reduced to the individuals with data for all three sample sources (n = 28). Of the three modeled measurements, the buffy coat data had the best fit (AIC = 246.3; dAIC = 0.0; wi = 0.720; ER = 1.38). When each was compared to the buffy coat model, the Oragene model performed better than the Oasis model (AIC = 248.5; dAIC = 2.2; wi = 0.237; ER = 3.04, and AIC = 251.9; dAIC = 5.6; wi = 0.043; ER = 16.74, respectively).
Quantifying MMQPCR Assay Variation
The intraclass correlation coefficient (ICC) was calculated using a one-way random effects model. This statistic provides a ratio of between plate variance to overall variance and avoids the issues of data heteroscedasticity and differing Y-intercepts between variables that can weaken the reliability of CV as a quality control measure for TL analyses (Eisenberg, 2016; Eisenberg et al., 2015; Verhulst et al., 2015).
Intra-assay CV and inter-assay CV were also calculated. These values represent the amount of variation among positive assay replicates on each plate, averaged across all 15 plates, and a comparison of average positive control T/S values run on all 15 plates, respectively.
A total of 49 samples were assayed more than once. Of these samples, 59.2% were assayed twice, 38.8% were assayed three times, and 2% were assayed four times, on separate plates. The ICC for all positive controls run across 15 plates was 0.91 (95% CI: 0.81, 0.96). Our ICC indicates substantial reproducibility of the TL assay results and is consistent with ICC values reported previously for the monochrome multiplex qPCR-based TL assay (Eisenberg et al., 2017a, 2017b, Lan et al., 2009, 2013; Shen et al., 2011). The mean intra-assay CV was 6.64% and the mean inter-assay CV was 11.74% for positive controls run across all 15 plates.
Statistical Analyses
Data were log-transformed to perform paired t-tests between buffy coat and Oragene, buffy coat and Oasis, and Oragene and Oasis datasets to assess the relative similarity of TL estimates yielded by these sample types. Bland-Altman plots were generated to assess the agreement in TL obtained from pairwise comparisons of buffy coat and Oragene T/S, buffy coat and Oasis T/S, and Oragene and Oasis T/S. Spearman’s rank correlation was used to determine if TL measurements from buffy coat, Oragene, and Oasis samples were correlated.
All statistical analyses were performed using RStudio: Integrated Development for R (2015). GraphPad Prism 5 was used to generate Bland-Altman agreement analysis plots. P values < 0.05 were considered significant.
Results
DNA Quantity and Quality Varied Across Sample Types
Results are summarized in Table 2. Oragene saliva DNA yield was the greatest of all sample types with a mean DNA concentration of 57.7 ng/μL [range: 2.48 – 308.0 ng/μL], as compared to 31.0 ng/μL [0.82 – 125.0 ng/μL], 12.5 ng/μL [2.72 – 41.0 ng/μL], 1.39 ng/μL [0.61 – 2.11 ng/μL] and 1.84 ng/μL [1.19 – 2.59 ng/μL] for buffy coat, Oasis saliva, vDBS and fDBS, respectively.
Table 2.
Mean DNA yield and percent standard error (SE) for each sample type tested.
| Sample type | Mean DNA yield (ng/μL) | Range (ng/μL) | Mean SE (%) | N |
|---|---|---|---|---|
| Buffy coat | 31.0 | 0.82 – 125.0 | 3.03 | 36 |
| Oragene | 57.7 | 2.48 – 308.0 | 2.21 | 40 |
| Oasis | 12.5 | 2.72 – 41.0 | 2.01 | 37 |
| vDBS | 1.39 | 0.61 – 2.11 | 46.8 | 21 |
| fDBS | 1.84 | 1.19 – 2.59 | 22.3 | 14 |
Low SE is indicative of greater measurement reliability. All extractions that were successfully amplified (i.e., passed data filtering steps 1 and 2 in Figure 2) are included in averages.
The mean standard error was 3.03% for buffy coat, 2.21% for Oragene saliva, 2.01% for Oasis saliva, 46.8% for vDBS, and 22.3% for fDBS for all samples that were successfully amplified (those that passed the first and second data filtering steps; see Figure 2).
Oragene Saliva and Buffy Coat TL were Correlated and Significantly Different
T/S ratios for buffy coat, Oragene, and Oasis sample sets were significantly different (Table 3, S2 and Figure 3A). Mean buffy coat T/S was greater than mean Oragene T/S by 0.13 (t(33) = 5.53, p = 3.86 × 10−6) and greater than mean Oasis T/S by 0.35 (t(31) = 10.62, p = 7.55 × 10−12). Mean Oragene T/S was greater than mean Oasis T/S by 0.18 (t(35) = 5.42, p = 4.6 × 10−6).
Table 3.
Paired t-test between sample types.
| Buffy coat - Oragene | Buffy coat - Oasis | Oragene - Oasis | |
|---|---|---|---|
| t-statistic | t(33) = 5.53 (p = 3.86 × 10−6) | t(31) = 10.62 (p = 7.55 × 10−12) | t(35) = 5.42 (p = 4.6 × 10−6) |
| Mean difference | 0.13 | 0.35 | 0.18 |
| 95% CI | 0.08 – 0.18 | 0.28 – 0.42 | 0.11 – 0.24 |
Figure 3. Comparison of TL in buffy coat, Oragene saliva, and Oasis saliva samples.
(A) Graphical representation of buffy coat (red), Oragene saliva (blue), and Oasis saliva (green) T/S ratios. The black solid line indicates the mean T/S ratio, and the error bars show the standard error of the mean. (B–D) Bland-Altman plots were generated to assess agreement in TL between (B) buffy coat and Oragene (bias ±SD: 0.26 ±0.31; limit of agreement (LoA), lower and upper: −0.34 – 0.86), (C) buffy coat and Oasis (bias ±SD: 0.66 ±0.45; LoA, lower and upper: −0.23 – 1.55), and (D) Oragene and Oasis (bias ±SD: 0.29 ±0.35; LoA, lower and upper: −0.40 – 0.97) T/S values. The dashed blue line indicates the bias, the dashed black lines indicate the 95% limits of agreement (mean ±1.96 SD), and the solid black line indicates zero bias. Strong bias and poor measurement precision are apparent in all three Bland-Altman plots.
Bland-Altman plots showed strong bias for all pairwise comparisons (Figure 3B – D). None of these data clustered around the zero line, as would be expected if TL measurements across these sample types were equivalent. Limits of agreement (LoA), which are used to indicate the relative precision of measurements, were broadest for the comparison between buffy coat and Oasis TL (bias ±SD: 0.66 ±0.45; LoA, lower and upper: −0.23 – 1.55) and narrowest for the comparison between buffy coat and Oragene TL (bias ±SD: 0.26 ±0.31; LoA, lower and upper: −0.34 – 0.86). Although the buffy coat – Oragene Bland-Altman plot showed greater measurement precision than the other pairwise comparisons, the agreement in TL between different sample types was ultimately poor.
Intra-individual buffy coat and Oragene saliva T/S ratios were significantly correlated (ρ = 0.48, p = 0.002, n = 34; Figure 4 and Table 4). Oasis saliva T/S was not correlated with either buffy coat or Oragene saliva T/S (Table 4). As noted, the DBS datasets were drastically reduced, which limited further analysis. As a result, these samples were not tested for correlation with other sample types.
Figure 4. Buffy coat-Oragene saliva TL correlation.
Spearman’s rank correlation for buffy coat TL and Oragene saliva TL. TL showed a significant positive correlation between these sample types (Spearman’s ρ = 0.48; p = 0.002).
Table 4.
Spearman’s rank correlation between sample types.
| Buffy coat | Oragene | Oasis | |
|---|---|---|---|
| Buffy coat | * | ||
| Oragene | ρ = 0.48 (p = 0.002) | * | |
| Oasis | ρ = 0.15 (p = 0.21) | ρ = 0.08 (p = 0.32) | * |
Bold value indicates a significant correlation.
Sample sizes for each comparison: Buffy coat-Oragene = 34; Buffy coat-Oasis = 32; Oragene-Oasis = 36.
Discussion
Currently, population-level studies of aging have only a few options for collecting high-quality biological samples in the field. To our knowledge, few studies have explicitly evaluated whether TL measured from minimally invasive sample types (e.g., saliva and DBS) is correlated with TL measured from matched venous blood (e.g., Mitchell et al., 2014) or characterized the magnitude of similarity in TL values. Based on the literature, we predicted that samples composed predominantly of leukocytes would yield TL values correlated with TL from buffy coat extracted from whole blood and evaluated this prediction using a dataset of TL measurements from matched buffy coat, saliva and DBS samples. Consistent with our prediction, we found that TL measured from buffy coat and Oragene samples was correlated within individuals; however, TL in buffy coat and Oragene samples differed substantially across the range of TL estimates for each sample type (Table 3 and Figure 3). This indicates that TL measured from Oragene-collected saliva should not be directly compared to TL measured from venous blood. Nevertheless, despite the poor agreement between buffy coat and Oragene TL in terms of actual T/S ratios, the correlation between these sample types as well as their confirmed negative association with age suggests each may serve as appropriate biomarkers of aging and age-related decline. Note, however, that the negative correlation between buffy coat TL and age was stronger than that between Oragene TL and age. Oasis saliva and DBS were not found to be suitable options, albeit for different reasons. On the one hand, although Oasis samples yielded adequate quantities of DNA, TL in this sample type showed no correlation with age or with TL in buffy coat or Oragene samples. On the other hand, DBS DNA yields were most often too low for reliable TL measurement using the MMQPCR assay.
There are several factors that may account for the differences in TL we observed between sample types. First, TL has been shown to be tissue- and cell-type specific (Daniali et al., 2013; de Lange et al., 1990; Lin et al., 2016; Rufer et al., 1998, 1999; Weng, 2001; Weng et al., 1995). Previous research has shown that the majority of cells found in samples collected using buccal swab, mouthwash, and cytobrush techniques are epithelial in origin (Endler et al., 1999; Thiede et al., 2000), while whole saliva samples most often contain a larger proportion of leukocytes (Garcıá-Closas et al., 2001; Rogers et al., 2007; Rylander-Rudqvist et al., 2006). Unlike leukocyte TL, buccal epithelial TL does not show a pattern of decline with age (Thomas et al., 2008). Therefore, correlation between TL measured in samples dominated by buccal cells and those composed primarily of leukocytes is not to be expected (Thomas et al., 2008). Second, DNA extraction method (Cunningham et al., 2013; Raschenberger et al., 2016), differences in execution of common laboratory techniques (e.g., pipetting), and differences in PCR sample preparation can all affect DNA quantification due to the vulnerability of telomeric DNA to degradation and the inherent sensitivity of the qPCR assay (Aubert et al., 2012; Aviv et al., 2011; Bustin et al., 2009; Eisenberg et al., 2015; Nussey et al., 2014; Olsen et al., 2012).
TL measured from Oragene Saliva
Saliva is an appealing option for field-based research because it is low-risk and easy to collect in non-clinical settings. The prospect of incorporating TL measurements from Oragene-collected saliva into large-scale epidemiological analyses is strengthened by numerous reports of consistently high DNA yields from Oragene samples (Hansen et al., 2007; Iwasiow et al., 2011; James et al., 2011; Nunes et al., 2012; Rogers et al., 2007; Rylander-Rudqvist et al., 2006), especially when compared to samples collected using buccal swabs or similar techniques (Hansen et al., 2007; Rogers et al., 2007). Additionally, multiple studies have shown that Oragene samples can be stored at room temperature for long periods of time without compromising DNA quality (Iwasiow et al., 2011; Nunes et al., 2012), although it is not known if this affects TL measurements. As measuring TL from Oragene-collected saliva has become more and more common (e.g., Chen et al., 2015; Gotlib et al., 2015; Lapham et al., 2015; Theall et al., 2013; Whisman et al., 2016), most likely due to the relative ease of collection (Chen et al., 2015) and assumption that salivary TL is comparable to BTL (Gotlib et al., 2015; Whisman et al., 2016), it has become increasingly important to not only confirm that buffy coat and Oragene-collected salivary TL are correlated, but also to determine the degree to which measurements from these two sample types are similar.
TL measured from Buffy Coat
Most telomere research has relied upon leukocyte DNA isolated from peripheral blood mononuclear cells (PBMCs) (Müezzinler et al., 2013). Our study is one of very few to use buffy coat as a primary source of leukocyte DNA. Our buffy coat DNA yields were, in general, lower than the DNA yields obtained from the Oragene saliva samples, and low DNA concentration likely caused some buffy coat samples to fail our quality control measures (Figure 2). This was surprising as buffy coat is routinely used as a source for high-quality DNA in qPCR-based research, including some TL studies (McDonnell et al., 2017; Mirabello et al., 2009; Wolkowitz et al., 2011). However, yields of DNA from frozen buffy coat have been found to be lower, have high inter-individual variability, and can be more difficult to process than the all-cell-pellet (ACP) of whole blood (Gail et al., 2013). Furthermore, participant variables (e.g., sex, age, BMI, health status, tobacco consumption), sample processing times or techniques, and manual versus automated DNA extraction have also been found to significantly affect DNA yields from buffy coat (Caboux et al., 2012). These factors, either alone or in combination, may have contributed to the low buffy coat DNA yields obtained in the present study and may also affect field-collected samples.
Despite these factors, we found a significant correlation consistent with the findings of Mitchell et al., (2014), namely that TL in Oragene-collected saliva and PBMC samples was significantly correlated (r = 0.72, p = 0.002). Because buffy coat is a less pure sample of WBCs as compared to PBMC isolate, a more moderate correlation than that of Mitchell et al., (2014) is to be expected.
TL Measured from Oasis-Collected Saliva is Not Correlated with Buffy Coat TL
Oasis TL measurements were significantly different from those derived from buffy coat or Oragene samples (Table 3 and Figure 3). We also found no significant correlation between Oasis TL and TL measured from buffy coat extracted from venous blood. This suggests Oasis sample cellular composition is substantially different from the other sample types tested in this study (Rogers et al., 2007). Because TL measurements from buccal cells show no correlation with age or with BTL (Thomas et al., 2008), and because samples collected using mouthwash, buccal swab, or similar oral sample collection techniques are typically dominated by cells from the buccal epithelium (Endler et al., 1999; Thiede et al., 2000), the absence of a correlation between Oasis TL measurements and those from the other two sample types may, in part, be the result of the mouthwash and/or buccal abrasion steps included in the Oasis collection protocol that could have introduced a substantial number of buccal epithelial cells into the samples. Oasis saliva samples may have also contained a disproportionate amount of pre-apoptotic, senescent or near-senescent buccal epithelial cells, resulting in lower mean TL values than might be observed in a tissue sample from oral mucosa (Thomas et al., 2008; Zayats et al., 2009).
Additionally, oral samples collected using methods that include epithelial abrasion (e.g., Oasis, buccal swab and cytobrush techniques) have been shown to yield lower quality DNA when compared to whole blood or saliva collected by passive drool (e.g., Oragene) (Garcıá-Closas et al., 2001; Hansen et al., 2007; Zayats et al., 2009). While TL in Oasis saliva showed no correlation with age or TL from other sample types, we encountered no major difficulties in obtaining adequate yields or amplifying DNA from this sample type (Table 2). This further indicates that real sample type-specific differences in cell composition may underlie the discrepancy between TL measured in Oasis samples as compared to those from buffy coat and Oragene saliva samples.
DBS DNA Yields Were Insufficient for qPCR Amplification
Unlike saliva samples, DBS samples yielded extremely low quantities of DNA, despite preliminary testing of several DBS-specific DNA extraction methods to establish which yielded the highest quality and quantity of DNA (Tables 1, 2). The drastic reduction in DBS datasets (90% and 85% of samples removed from vDBS and fDBS datasets, respectively; Figure 2) following quality control measures precluded further statistical analysis. The failure to amplify was likely due to the extremely low DNA concentrations observed in these samples.
Whatman 903 protein saver cards were chosen because they are widely used for large-scale DBS sample collection, broadening the utility and possible application of our findings. While FTA cards are specifically designed for DNA collection, they have been used less frequently in field-based research and may pose technical issues similar to the ones encountered in this study when amplifying extracted DNA using qPCR (see Nattrass et al., 2011).
A drop of blood collected on a DBS card typically only contains approximately 50 μL of whole blood (McDade et al., 2007): to account for possible limitations imposed by small sample volume, we tested two different extraction protocols using two, three or four 6-mm DBS punches. DNA yields for two- and three-punch samples were low but essentially equal, while four-punch samples showed lower DNA concentrations and were more difficult to work with due to the larger amount of filter paper in the sample.
Finally, it is of note that the DBS samples used in our study were collected, dried and stored under optimal conditions for sample preservation. Field-based collections are typically carried out in settings where ideal preservation measures are not possible; this may result in even greater difficulty when attempting DNA extractions from DBS collected in the field. While alternative methods of blood collection (e.g., capillary tube from finger prick) may yield sufficient quantities of DNA for PCR amplification, such methods were not included in our study due to sample processing and cold storage requirements that preclude their widespread use in field-based research.
Considerations for Future Research Using Minimally Invasive Sample Types
Minimally invasive samples are an appealing option for use in biomarker analyses, particularly for collecting large numbers of samples in the field or other non-clinical settings. However, experimental design and methodology must be considered if TL is to be effectively incorporated as a biomarker of aging and disease in population- and field-based research. Our results demonstrate that TL is not consistent across all biological sample types or methods of sample collection. Given that shorter BTL has often been used as evidence of the cumulative impact of environmental, behavioral or psychosocial stressors on health, it is important that all values used to identify such effects are derived from sample types that display the negative association between TL and age upon which such hypotheses rely (Eisenberg, 2016; Eisenberg et al., 2015), and also contain comparable cell population ratios.
Although buccal swab and abrasion techniques are commonly used and cost-effective methods of sample collection, DNA from samples collected by these means does not appear suitable for measuring TL as a biomarker of aging. Similarly, while DBS are routinely collected from neonates in over a dozen countries and allow for simple and rapid testing for metabolic disorders, HIV infection and other conditions, our results demonstrate that DNA from blood stored on Whatman 903 DBS cards may be an impractical choice for TL analysis until extraction protocols are optimized to consistently yield greater quantities of DNA. DNA cleanup methods might enhance DBS extraction quality and improve the likelihood of successful qPCR amplification; however, such protocols will remain infeasible until greater DNA yields can be consistently produced from this sample type due to the expected loss of some DNA during clean-up.
The advantages of minimally invasive collection methods over venipuncture include both the relative ease of collection, and, generally, the lower per-participant cost. While we observed a significant correlation between Oragene and buffy coat TL, the Bland-Altman plot for this comparison displayed strong bias, indicating that TL measurements derived from these two sample types are substantially different. These findings should be verified in larger datasets. Future research should aim to identify the source of measurement variability by employing cell-sorting techniques to characterize the composition of blood and saliva samples. This will help to determine the relative effect of sample type as compared to extraction method on TL estimates.
Potential Study Limitations
Our study had several potential limitations. First, while we suspect cellular composition differed by sample type, we were unable to evaluate cell type frequencies in our samples to test this supposition. Previous genetic analyses have found no correlation between buccal cell TL and age in adults (Küffer et al., 2016; Thomas et al., 2008). Furthermore, the predictive capacity of epigenetic models of age using saliva and buccal swab samples deteriorates in direct proportion to the amount of buccal cells present in the sample (Eipel et al., 2016). In the future, characterization of each sample type’s cellular makeup could be used to test the presumed effect of sample type on TL estimates (Eipel et al., 2016; Thiede et al., 2000) to more definitively determine which sample types are truly suitable for use in age-related telomere research.
Second, sample types from the same individual were positioned randomly on plates with respect to the well positions of that sample on other plates. Because previous studies have shown that well position can significantly affect qPCR-based TL estimates (Eisenberg et al., 2015), the random positioning of our samples may have decreased our power to detect correlations between the various sample types. Therefore, inter-sample TL correlation in buffy coat and Oragene samples may in fact be stronger than reported here.
While Eisenberg et al., (2015) proposed a well-correction factor to account for well-to-well variation, we considered application of this correction factor to our dataset inappropriate due to the considerable differences in TL distribution, mean and variance in different sample types from the sample individual (see Table S2 and Figure 3A). Furthermore, this correction factor was originally developed for correction of T/S values from the same sample type and across a much larger number of samples that allowed for a better estimate of well-specific effect.
Third, systematic differences in sample collection and DNA extraction protocols may have influenced the disparity observed in TL values between sample types. Salting-out, ethanol precipitation and column-based extraction methods rely on different chemical reactions to isolate DNA, and these methodological distinctions may lead to differences in TL estimates (Aubert et al., 2012; Aviv et al., 2011, 2006; Cunningham et al., 2013; Eisenberg et al., 2015; Olsen et al., 2012). As noted previously, we chose to tailor methods of DNA extraction to each sample type to ensure the highest possible accuracy of TL estimates produced by the qPCR assay. Quantitative PCR is a powerful and robust analytical method, but it requires adequate quantities of high-quality DNA to yield accurate data (Aubert et al., 2012; Eisenberg et al., 2015): although extraction method may change the mean T/S ratio, it is not intrinsically expected to affect correlations between samples extracted by different methods, while DNA samples of insufficient quantity or quality would have prevented us from obtaining reliable TL estimates. However, because we could not account for the influence of methodological differences in DNA extraction technique on TL measurement variation, we are unable to establish whether sample type or extraction method was the primary determinant of differences in TL measurements between sample types.
Conclusions
Understanding how populations age and the factors that contribute to the process of aging and age-related decline requires the evaluation of biomarkers that can be analyzed from biological samples collected from large-scale, field-based studies. Using a matched dataset we demonstrate that TL measured from different sample types using the MMQPCR assay is indeed significantly different. Future studies that use TL as a biomarker should be carried out judiciously using verified sample types, and longitudinal studies should ideally be limited to comparisons within the same type of biological sample.
Supplementary Material
Acknowledgments
Grant sponsorship
This research was funded by an NIH NIA Interagency Agreement with the World Health Organization (Grant Number: YA1323-08-CN-0020); NIH (Grant Number: R01-AG034479); and the University of Oregon.
We would like to thank the residents of Eugene, OR who participated in this study and all those who helped to collect data for the Eugene200 Validation Sample Set. We also wish to thank Dr. Hans Dreyer for the use of the BioRad CFX96 Thermocycler for the MMQPCR TL assay, Dr. Ryan Raaum for his assistance with data analysis, and comments from two anonymous reviewers that helped to strengthen the manuscript.
Footnotes
Institution from which paper emanated
University of Oregon, Eugene, OR 97403, USA
Authors’ Contributions
GNE, PK, JJS and KNS designed the Eugene200 study; EAG, GNE, DTAE, and KNS designed the telomere study; EAG, GNE, and DC performed the DNA extractions and telomere assays; EAG and GNE analyzed data; EAG, GNE and KNS wrote the manuscript with intellectual contributions from JJS and DTAE. All authors read and approved the final manuscript.
Statement of Informed Consent
IRB approval for the collection was obtained from the University of Oregon Committee for the Protection of Human Subjects (CPHS), IRB# 08202014.022, entitled “Bioassay Validation for the World Health Organization’s Study on Global AGEing and Adult Health (SAGE).” All participants provided informed consent.
Statement of Competing Interests
The authors wish to acknowledge Oasis Diagnostics Corporation for providing collection kits for the Eugene200 and DNA Genotek for providing collection kits for an earlier Wave of SAGE. Oasis and DNA Genotek were not involved in any aspect of the study design or the data collection, analyses or interpretation.
Literature Cited
- Aubert G, Hills M, Lansdorp PM. Telomere length measurement—Caveats and a critical assessment of the available technologies and tools. Mutat Res Mol Mech Mutagen. 2012;730(1–2):59–67. doi: 10.1016/j.mrfmmm.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv A, Chen W, Gardner JP, Kimura M, Brimacombe M, Cao X, … Berenson GS. Leukocyte Telomere Dynamics: Longitudinal Findings Among Young Adults in the Bogalusa Heart Study. Am J Epidemiol. 2008;169(3):323–329. doi: 10.1093/aje/kwn338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv A, Hunt SC, Lin J, Cao X, Kimura M, Blackburn E. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 2011;39:20, e134. doi: 10.1093/nar/gkr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv A, Valdes AM, Spector TD. Human telomere biology: pitfalls of moving from the laboratory to epidemiology. Int J Epidemiol. 2006;35(6):1424–1429. doi: 10.1093/ije/dyl169. [DOI] [PubMed] [Google Scholar]
- Baird DM. Telomeres. Exp Gerontol. 2006;41(12):1223–1227. doi: 10.1016/j.exger.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Baker GT, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23(5):4. 223–239. doi: 10.1016/0531-5565(88)90025-3. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, … Wittwer CT. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Caboux E, Lallemand C, Ferro G, Hémon B, Mendy M, Biessy C, … Hainaut P. Sources of pre-analytical variations in yield of DNA extracted from blood samples: analysis of 50,000 DNA samples in EPIC. PloS One. 2012;7(7):e39821. doi: 10.1371/journal.pone.0039821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21–e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae DH, Nuru-Jeter AM, Adler NE, Brody GH, Lin J, Blackburn EH, Epel ES. Discrimination, Racial Bias, and Telomere Length in African-American Men. Am J Prev Med. 2014;46(2):103–111. doi: 10.1016/j.amepre.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Velez JC, Barbosa C, Pepper M, Andrade A, Stoner L, … Williams MA. Smoking and perceived stress in relation to short salivary telomere length among caregivers of children with disabilities. Stress. 2015;18(1):20–28. doi: 10.3109/10253890.2014.969704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Johnson RA, Litzelman K, Skinner HG, Seo S, Engelman CD, … Boardman LA. Telomere Length Varies By DNA Extraction Method: Implications for Epidemiologic Research. Cancer Epidemiol Biomarkers Prev. 2013;22(11):2047–2054. doi: 10.1158/1055-9965.EPI-13-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daae LN, Halvorsen S, Mathisen PM, Mironska K. A comparison between haematological parameters in “capillary” and venous blood from healthy adults. Scand J Clin Lab Invest. 1988;48(7):723–726. doi: 10.1080/00365518809085796. [DOI] [PubMed] [Google Scholar]
- Daniali L, Benetos A, Susser E, Kark JD, Labat C, Kimura M, … Aviv A. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013;4:1597. doi: 10.1038/ncomms2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10(2):518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds GW, Côté HCF, Hampson SE. Childhood Conscientiousness and Leukocyte Telomere Length 40 Years Later in Adult Women—Preliminary Findings of a Prospective Association. PLOS ONE. 2015;10(7):e0134077. doi: 10.1371/journal.pone.0134077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick G, Urlacher SS, McDade TW, Kowal P, Snodgrass JJ. Validation of an Optimized ELISA for Quantitative Assessment of Epstein-Barr Virus Antibodies from Dried Blood Spots. Biodemography Soc Biol. 2016;62(2):222–233. doi: 10.1080/19485565.2016.1169396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipel M, Mayer F, Arent T, Ferreira MR, Birkhofer C, Gerstenmaier U, … Wagner W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging. 2016;8(5):1034–1048. doi: 10.18632/aging.100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DTA. Telomere length measurement validity: the coefficient of variation is invalid and cannot be used to compare quantitative polymerase chain reaction and Southern blot telomere length measurement techniques. Int J Epidemiol. 2016;45(4):1295–1298. doi: 10.1093/ije/dyw191. [DOI] [PubMed] [Google Scholar]
- Eisenberg DTA, Borja JB, Hayes MG, Kuzawa CW. Early life infection, but not breastfeeding, predicts adult blood telomere lengths in the Philippines. Am J Hum Biol. 2017a;29(4):e22962. doi: 10.1002/ajhb.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DTA, Kuzawa CW, Hayes MG. Improving qPCR telomere length assays: Controlling for well position effects increases statistical power. Am J Hum Biol. 2015;27(4):570–575. doi: 10.1002/ajhb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DTA, Tackney J, Cawthon RM, Cloutier CT, Hawkes K. Paternal and grandpaternal ages at conception and descendant telomere lengths in chimpanzees and humans. Am J Phys Anthropol. 2017b;162(2):201–207. doi: 10.1002/ajpa.23109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler G, Greinix H, Winkler K, Mitterbauer G, Mannhalter C. Genetic fingerprinting in mouthwashes of patients after allogeneic bone marrow transplantation. Bone Marrow Transplant. 1999;24(1):95–98. doi: 10.1038/sj.bmt.1701815. [DOI] [PubMed] [Google Scholar]
- Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci. 2004;101(49):17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gail MH, Sheehy T, Cosentino M, Pee D, Diaz-Mayoral NA, Garcia-Closas M, … Ziegler RG. Maximizing DNA Yield for Epidemiologic Studies: No More Buffy Coats? Am J Epidemiol. 2013;178(7):1170–1176. doi: 10.1093/aje/kwt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcıá-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, … Rothman N. Collection of Genomic DNA from Adults in Epidemiological Studies by Buccal Cytobrush and Mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–696. [PubMed] [Google Scholar]
- Gotlib IH, LeMoult J, Colich NL, Foland-Ross LC, Hallmayer J, Joormann J, … Wolkowitz OM. Telomere length and cortisol reactivity in children of depressed mothers. Mol Psychiatry. 2015;20(5):615–620. doi: 10.1038/mp.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TvO, Simonsen MK, Nielsen FC, Hundrup YA. Collection of Blood, Saliva, and Buccal Cell Samples in a Pilot Study on the Danish Nurse Cohort: Comparison of the Response Rate and Quality of Genomic DNA. Cancer Epidemiol Biomark Amp Prev. 2007;16(10):2072–2076. doi: 10.1158/1055-9965.EPI-07-0611. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hornsby PJ. Telomerase and the aging process. Exp Gerontol. 2007;42(7):575–581. doi: 10.1016/j.exger.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama H, Ohyashiki K, Ohyashiki JH, Hayashi S, Yahata N, Ando K, … Shay JW. Telomeric length and telomerase activity vary with age in peripheral blood cells obtained from normal individuals. Hum Genet. 1998;102(4):397–402. doi: 10.1007/s004390050711. [DOI] [PubMed] [Google Scholar]
- Iwasiow RM, Desbois A, Birnboim HC. Long-term stability of DNA from saliva samples stored in Oragene® DNA. 2011 White paper retrieved from http://www.dnagenotek.com/ROW/pdf/PD-WP-005.pdf.
- James C, Iwasiow RM, Birnboim HC. Human genomic DNA content of saliva samples collected with the Oragene® self-collection kit. 2011 White paper retrieved from http://www.dnagenotek.com/ROW/pdf/PD-WP-011.pdf.
- Kowal P, Chatterji S, Naidoo N, Biritwum R, Fan W, Lopez Ridaura R, … Newell M-L. Data Resource Profile: The World Health Organization Study on global AGEing and adult health (SAGE) Int J Epidemiol. 2012;41(6):1639–1649. doi: 10.1093/ije/dys210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffer AL, O’Donovan A, Burri A, Maercker Andreas. Posttraumatic Stress Disorder, Adverse Childhood Events, and Buccal Cell Telomere Length in Elderly Swiss Former Indentured Child Laborers. Front Psychiatry. 2016;7:147. doi: 10.3389/fpsyt.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Cawthon R, Gao Y, Hu W, Hosgood HD, Barone-Adesi F, … Rothman N. Longer Telomere Length in Peripheral White Blood Cells Is Associated with Risk of Lung Cancer and the rs2736100 (CLPTM1L-TERT) Polymorphism in a Prospective Cohort Study among Women in China. PLoS ONE. 2013;8(3):e59230. doi: 10.1371/journal.pone.0059230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Cawthon R, Shen M, Weinstein SJ, Virtamo J, Lim U, … Rothman N. A Prospective Study of Telomere Length Measured by Monochrome Multiplex Quantitative PCR and Risk of Non-Hodgkin Lymphoma. Clin Cancer Res. 2009;15(23):7429–7433. doi: 10.1158/1078-0432.CCR-09-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapham K, Kvale MN, Lin J, Connell S, Croen LA, Dispensa BP, … Blackburn EH. Automated Assay of Telomere Length Measurement and Informatics for 100, 000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1061–1072. doi: 10.1534/genetics.115.178624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Cheon J, Brown R, Coccia M, Puterman E, Aschbacher K, … Blackburn EH. Systematic and Cell Type-Specific Telomere Length Changes in Subsets of Lymphocytes. J Immunol Res. 2016;2016:1–9. doi: 10.1155/2016/5371050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44(4):899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- McDonnell BJ, Yasmin, Butcher L, Cockcroft JR, Wilkinson IB, Erusalimsky JD, McEniery CM. The age-dependent association between aortic pulse wave velocity and telomere length. J Physiol. 2017;595(5):1627–1635. doi: 10.1113/JP273689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Huang W-Y, Wong JYY, Chatterjee N, Reding D, Crawford ED, … Savage SA. The association between leukocyte telomere length and cigarette smoking, dietary and physical variables, and risk of prostate cancer. Aging Cell. 2009;8(4):405–413. doi: 10.1111/j.1474-9726.2009.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Hobcraft J, McLanahan SS, Siegel SR, Berg A, Brooks-Gunn J, … Notterman D. Social disadvantage, genetic sensitivity, and children’s telomere length. Proc Natl Acad Sci. 2014;111(16):5944–5949. doi: 10.1073/pnas.1404293111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müezzinler A, Zaineddin AK, Brenner H. A systematic review of leukocyte telomere length and age in adults. Ageing Res Rev. 2013;12(2):509–519. doi: 10.1016/j.arr.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Nattrass GS, Banks RG, Pitchford WS. The effect of telomere length variation on lifetime productivity traits in sheep. In Proc Assoc Advmt Anim Breed Genet. 2011;19:247–250. Retrieved from http://www.aaabg.org/livestocklibrary/2011/nattrass247.pdf. [Google Scholar]
- Needham BL, Adler N, Gregorich S, Rehkopf D, Lin J, Blackburn EH, Epel ES. Corrigendum to “Socioeconomic status, health behavior, and leukocyte telomere length in the National Health and Nutrition Examination Survey, 1999–2002” [Soc. Sci. Med. 85 (2013) 1–8] Soc Sci Med. 2015;133:101. doi: 10.1016/j.socscimed.2015.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes AP, Oliveira IO, Santos BR, Millech C, Silva LP, González DA, … Barros FC. Quality of DNA extracted from saliva samples collected with the Oragene™ DNA self-collection kit. BMC Med Res Methodol. 2012;12(65) doi: 10.1186/1471-2288-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey DH, Baird D, Barrett E, Boner W, Fairlie J, Gemmell N, … Monaghan P. Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol Evol. 2014;5(4):299–310. doi: 10.1111/2041-210X.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen MT, Bérubé M, Robbins J, Palsbøll PJ. Empirical evaluation of humpback whale telomere length estimates; quality control and factors causing variability in the singleplex and multiplex qPCR methods. BMC Genet. 2012;13:77. doi: 10.1186/1471-2156-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschenberger J, Lamina C, Haun M, Kollerits B, Coassin S, Boes E, … Kronenberg F. Influence of DNA extraction methods on relative telomere length measurements and its impact on epidemiological studies. Sci Rep. 2016;6:25398. doi: 10.1038/srep25398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers NL, Cole SA, Lan H-C, Crossa A, Demerath EW. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;193:319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufer N, Brümmendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L, … Lansdorp PM. Telomere Fluorescence Measurements in Granulocytes and T Lymphocyte Subsets Point to a High Turnover of Hematopoietic Stem Cells and Memory T Cells in Early Childhood. J Exp Med. 1999;1902:157–168. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 1998;16(8):743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- Rylander-Rudqvist T, Hakansson N, Tybring G, Wolk A. Quality and Quantity of Saliva DNA Obtained from the Self-administrated Oragene Method--A Pilot Study on the Cohort of Swedish Men. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1742–1745. doi: 10.1158/1055-9965.EPI-05-0706. [DOI] [PubMed] [Google Scholar]
- Shen M, Cawthon R, Rothman N, Weinstein SJ, Virtamo J, Hosgood HD, … Lan Q. A prospective study of telomere length measured by monochrome multiplex quantitative PCR and risk of lung cancer. Lung Cancer. 2011;73(2):133–137. doi: 10.1016/j.lungcan.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theall KP, Brett ZH, Shirtcliff EA, Dunn EC, Drury SS. Neighborhood disorder and telomeres: Connecting children’s exposure to community level stress and cellular response. Soc Sci Med. 2013;85:50–58. doi: 10.1016/j.socscimed.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C, Prange-Krex G, Freiberg-Richter J, Bornhäuser M, Ehninger G. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 2000;25(5):575–577. doi: 10.1038/sj.bmt.1702170. [DOI] [PubMed] [Google Scholar]
- Thomas P, O’ Callaghan NJ, Fenech M. Telomere length in white blood cells, buccal cells and brain tissue and its variation with ageing and Alzheimer’s disease. Mech Ageing Dev. 2008;129(4):183–190. doi: 10.1016/j.mad.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci U S A. 1994;91(21):9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst S, Susser E, Factor-Litvak PR, Simons MJ, Benetos A, Steenstrup T, … Aviv A. Commentary: The reliability of telomere length measurements. Int J Epidemiol. 2015;44(5):1683–1686. doi: 10.1093/ije/dyv166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, Martin-Ruiz CM. Telomeres as biomarkers for ageing and age-related diseases. Curr Mol Med. 2005;5(2):197–203. doi: 10.2174/1566524053586545. [DOI] [PubMed] [Google Scholar]
- Weng N. Interplay between telomere length and telomerase in human leukocyte differentiation and aging. J Leukoc Biol. 2001;70(6):861–867. [PubMed] [Google Scholar]
- Weng N-P, Levine BL, June CH, Hodes RJ. Human naive and memory T lymphocytes differ in telomeric length and replicative potential. Proc Natl Acad Sci. 1995;92(24):11091–11094. doi: 10.1073/pnas.92.24.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisman MA, Robustelli BL, Sbarra DA. Marital disruption is associated with shorter salivary telomere length in a probability sample of older adults. Soc Sci Med. 2016;157:60–67. doi: 10.1016/j.socscimed.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowitz OM, Mellon SH, Epel ES, Lin J, Dhabhar FS, Su Y, … Blackburn EH. Leukocyte telomere length in major depression: correlations with chronicity, inflammation and oxidative stress--preliminary findings. PloS One. 2011;6(3):e17837. doi: 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet DL, Saberi S, Oliveira L, Sattha B, Gadawski I, Côté HCF. Blood and Dried Blood Spot Telomere Length Measurement by qPCR: Assay Considerations. PLoS ONE. 2013;8(2):e57787. doi: 10.1371/journal.pone.0057787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayats T, Young TL, Mackey DA, Malecaze F, Calvas P, Guggenheim JA. Quality of DNA Extracted from Mouthwashes. PLoS ONE. 2009;4(7):e6165. doi: 10.1371/journal.pone.0006165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.