Abstract
Natural habitats are exposed to an increasing number of environmental stressors that cause important ecological consequences. However, the multifarious nature of environmental change, the strength and the relative timing of each stressor largely limit our understanding of biological responses to environmental change. In particular early response to unpredictable environmental change, critical to survival and fitness in later life stages, is largely uncharacterized. Here, we characterize the early transcriptional response of the keystone species Daphnia magna to twelve environmental perturbations, including biotic and abiotic stressors. We first perform a differential expression analysis aimed at identifying differential regulation of individual genes in response to stress. This preliminary analysis revealed that a few individual genes were responsive to environmental perturbations and they were modulated in a stressor and genotype-specific manner. Given the limited number of differentially regulated genes we were unable to identify pathways involved in stress response. Hence, to gain a better understanding of the genetic and functional foundation of tolerance to multiple environmental stressors we leveraged the correlative nature of networks and performed a weighted gene co-expression network analysis. We discovered that approximately one third of the Daphnia genes, enriched for metabolism, cell signalling and general stress response, drives transcriptional early response to environmental stress and it is shared among genetic backgrounds. This initial response is followed by a genotype and/or condition-specific transcriptional response with a strong genotype by environment interaction. Intriguingly, genotype and condition- specific transcriptional response is found in genes not conserved beyond crustaceans, suggesting niche-specific adaptation.
Keywords: waterflea, differential co-expression networks, differential gene expression, biotic stressors, abiotic stressors, ecoresponsive genes, ecological gene annotation
Introduction
Natural habitats are under increasing threat from human activity, with pronounced ecological consequences (Hoffmann & Sgro 2011; Hofmann & Todgham 2010). However, the multifarious nature of environmental change (Gunderson et al. 2016) as well as the relative timing of each stressor (Vincenzi 2014) represent major obstacles to our understanding of biological responses to environmental change. Traditionally, biological studies subject organisms to constant and severe experimental stress conditions (Hofmann & Todgham 2010; Somero 2012). However, environmental parameters fluctuate on multiple timescales, from hours to months and years. Moreover, the intensity of stress events may vary considerably, especially between biotic and abiotic perturbations. The evolutionary success of organisms resides is their capacity to keep pace with environmental conditions that change over short as well as long periods (Vincenzi 2014), and in their ability to cope with multiple stressors, which depends on the magnitude and relative timing of each stressor. Understanding these complex dynamics is essential for assessing organismal performance under prevailing conditions and under human-induced environmental change (e.g. Boyd et al. 2015)
To cope with changes in the environment, organisms must respond during and immediately after environmental perturbations (Wingfield 2013). Especially for short term responses ranging from hours to days, early response is critical to survival and fitness in later life stages (Brooks et al. 2011). Early response to environmental perturbations requires a fine tuning of the molecular machinery regulating physiological and behavioural responses. Early response genes maximize resource utilization while maintaining structural and genetic integrity by repairing and minimizing damage to cellular structure (Huisman & Kolter 1994; Ram et al. 2005). Genes interacting with the environment generally return to their original expression level after an initial acclimation phase (Eng et al. 2010). Because a large proportion of studies does not focus on this early modulation phase (but see studies on temperature of maximum tolerance: Geerts et al. 2015; Hofmann & Todgham 2010; Somero 2010), the modulation of mRNA in early response to environmental stress is largely uncharacterized. Conversely, environmental perturbations are most commonly studied in the context of organisms life cycle [e.g. tadpoles (Leduc et al. 2015), mice (Jangiam et al. 2015), marine algae (Zou et al. 2015), and plants (Kim et al. 2015)] or to uncover the mode of action of lethal concentrations of toxicants [e.g. fish (Techer et al. 2015), frogs (Mardirosian et al. 2015), bivalves (Larguinho et al. 2014), copepods (Overjordet et al. 2014), snails (Khalil 2015) crustaceans (Tang et al. 2015), and plants (Keunen et al. 2015), (Fu et al. 2014)].
Although the response to simultaneous stressors generally leads to more complex scenarios than responses to single stressors (Holmstrup et al. 2010; Rejeb et al. 2014), the analysis of a wide range of single stressors is the first critical step in the identification of defence mechanisms pathways that may be shared among stressors, leading to a better understanding of cross-tolerance mechanisms (Perez & Brown 2014). In addition, stress responses can vary substantially between genotypes as plasticity in gene expression, i.e. variation in expression of a gene in response to stress, is known to be at least partly heritable and can be affected by natural selection (Whitehead & Crawford 2006). Understanding variation in stress response among genotypes is a prerequisite to our understanding of adaptive evolution in nature.
Here, we investigate the early transcriptional response of three genotypes of the branchiopod crustacean Daphnia magna to a suite of environmental perturbations, including six biotic and six abiotic stressors. These stressors and their intensity represent either biotic ecologically relevant perturbations encountered in the natural environment or abiotic perturbations found in human-impacted environments.
Daphnia are freshwater grazers renowned as ecotoxicological models (Colbourne et al. 2005; Miner et al. 2012) and central to virtually all inland lentic aquatic habitats (Miner et al. 2012). As filter feeders these small crustaceans are exposed to numerous environmental insults to which they respond via physiological, microevolutionary or genetic mechanisms (e.g. Decaestecker et al. 2007; Latta et al. 2012; Orsini et al. 2012; Yampolsky et al. 2014). Daphnia has a parthenogenetic life cycle that allows the rearing of populations of genetically identical individuals (clones) from a single genotype, providing the advantages of isogenic model organisms while retaining the natural genetic variation (Miner et al. 2012). Capitalizing on the ability to maintain isoclonal lines in the laboratory, we exposed populations of three D. magna genotypes to twelve environmental perturbations lasting maximum 24 hours to capture the early transcriptional response to environmental perturbations via genome-wide transcriptional profiling.
We identified a few individual genes responsive to environmental perturbations and these were modulated in a genotype-specific manner. Leveraging a gene co-expression network analysis, we were able to link clusters of genes of unknown function to genes with known function and a range of environmental perturbations. Many of the identified clusters of genes were co-expressed across the environmental conditions surveyed and/or the genotypes analysed. With evidence of shared networks among conditions and/or genotypes we identified the biological roles of hundreds of genes in stress response of a key grazer of the aquatic community. Intriguingly, highly responsive networks of genes to environmental perturbations were less conserved beyond crustaceans than genes overall. This finding indicates that these genes may be broadly associated with crustacean-specific adaptation to environmental stress.
Materials and Methods
Study species
The cladoceran D. magna is a keystone species present in virtually all lentic ecosystems. This species has a parthenogenetic life cycle that allows the rearing of populations of genetically identical individuals (clones) from a single genotype. For the present study we used two natural and a recombinant genotype obtained from the crossing of the first two. The two natural genotypes were collected from a system of ephemeral rock pools from the northern distributional range of the species (Xinb3, South west Finland 59.833183, 23.260387) and a fish-rearing pond in Southern Germany (Iinb1, Germany, 48.206375, 11.709727), respectively. The Xinb3 genotype was the result of three generations of selfing, and the Iinb1 strain was selfed for one generation, leading to a predicted 87.5 and 50% reduction in their original level of heterozygosity, respectively. The recombinant line is an F2 laboratory strain part of a mapping panel supporting research on the genetic basis of adaptive traits in D. magna (Routtu et al. 2014). The genotypes will be hereafter referred to as: X - Xinb3, I - Iinb1 and XI – recombinant. The X and I genotypes were previously used to generate ‘omics’ resources for D. magna. More specifically, the X genotype was used to generate a reference genome (NCBI accession number: LRGB00000000), whereas the X and I RNA-Seq data were previously used to generate a reference transcriptome (Orsini et al. 2016). Here, we use the available RNA-Seq data previously for X and I as well as the newly generated RNA-Seq data for the XI genotype to investigate transcriptional response across stressors and genotypes.
Experimental design
The three genotypes were distributed for environmental exposures between two laboratories of the Stressflea consortium (ESF EUROCORES Programme EuroEEFG, Grant 09-EEFGFP-040) that studies mechanisms of adaptation to environmental stress using Daphnia as model species. X and I genotypes were exposed to five biotic and one abiotic perturbations: vertebrate (FI, 19 sticklebacks in 100L water) and invertebrate predation (TR, 1 adult Triops in 2L water), parasites (PA, 40,000 Pasteuria ramosa spores/mL), crowding (CR, 100 individuals/250 mL), the methylcarbamate insecticide Carbaryl (CA, 8µg/L), microcystin-producing (BX) and microcystin - free (BN) cyanobacteria. Perturbation from cyanobacteria was obtained by feeding Daphnia with a toxic (Cyanobacteria, strain MT50) and a non-toxic strain of Microcystis aeruginosa (strain CCAP 1450/1) (Lemaire et al. 2012). The recombinant genotype XI was exposed to five abiotic perturbations: Cadmium (CD, 6 µg/L), Lead (PB, 278 µg/L), low pH (5.5), Sodium Chloride (NaCl, 5g/L), and UV radiation (UV, 30 W, 36-inch Reptisun 5.0 UV-B fluorescent light bulbs). Overall the exposures cover a wide range of environmental perturbations that Daphnia is exposed to in the natural environment, including human-impacted habitats.
The exposures of the X and I genotypes were completed over two days. For each day a control (no stress imposed) was run in parallel to the environmental perturbations. Each treatment, including controls, was performed on three biological replicates and for each replica we obtained genome-wide transcription profiling. The environmental perturbations for the X and I genotypes and the UV exposures for the XI genotype were 4-hours long, whereas the remaining exposures were 24 hours long. The length of exposure varied with the environmental stress tested driven by previous pilot experiments designed to identify realistic environmental perturbations encountered in the natural environment. Prior to the exposures to environmental perturbations, clonal populations of the three genotypes were synchronized in common garden conditions – controlled climate chambers with a fixed long day photoperiod (16h light/8h dark) at 20°C- for at least two generations to reduce interference from maternal effect. ADaM medium (Aachener Daphnien Medium: Klüttgen et al. 1994) was used as growth medium and for the environmental perturbations. The animals were fed daily with 150,000 cells Scenedesmus obliquus/ml. The first generation was cultured at a density of 10 individuals/L, and increased to 50 individuals/L in large aquaria in the second generation to enable the harvesting of enough animals for the environmental perturbation exposures. The second clutch of the second generation was used for exposures to environmental perturbations. Five-day old female juveniles randomly chosen from the offspring of the second generation of the synchronized animals at a density of 100 juveniles/L were exposed to the different environmental treatments. The animal density for the exposures was determined following prior literature studies on Daphnia exposures (e.g. Jansen et al. 2011).
Up to three investigators performed the environmental exposures and randomly distributed the animals from the aquaria to the experimental vials. Exposures to environmental perturbations for the two natural strains were conducted at the University of Leuven, Belgium. The sequencing for this experiment was performed at the Finnish Institute of Molecular Medicine (FIMM, Technology Centre, Sequencing unit) at the University of Helsinki. Exposures of the recombinant line were completed at the University of Notre Dame, IN, USA. The sequencing data from this experiment were obtained at the JP Sulzberger Columbia Genome Center (https://systemsbiology.columbia.edu/genome-center). RNA-Seq data were obtained for each treatment and control in triplicates. The quality assessment of the RNA-Seq data in terms of reproducibility across the biological replicates identified the sample I_BN_r3 (toxic Microcystis treatment for strain I) as an outlier (Orsini et al. 2016). I_BN_r3 was excluded from downstream analyses as it obscured any signal from both the genotype and the treatment. Once this treatment was excluded replicates per treatment clustered as expected (Orsini et al. 2016).
RNA-Seq
Library construction was performed on three biological replicates following Nextera workflow (Illumina) with minor modifications. TruSeq PE Cluster Kit v3 (Illumina, San Diego, CA, USA) was used for paired-end sequencing with 101 bp read length and sequenced on an Illumina HiSeq2000 platform (TruSeq SBS Kit v3 reagent kit).
Read sequences were subjected to adapter trimming and quality filtering using Trimmomatic ver.0.33 (Bolger et al. 2014). RNA-Seq reads were checked for foreign RNA contamination. Human and mouse contaminant sequences were screened and removed by mapping D. magna reads onto ncbigno2014-human.rna and ncbigno2014-mouse.rna using bowtie2 ver.2.1.0 (Langmead & Salzberg 2012). Finally, 80% of the reads for the inbred genotypes and 99% of the reads for the recombinant genotype were retained (Q>20). The cleaned reads were mapped onto the reference transcriptome of D. magna obtained from de novo assembly of RNA-Seq data (Orsini et al. 2016). These data consisted mostly of the Xinb3 inbred genotype data, but also included a subset of data from the Iinb1 genotype and RNA-Seq available in public databases for D. magna at the time of the analysis (mostly, Labbe et al. 2009). Details on the generation of reference transcriptome and gene models are provided in (Orsini et al. 2016).
Differential expression analysis
Reads mapping uniquely to primary gene transcripts were used to measure differential expression of individual genes. This approach is suitable also for genes with no location or poor match to the D. magna draft chromosome assembly (NCBI accession number: LRGB00000000). Differential gene expression was calculated on normalized read counts per treatment as compared to a control (no stress) using DEseq2.0 by use of negative binomial generalized linear models (Padj=0.01; Love et al. 2014).
We assessed the conservation beyond crustaceans of genes differentially expressed in response to environmental perturbation by quantifying the correlation between fold change - no differential expression, 2-fold, 4-fold and 16 - fold expression- and the percentage of orthologous genes found in other model species for each category. By partitioning genes in categories based on their magnitude of expression after environmental perturbations, we separated genes that showed basal expression - no differential expression after any perturbation - from genes showing different magnitude of expression under at least one environmental perturbation. For each category we identified orthologs in six other model species -Daphnia pulex (dpul), Danio rerio (drer), Caenorhabditis elegans (cele), Drosophila melanogaster (dmel), Mus musculus (mmus), and Homo sapiens (hsap). This was done using an ad hoc developed orthologue detection pipeline (Appendix 1). This pipeline uses six orthologues detection engines to produce a list of annotated orthologous genes. The combined use of these engines provided robust results as compared to the use of single search engines as orthologous genes function is assigned bases on different criteria, including sequence similarity, domain architecture, and phylogenetic relationship among species. Our pipeline combines the following orthologous genes assignment tools: DODO (Chen et al. 2010), OMA (Altenhoff et al. 2015), InParanoid8 (Sonnhammer & Ostlund 2015), TreeFam (Ruan et al. 2008), Proteinortho (Lechner et al. 2011), and OrthoMCL (Li et al. 2003).
Weighted gene co-expression network analysis
To gain insights into the regulatory patterns of uncharacterized genes we performed a weighted gene co-expression network analysis with MODA (Li et al. 2016) using the normalized expression data for all genes. MODA performs differential expression analysis of gene clusters. The clusters are identified using the WGCNA Topological Overlap Matrices (TOM; Langfelder & Horvath 2008). However, differently than WGCNA, MODA uses a cut-off threshold based on optimal average density to identify clusters of genes (also called modules; Li et al. 2016). The cut-off threshold for the clusters identified by the hierarchical clustering is determined based on the optimal average density of modules defined as:
where aij is the similarity of gene i and gene j in module A. After the clusters or modules are identified, differential expression is performed using the Jaccard similarity index. The Jaccard index measures similarity between finite sample sets, and is defined as the size of the intersection divided by the size of the union of the sample sets (Jaccard 1902). When two modules appear in networks N1 and N2 (each network is represented by several gene clusters or modules), a similarity matrix B is calculated, in which each entry Bij is the similarity between the ith module in network N1 (denoted by N1(Ai)) and jth module in network N2 (denoted by N2(Aj)). The similarity index Bij is estimated as:
In our analysis N1 is the background gene network including genes expressed in all samples and biological replicates whereas N2 is the network obtained after subtracting gene clusters specific to a treatment or a genotype. We used MODA to identify gene clusters shared among genotypes as well as gene clusters shared among either the biotic or the abiotic treatments. The frequency with which each gene cluster was identified in the treatments and the genotypes is visualized in a bar plot. This visualization enables the identification of condition-specific and shared modules among genotypes and/or environmental perturbations.
Modules identified by MODA in the treatment and genotype analysis were used for gene enrichment analysis. The genes found in each module were analysed using DAVID (Huang et al. 2009) and enriched GO terms and enriched functional-related gene groups were identified based on similarity to the Drosophila Uniprot database.
Results and Discussion
Ecoresponsive gene clusters are not conserved beyond crustaceans
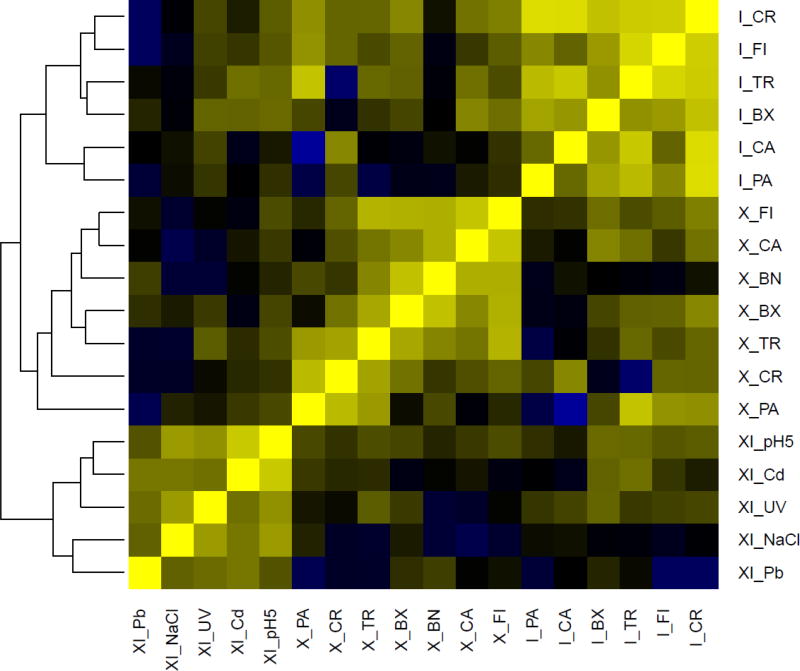
A genome wide differential expression analysis revealed that most differentially expressed genes (DEseq p-adj < 0.01) were modulated in a genotype-specific manner, with individual treatment effects obscured by genotype effects (Fig. 1). This early transcriptional response is often mediated by functionally uncharacterized genes (Table S1).
Figure 1. Genome-wide differential expression analysis.
Heat map of the most genome-wide scale differentially expressed genes (DEseq p-adj < 0.01) in the three genotypes: X, I and XI. The data are obtained from three biological replicates per genotype. Yellow indicates high similarity whereas blue indicates low similarity. The abbreviations for the environmental perturbations are as follows: FI: Vertebrate predation; TR: Invertebrate predation; PA: exposure to the parasite Pasteuria ramosa; CR: crowding; BX: microcystin-producing cyanobacteria; BN: microcystin-free cyanobacteria; CA: insecticide Carbaryl; CO: control. The environmental perturbations imposed on the recombinant genotype are as follows: CD: Cadmium; PB: Lead; pH 5.5: low pH; UV: UV-B light; NaCl: osmotic stress. The treatment BN for the I strain was not included in this analysis as a PCA plot inclusive of all data identified this treatment as an outlier (Orsini et al. 2016).
To better contextualize genes of unknown function, we conducted gene modules differential analysis on weighted gene co-expression networks using MODA (https://bioconductor.org/packages/release/bioc/html/MODA.html; Li et al. 2016) which enables the association of individual genes in co-responsive modules (gene clusters). Using this approach we identified numerous unannotated genes tightly linked to our differentially expressed genes and gained insights into the pathways and the regulatory mechanisms in which novel genes participate. This association points to the power of exposure biology for the annotation of unknown genes in an ecological context.
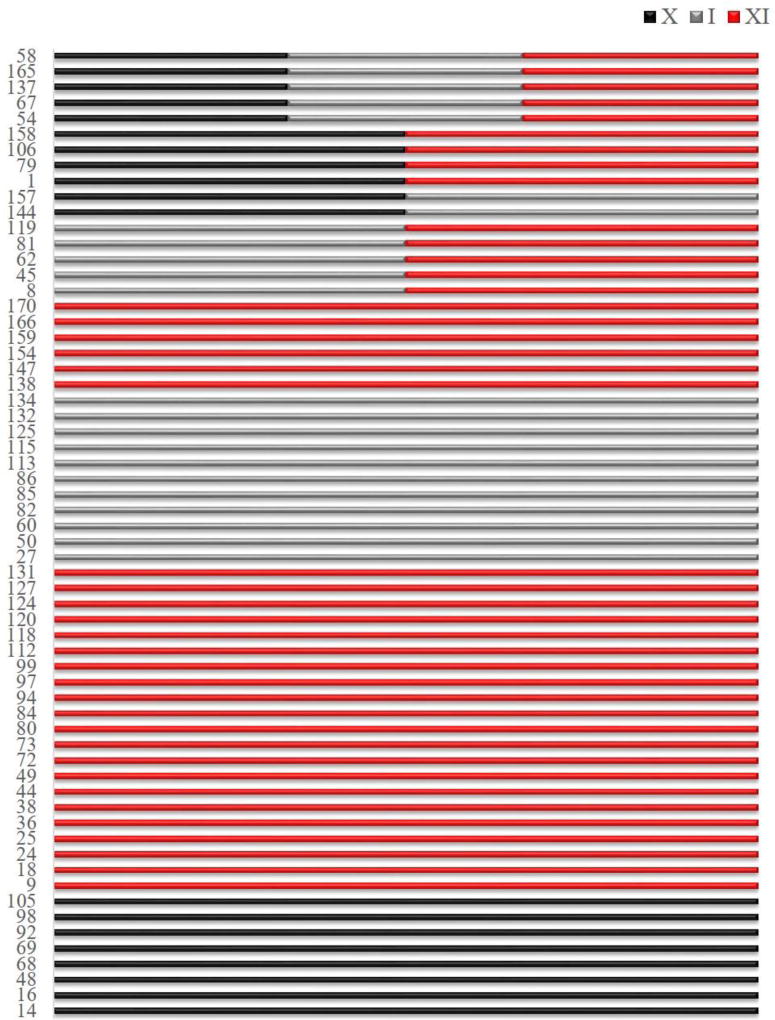
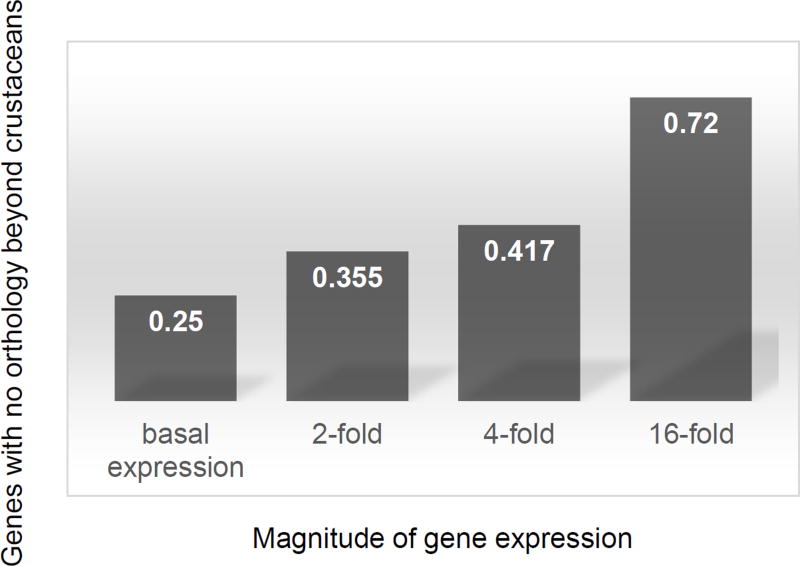
Across all genotypes we discovered 62 modules (Table S2). Of these, 5 (6.45%) are shared among the three genotypes whereas the remainder is found uniquely in one of the genotypes (Fig. 2). The shared modules include overall 11,698 genes, approximately a third of the total genes in D. magna (Orsini et al. 2016), with shared Module #58 (Table S2) including 11,537 genes. This module is enriched for morphogenesis, cell signalling, development and metabolic genes as well as by defence response genes, including oxidative response (Table S3). There are in total 47 genotype-specific modules; of these 28 are unique to XI, 11 to I and 8 to X (Fig. 2). The genotype-specific modules are generally small (< 100 genes) and include many unannotated genes (Table S4). Finally, 11 modules (17.7%) are shared between pairs of genotypes as follows: Modules #144 and #157 are shared between X and I; Modules #1, #79, #106 and #158 are shared between X and XI; Modules #8, #45, #62, #81 and #119 are shared between I and XI (Fig. 2). These findings suggest that there may be a hierarchical activation of stress response with general mechanisms of response at metabolic and cellular level activated as first defence regardless of the genetic background, followed by more specific responses that is genotype or treatment specific. We discuss below about the treatment-specific responses. This implies that whereas a proportion of the transcriptome is shared among genotypes, there is an important component of the transcriptome displaying a genotype-by environment (GxE) interaction. Previous studies on Daphnia spp identified a large number of responsive genes to environmental perturbations with a significant GxE interaction (Colbourne et al. 2011; Yampolsky et al. 2014). Remarkably, more than 60% of these ecoresponsive genes were lineage specific and over-represented in the transcriptome of the congeneric species D. pulex (Colbourne et al. 2011). With the rapid growth of sequencing technology overrepresentation of lineage specific genes has been reported in other species. For example, it is found in Caenorhabditis elegans in response to extreme environments (Zhou et al. 2015). In general, lineage specific genes have been suggested to be one of the principal means of adaptation and one of the most important sources of organizational and regulatory diversity in eukaryotes (Lespinet et al. 2002). Intriguingly, we find that responsive genes to ecologically relevant environmental perturbations are systematically less conserved than genes in general. Around half the genes with strong early transcriptional responses to one or more stressors are specific to crustaceans (Fig. 3), and are as-yet functionally uncharacterized (Table S4). Of the 1,396 differentially expressed genes identified in our study, only 612 (44%) were conserved outside of the Daphnia lineage and possibly crustaceans (Fig. 3). This is a small number compared to other metazoans where up to 67% of genes have known orthologs in other species (Brown et al. 2014). Hence, our findings represent an overall 17 standard deviations from expectation based on other metazoan (normal approximation to binomial p-value < 1e-100). We cannot definitively conclude that these genes are newly evolved or niche specific as it is likely that any error in the transcriptome assembly enriches for the appearance of unconserved genes. However, if we condition on genes that are detectably conserved between D. pulex and D. magna using information available from the assembled D. magna transcriptome (Orsini et al. 2016) to mitigate this source of statistical confounding, we see a striking pattern: the degree of induction or suppression of transcript abundance in response to environmental perturbations is inversely correlated with the likelihood of conservation beyond crustaceans (Fig. 3). That is, more strongly induced or suppressed genes are systematically less conserved. Remarkably, 72% of genes that respond 16 fold to at least one assayed perturbation have no orthologues outside crustaceans – a dramatic difference compared to the 25% of genes overall having known orthologues in other species (p-value < 1e−100; Fig. 3). In summary, our analysis of transcriptional response in multiple genotypes of the same species provides insights into stress response in different genetic backgrounds, critical to our understanding of tolerance and performance in nature (Hofmann & Todgham 2010; Latta et al. 2012; Ramu et al. 2016). Our results also confirm previous findings about the overrepresentation of lineage specific genes in crustaceans as a potential mechanisms of adaptive response to changing environments (Colbourne et al. 2011).
Figure 2. Co-expressed gene networks identified in the genotype analysis.
Significantly co-expressed gene networks (numbered on y-axis) shared among genotypes: X, I and XI. From top to bottom the networks are ordered starting from shared among all genotypes, shared between two genotypes and genotype-specific. The genotypes are color-coded as follows: X - black; I - grey; XI - red. Numbers on the y-axis refer to the module number listed in Supplementary Table S2
Figure 3. Conservation of genes responsive to environmental perturbations.
Genes responsive to environmental perturbations (N = 1,396 genes) are partitioned based on their magnitude of differential expression, ranging from basal expression (no differential expression under any perturbation) to 16-fold differential expression under at least one assayed environmental perturbation. For each category of differentially expressed genes the percentage of non-orthologous genes to other model species outside crustaceans is shown. The following model species were used for orthology searches: Danio rerio, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens. The two crustacean species Daphnia magna and Daphnia pulex were used in orthology searches to identify genes conserved within crustaceans. The percentage of genes with no orthology beyond crustaceans is indicated as percentage for each category of differentially expresses genes.
Predictable networks of stress-response genes
Our first approach to identify stress-specific early transcriptional response was an analysis of differential gene regulation. This analysis revealed that only a few individual genes were differentially expressed, many of which under abiotic perturbations (Table S1). In the following, we first discuss the results of this analysis and then present the results obtained leveraging the correlative structure of networks that allowed us to identify co-responsive gene clusters shared among and unique to environmental perturbations.
In total, 1,396 genes were differentially expressed across all treatments and genotypes (Table S1). Generally, very few of these genes were shared among environmental perturbations (Table S5). More specifically, 75 genes (5.4%) were differentially expressed in more than one abiotic treatment (Table S5), whereas only 10 genes (0.7%) were differentially expressed in at least one biotic and one abiotic treatment (Table S5). Genes that were differentially expressed in multiple treatments did not necessarily change in the same direction across abiotic treatments but consistently changed in the same direction in biotic treatments (Table S5).
Under sublethal heavy metal exposure (Pb and Cd) and UV treatments, genes involved in nucleotide excision and transcriptionally coupled DNA repair pathways were strongly down-regulated, and germ cell proliferation was pervasively shut down (Table S1). The most strongly induced genes under heavy metal exposure and low pH include glutathione S-transferases and synthases. Interestingly, the glutathione S-transferases are upregulated under heavy metals but appear to be both up and down-regulated under osmotic stress (Dapma7bEVm002921t1, Dapma7bEVm010893t1; Table S1). In addition to the well-known chitin remodelling response to heavy metals (Bekesiova et al. 2008; Lanfranco et al. 2004; Poynton et al. 2007; Wang et al. 2015), mediated by strong up-regulation of genes encoding chitinases, cuticle proteins, and cuticle binding proteins (Table S1), we see the activation and repression of numerous trypsins and trypsin inhibitors under heavy metals and UV treatments. These differentially expressed genes indicate regulation of digestive enzymes and associated processes, similarly to reported response in Drosophila melanogaster under heavy metal exposure (Brown et al. 2014).
Under metal stress we also observe the down-regulation of early developmental factors: several transcription factors and RNA binding proteins. These include the Daphnia ortholog of paired (prd) (Dapma7bEVm010730t1, Dapma7bEVm023055t1, Table S1, XI genotype), also down-regulated under UV treatment, and the nuclear receptor coactivator, homologous to the splicing factor Neosin (Neos) in Drosophila (Dapma7bEVm029191t1, Table S1), indicating that both transcriptional and post-transcriptional regulation play a role in adaptation to heavy metal exposure. Further, the gene homologous to steroid dehydrogenase (Dapma7bEVm002436t1) linked to the production of 20-Hydroxyecdysone (20E or ecdysone) in arthropods (Masuoka & Maekawa 2016) also responds, possibly preceding cuticle remodelling, and hence playing a potentially important role in moulting and growth. The activation of larval cuticle proteins in insects, including the model organism D. melanogaster, has been reported in response to environmental perturbations (Brown et al. 2014). In Daphnia spp, body remodelling is a known response to predator cues, in the presence of which they become bulkier or produce neck teeth (Laforsch & Tollrian 2004; Stoks et al. 2016). The pervasive activation of cuticle protein precursors in arthropods in response to environmental perturbations prompted us to further examine similarities between genes responsive under environmental perturbations in D. magna (present study) and those in D. melanogaster (Brown et al. 2014; Stoiber et al. 2016). The fly studies involved chronic exposures (48 hours) generally at much higher (highly toxic) doses; hence a large overlap was not expected. Consistently, we find a small, but intriguing overlap between the fly studies and the present study, including Amylase proximal (Amy-p) and PAR-domain protein 1 (Pdp1), which were repressed in both species under heavy metal treatments (Table S6). The steroid dehydrogenase, repressed in Drosophila, was either repressed under microcystin-producing cyanobacteria or induced under heavy metals and UV treatment in Daphnia (Table S6, CG32369). The overlap in gene expression between Drosophila and Daphnia is suggestive of similarity in mechanisms of response to stress. However, functional studies are needed to validate these findings and confirm similarities between the two species.
We found strong down-regulation of SOD1-vtg fusion in response to osmotic (NaCl) and oxidative stress (UV-B light), whereas we observed up-regulation of SOD-vtg under Cd, carbaryl and both microcystin treatments (Table S1). SOD-vtg is generally expressed in diapausing insects (Bi et al. 2014) and crustaceans (Acton 2012) ); hence, the regulation of this gene under environmental perturbation indicates a potential alteration of the diapause process by environmental perturbations (e.g. Slusarczyk & Rybicka 2011). In summary, early response to abiotic perturbations (UV-B, acidification and osmotic stress) was characterized by strong down-regulation of germ cell proliferation and activation of genes regulating either cuticle formation involved in moulting and growth or egg formation. These responses indicate that, despite the sub-lethal concentrations used reflecting current or realistic contamination of human-impacted inland waters, the environmental perturbations are severe enough to potentially affect growth, moulting and reproduction. The observation that chitinase and cuticle protein synthesis is upregulated under heavy metals suggest that short term exposures to metals affect animal fitness via their ability to moult. Further, the silencing of SOD1-vtg under osmotic and oxidative stress might indicate that the production of resting eggs is impaired or that maturation of the animals is severely delayed, as the production of eggs in Daphnia is tightly linked to moulting (Raborn et al. 2016). Further experimental work is needed to validate these hypotheses.
As opposed to the abiotic perturbations that provided a number of differentially expressed genes with homology in other species, the biotic responses were challenging to interpret, as the vast majority of responsive genes to biotic stress lack known conserved functional domains (Table S1, X genotype, I genotype). Among the biotic treatments, an interesting response was the one to microcystin treatment. Under this treatment, the suppression of germ cell proliferation and activation of heat shock factors and antioxidant transporters, expected in presence of cell toxicity, was not observed (Table S1). In our environmental exposures to microcystin - producing and microcystin - free cyanobacteria, we observe a typical stress response caused by poor food quality rather than by the presence of toxins; we observe the activation of digestive enzymes (Table S1). The absence of a toxic response is corroborated by the absence of signatures of germ cell suppression and stress proteins in the presence of cyanobacteria coupled with the activation of metabolic stress response genes (e.g. glucose metabolism, trehalose transporter and cellular response to starvation). Recent studies have shown genotype-based resistance of Daphnia to microcystin (Jiang et al. 2013; Lemaire et al. 2012). In line with previous studies (Lemaire et al. 2012), our results point to different response to mycrocystin dependent on genetic background (genotype) and/or prior environmental exposures, as the transcriptional response to cyanobacteria treatments was more severe in the X than in the I genotype. Indeed, the X genotype was sampled from a temporary rock pool system in which cyanobacteria blooms were never reported. Conversely, the I genotype was sampled from a typical eutrophic lake in central Europe that has likely experienced cyanobacteria blooms (O'Neil et al. 2012). The accumulation of cyanotoxins in freshwater environments due to harmful algal blooms is an increasing concern for environmental and governmental agencies because of the potent adverse effect they can have on human health and livestock (Backer et al. 2015; Hudnell 2008). The use of Daphnia to remediate watersheds and drinking water supplies affected by toxic algal blooms is an intriguing possibility for freshwater restoration (see Peretyatko et al. 2012; Sarnelle 2007 for applications) and may be envisioned considering the observed response to the toxic cyanobacteria treatment we observed.
The differential expression analysis enabled us to identify key genes regulated in specific environmental perturbations, in particular in abiotic ones. However, given the limited number of differentially regulated genes, we were unable to identify shared pathways among treatments leading to the identification of predictive pathways activated by the environmental stressors studied here. To gain a better understanding of the genetic and functional foundation of tolerance to multiple environmental stressors we leveraged the correlative nature of networks using MODA (Li et al. 2016) and identifying modules shared among environmental perturbations. The discovery of gene networks predictive of stress-response in the keystone species Daphnia could be used as a monitoring tool of freshwater ecosystems health and eventually lead to an anticipation strategy of response to stress.
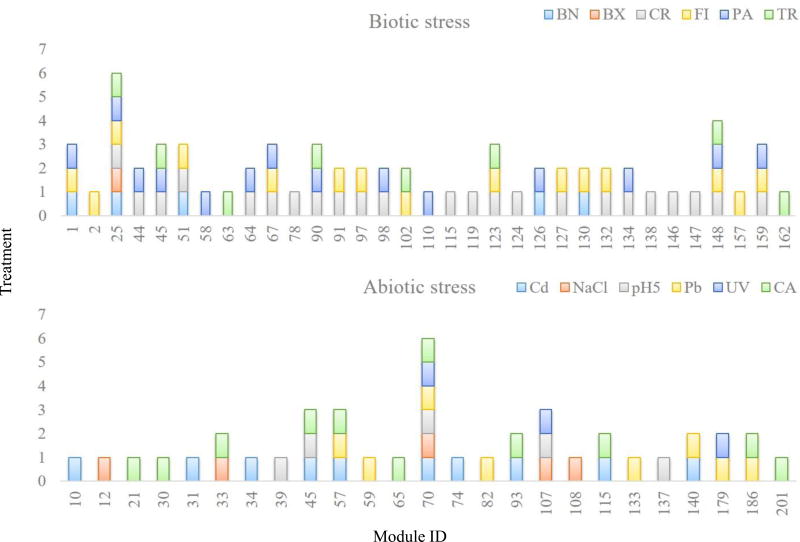
We discovered 33 co-expression modules across biotic and 25 across abiotic treatments (Fig. 4 and Table S7 for all genes co-expressed in each module). Intriguingly, only two modules are shared across all conditions – biotic and abiotic (Table S7, bio_abio). One contains essentially all housekeeping genes (ca. 9,000 genes, Module #54). The other (Module #154) includes 531 genes (Table S7, bio_abio), 280 of which have no conserved domain, no known orthologues outside crustaceans, and no known functions. This module is strongly enriched for a variety of P450 cytochromes (40 genes, GO terms: 0004497, 0020037, 0009055, 0005506, max Fisher’s exact test FDR-corrected p-value = 8e-24, Table 8), cell membrane and structural components (84 genes, GO terms: 0042598, 0005792, 0005624, 0005626, 0000267, 0019898, 0016020, 0005783, max FDR-corrected p-value = 1e-9, Table S8), and genes involved in cuticle formation, which is critical for growth and moulting (17 genes, GO terms: 0005214, 0042302, max FDR-corrected p-value = 5e-9, Table S8). Collectively (141 genes), these enrichments indicate that detoxification and cell proliferation are common features of Daphnia’s early response to stress. Whether cytochrome expression or proliferating cells are localized to a single physiological compartment, e.g. the haemolymph (inferred from the respiration signal) or epithelia (from the cuticle signal), or more broadly distributed across multiple organs will require additional transcriptomics analysis in dissected tissues or isolated cells. The association of this large collection of genes of unknown function with genes involved in respiration and cell proliferation may help to focus targeted studies to uncover their functions. For now, all we know about these 280 genes is that they are co-responsive with the genes of known function (see Table S7 for all genes in each module).
Figure 4. Co-expression networks under environmental perturbations.
Significantly co-expressed gene networks (numbered on x-axis) shared among the biotic (N = 6 treatments) and abiotic treatments (N = 6 treatments). Abbreviations for environmental perturbations are as in Figure 1. Colours and cluster numbers in the bar plots are randomly generated, hence identical colours and numbers in the two plots do not indicate the same network. Numbers on the x-axis refer to the module number listed in Supplementary Table S7.
Co-expression Module #70 is shared among abiotic conditions (Fig. 4). This module includes ca. 6,800 genes (Table S7) and is enriched for membrane and transporter activity, cell signalling, morphogenesis and redox activity genes; it also contains developmental and growth genes (Table S9). Module #107 shared among UV, pH and osmotic stress contains 45 genes (Table S7) enriched for ion transport activity and metabolic processes as it can be predicted under these stress conditions, suggesting that our unbiased approach reveals biological relevant mechanisms of response to environmental stress.
Module #25 is shared across all biotic conditions. It contains 37 genes (Table S7), 18 of which have no known function, and 10 encode vitellogenin lipoproteins (FDR-corrected p-value < 1e-9). Notably, these vitellogenins are fused to a superoxide dismutase domain (SOD1-vtg). The remaining seven genes of known function are likely lipid or lipoprotein transporters, including Dapma7bEVm010067t1 and Dapma7bEVm019273t1, two CRAL/TRIO-domain containing proteins and yeast Sec14p orthologs; two probable haemolymph coagulation co-factors, one encoding a von Willebrand factor (vWF, Dapma7bEVm022355t1) and a BTB/POZ domain containing gene (Dapma7bEVm022613t1); and one uncharacterized, but conserved peptide (Dapma7bEVm020225t1) which encodes the conserved domain of unknown function pfam09172, DUF1943. While changes in vitellogenins are not statistically significant in each treatment, the pervasive presence of these genes across treatments indicates that their modulation is linked to numerous environmental perturbations.
Four modules were shared between the vertebrate and invertebrate predator treatments (Modules #25, #102, #123, and #148) and enriched for canonical signatures of body remodelling (chitinase activation) and activation of cuticle proteins, including larval cuticle proteins (Table S10). Discussing each of the modules in turn is beyond the scope of this manuscript, and hence all modules (Table S7), as well as their GO term analyses (Tables S8, S9, and S10), are provided in Supplementary material as a community resource.
Conclusions
We discovered that approximately one third of the Daphnia genes, enriched for metabolism, cell signalling and general stress response, is responsible for early transcriptional response to environmental stress and it is shared among genotypes. We also discovered that a large proportion of the transcriptome responds to environmental perturbations with a strong GxE component. This suggests a sequential activation of transcriptional response to environmental stress, which first involves a general stress response, and then activates a genotype and/or condition-specific transcriptional response. The latter is driven by smaller modules of genes enriched for more specific functions - e.g. body remodelling to predator cues or ion transporter for oxidative and osmotic stress. Intriguingly, genotype and condition- specific transcriptional response is found in genes not conserved beyond crustaceans. These genes may be associated with niche-specific adaptation. Finally, our study associated hundreds of uncharacterized genes within co-responsive modules to genes of known function and to specific environmental perturbations. This link represents a fundamental basis for our understanding of the genetic and functional foundation of tolerance to multiple environmental stressors. Establishing these associations is generally challenging, even in the best studied model species [e.g. yeast, (Pena-Castillo & Hughes 2007) and fly, (Brown & Celniker 2015)].
Supplementary Material
Acknowledgments
This research was supported by the ESF EUROCORES Programme EuroEEFG, Grant 09-EEFGFP-040. The KU Leuven team was supported by FWO project STRESSFLEA-B (G061411N) and by the KU Leuven Research Fund (coordination grant and grant PF/2010/07). The University of Notre Dame team was supported by NIH grant R24-GM078274 to MEP. The University of Helsinki Team was supported by Academy of Finland grants 250444, 284601 and 135291. Donald Gilbert has been supported by the National Science Foundation (grant No. 0640462 to DGG), including genomics computational resources via TeraGrid amd XSEDE. The KU Leuven team was supported by a PhD fellowship of the Agency for Innovation by Science and Technology in Flanders (IWT) to Katina I. Spanier (SB/101575) and a postdoctoral grant to Mieke Jansen (12B6513N).
Footnotes
Author contribution
LO and LDM conceived the study and the experimental design. LO coordinated the research activities, data analysis, and manuscript writing with input from all co-authors.
MJ coordinated and performed the exposures of the two natural genotypes.
MEP coordinated and performed the exposures of the inbred genotype.
JBB and OSS coordinated and performed the DE analyses.
SH, JBB and DL coordinated and performed the network analysis.
RP contributed to DE analysis, and quality checks.
KIS, and RP performed manual functional gene annotations.
MS performed functional analysis across species.
All authors contributed to manuscript editing and discussion.
Data accessibility:
The transcriptome assembly is available from NCBI Sequence Read Archive SRP059260 (2015): http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA284518
A searchable data base of functional annotations for D. magna genes against other model species including Daphnia pulex (dpul), Danio rerio (drer), Caenorhabditis elegans (cele), Drosophila melanogaster (dmel), Mus musculus (mmus), and Homo sapiens (hsap) can be found here: http://merlot.lbl.gov/omid/Daphnia/deliverables-v2.2/. D. magna genes are also deposited in OrthoDB (Zdobnov et al. 2017). The scripts used to combine methods of ortholog detection are provided in supplementary material (Appendix 1).
NCBI accession numbers to RNASeq data across the 12 environmental conditions and genotypes are provided in Appendix 2.
References
- Acton AQ. Vitellins—Advances in Research and Application Scolarly Editions. Atlanta, Georgia: 2012. [Google Scholar]
- Altenhoff AM, Skunca N, Glover N, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Research. 2015;43:D240–249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer LC, Manassaram-Baptiste D, LePrell R, Bolton B. Cyanobacteria and Algae Blooms: Review of Health and Environmental Data from the Harmful Algal Bloom-Related Illness Surveillance System (HABISS) 2007–2011. Toxins. 2015;7:1048–1064. doi: 10.3390/toxins7041048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekesiova B, Hraska S, Libantova J, Moravcikova J, Matusikova I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Molecular Biology Reports. 2008;35:579–588. doi: 10.1007/s11033-007-9127-x. [DOI] [PubMed] [Google Scholar]
- Bi ZL, Yang XL, Yu W, Shu JH, Zhang YZ. Diapause-Associated Protein3 Functions as Cu/Zn Superoxide Dismutase in the Chinese Oak Silkworm (Antheraea pernyi) PLoS One. 2014;9 doi: 10.1371/journal.pone.0090435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd PW, Lennartz ST, Glover DM, Doney SC. Biological ramifications of climate-change-mediated oceanic multi-stressors. Nature Climate Change. 2015;5:71–79. [Google Scholar]
- Brooks AN, Turkarslan S, Beer KD, Lo FY, Baliga NS. Adaptation of cells to new environments. Wiley Interdisciplinary Review System Biology and Medicine. 2011;3:544–561. doi: 10.1002/wsbm.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Celniker SE. Lessons from modENCODE. Annual Review Genomics Human Genetics. 2015;16:31–53. doi: 10.1146/annurev-genom-090413-025448. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wu TH, Ng WV, Lin WC. DODO: an efficient orthologous genes assignment tool based on domain architectures. Domain based ortholog detection. BMC Bioinformatics. 2010;11(Suppl 7):S6. doi: 10.1186/1471-2105-11-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Singan VRa, Gilbert D. wFleaBase: the Daphnia genome database. BMC Bioinformatics. 2005;6:45. doi: 10.1186/1471-2105-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers J, et al. Host-parasite Red Queen dynamics archived in pond sediment. Nature. 2007;450:870–874. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- Eng KH, Kvitek DJ, Keles S, Gasch AP. Transient genotype-by-environment interactions following environmental shock provide a source of expression variation for essential genes. Genetics. 2010;184:587–593. doi: 10.1534/genetics.109.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SF, Chen PY, Nguyen QTT, et al. Transcriptome profiling of genes and pathways associated with arsenic toxicity and tolerance in Arabidopsis. Bmc Plant Biology. 2014;14 doi: 10.1186/1471-2229-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts AN, Vanoverbeke J, Vanschoenwinkel B, et al. Rapid evolution of thermal tolerance in the water flea Daphnia. Nature Climate Change. 2015;5:665–668. [Google Scholar]
- Gunderson AR, Armstrong EJ, Stillman JH. Multiple Stressors in a Changing World: The Need for an Improved Perspective on Physiological Responses to the Dynamic Marine Environment. Annual Review of Marine Science, Vol 8. 2016;8 doi: 10.1146/annurev-marine-122414-033953. 357-+ [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hofmann GE, Todgham AE. Living in the Now: Physiological Mechanisms to Tolerate a Rapidly Changing Environment. Annual Review of Physiology. 2010;72:127–145. doi: 10.1146/annurev-physiol-021909-135900. [DOI] [PubMed] [Google Scholar]
- Holmstrup M, Bindesbol AM, Oostingh GJ, et al. Interactions between effects of environmental chemicals and natural stressors: A review. Science of the Total Environment. 2010;408:3746–3762. doi: 10.1016/j.scitotenv.2009.10.067. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudnell HK. Cyanobacterial Harmful Algal Blooms: State of the Science and Research Needs 2008 [Google Scholar]
- Huisman GW, Kolter R. Sensing starvation: a homoserine lactone--dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- Jaccard P. Lois de distribution florale. Bulletin de la Socíeté Vaudoise des Sciences Naturelles. 1902;38:67–130. [Google Scholar]
- Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. Int J Radiation Biology. 2015;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- Jansen M, De Meester L, Cielen A, Buser CC, Stoks R. The interplay of past and current stress exposure on the water flea Daphnia. Functional Ecology. 2011;25:974–982. [Google Scholar]
- Jiang XD, Zhang Lihua, Liang H, et al. Resistance variation within a Daphnia pulex population against toxic cyanobacteria. Journal of Plankton Research. 2013;0:1–5. [Google Scholar]
- Keunen E, Schellingen K, Van der Straeten D, et al. ALTERNATIVE OXIDASE1a modulates the oxidative challenge during moderate Cd exposure in Arabidopsis thaliana leaves. Journal of Experimental Botany. 2015;66:2967–2977. doi: 10.1093/jxb/erv035. [DOI] [PubMed] [Google Scholar]
- Khalil AM. Toxicological effects and oxidative stress responses in freshwater snail, Lanistes carinatus, following exposure to chlorpyrifos. Ecotoxicology and Environmental Safety. 2015;116:137–142. doi: 10.1016/j.ecoenv.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Kim DY, Hong MJ, Park CS, Seo YW. The effects of chronic radiation of gamma ray on protein expression and oxidative stress in Brachypodium distachyon. International Journal of Radiation Biology. 2015;91:407–419. doi: 10.3109/09553002.2015.1012307. [DOI] [PubMed] [Google Scholar]
- Klüttgen B, U D, Engels M, T RH. Combined effects of 3,4-dichloroaniune and food concentration on life-table data of two related cladocerans, Daphnia magna and Ceriodaphnia Quadrangula. Chemosphere. 1994;32:2015–2028. [Google Scholar]
- Labbe P, McTaggart SJ, Little TJ. An ancient immunity gene duplication in Daphnia magna: RNA expression and sequence analysis of two nitric oxide synthase genes. Developmental and Comparative Immunology. 2009;33:1000–1010. doi: 10.1016/j.dci.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforsch C, Tollrian R. Extreme helmet formation in Daphnia cucullata induced by small-scale turbulence. Journal of Plankton Research. 2004;26:81–87. [Google Scholar]
- Lanfranco L, Balsamo R, Martino E, Bonfante P, Perotto S. Zinc ions differentially affect chitin synthase gene expression in an ericoid mycorrhizal fungus. Plant Biosystems. 2004;138:271–277. [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larguinho M, Cordeiro A, Diniz MS, Costa PM, Baptista PV. Metabolic and histopathological alterations in the marine bivalve Mytilus galloprovincialis induced by chronic exposure to acrylamide. Environ Res. 2014;135:55–62. doi: 10.1016/j.envres.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Latta LC, Weider LJ, Colbourne JK, Pfrender ME. The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecol Lett. 2012;15:794–802. doi: 10.1111/j.1461-0248.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, et al. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc J, Echaubard P, Trudeau V, Lesbarreres D. Copper and nickel effects on survival and growth of Northern Leopard Frog (Lithobates pipiens) tadpoles in field-collected smelting effluent water. Environmental Toxicology and Chemistry. 2015 doi: 10.1002/etc.3227. [DOI] [PubMed] [Google Scholar]
- Lemaire V, Brusciotti S, van Gremberghe I, et al. Genotype x genotype interactions between the toxic cyanobacterium Microcystis and its grazer, the waterflea Daphnia. Evolutionary Applications. 2012;5:168–182. doi: 10.1111/j.1752-4571.2011.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Research. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Brown JB, Orsini L, Hu G, Pan Z, He S. MODA: MOdule Differential Analysis for weighted gene co-expression network. bioRxiv 053496 2016 [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber w, Anders s. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardirosian MN, Lascano CI, Ferrari A, Bongiovanni GA, Venturino A. Acute toxicity of arsenic and oxidative stress responses in the embryonic development of the common South American toad Rhinella arenarum. Environmental Toxicology and Chemistry. 2015;34:1009–1014. doi: 10.1002/etc.2856. [DOI] [PubMed] [Google Scholar]
- Masuoka Y, Maekawa K. Ecdysone signaling regulates soldier-specific cuticular pigmentation in the termite Zootermopsis nevadensis. FEBS Letters. 2016;590:1694–1703. doi: 10.1002/1873-3468.12219. [DOI] [PubMed] [Google Scholar]
- Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG. Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proceedings of the Royal Society B-Biological Sciences. 2012;279:1873–1882. doi: 10.1098/rspb.2011.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil JM, Davis TW, Burford MA, Gobler CJ. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae. 2012;14:313–334. [Google Scholar]
- Orsini L, Gilbert D, Podicheti R, et al. Daphnia magna transcriptome by RNA-Seq across 12 environmental stressors. Scientific Data. 2016;3:160030. doi: 10.1038/sdata.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini L, Spanier KI, De Meester L. Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Molecular Ecology. 2012;21:2160–2175. doi: 10.1111/j.1365-294X.2011.05429.x. [DOI] [PubMed] [Google Scholar]
- Overjordet IB, Altin D, Berg T, et al. Acute and sub-lethal response to mercury in Arctic and boreal calanoid copepods. Aquatic Toxicology. 2014;155:160–165. doi: 10.1016/j.aquatox.2014.06.019. [DOI] [PubMed] [Google Scholar]
- Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretyatko A, Teissier S, De Backer S, Triest L. Biomanipulation of hypereutrophic ponds: when it works and why it fails. Environmental Monitoring and Assessment. 2012;184:1517–1531. doi: 10.1007/s10661-011-2057-z. [DOI] [PubMed] [Google Scholar]
- Perez IB, Brown PJ. The role of ROS signaling in cross-tolerance: from model to crop. Frontiers in Plant Science. 2014;5:754. doi: 10.3389/fpls.2014.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton HC, Varshavsky JR, Chang B, et al. Daphnia magna ecotoxicogenomics provides mechanistic insights into metal toxicity. Environmental Science & Technology. 2007;41:1044–1050. doi: 10.1021/es0615573. [DOI] [PubMed] [Google Scholar]
- Raborn RT, Spitze K, Brendel VP, Lynch M. Promoter Architecture and Sex-Specific Gene Expression in Daphnia pulex. Genetics. 2016;204:593–612. doi: 10.1534/genetics.116.193334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram RJ, Verberkmoes NC, Thelen MP, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- Ramu VS, Paramanantham A, Ramegowda V, et al. Transcriptome Analysis of Sunflower Genotypes with Contrasting Oxidative Stress Tolerance Reveals Individual- and Combined-Biotic and Abiotic Stress Tolerance Mechanisms. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejeb IB, Pastor V, Mauch-Mani B. Plant Responses to Simultaneous Biotic and Abiotic Stress: Molecular Mechanisms. Plants (Basel) 2014;3:458–475. doi: 10.3390/plants3040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routtu J, Hall MD, Albere B, et al. An SNP-based second-generation genetic map of Daphnia magna and its application to QTL analysis of phenotypic traits. BMC Genomics. 2014;15(1033) doi: 10.1186/1471-2164-15-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, et al. TreeFam: 2008 Update. Nucleic Acids Research. 2008;36:D735–740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnelle O. Initial conditions mediate the interaction between Daphnia and bloom-forming cyanobacteria. Limnology and Oceanography. 2007;52:2120–2127. [Google Scholar]
- Slusarczyk M, Rybicka B. Role of temperature in diapause response to fish kairomones in crustacean Daphnia. Journal of Insect Physiology. 2011;57:676–680. doi: 10.1016/j.jinsphys.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Somero GN. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine 'winners' and 'losers'. Journal of Experimental Biology. 2010;213:912–920. doi: 10.1242/jeb.037473. [DOI] [PubMed] [Google Scholar]
- Somero GN. The Physiology of Global Change: Linking Patterns to Mechanisms. Annual Review of Marine Science, Vol 4. 2012;4:39–61. doi: 10.1146/annurev-marine-120710-100935. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Research. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber M, Celniker S, Cherbas L, Brown B, Cherbas P. Diverse Hormone Response Networks in 41 Independent Drosophila Cell Lines. G3 (Bethesda) 2016 doi: 10.1534/g3.115.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoks R, Govaert L, Pauwels K, Jansen B, De Meester L. Resurrecting complexity: the interplay of plasticity and rapid evolution in the multiple trait response to strong changes in predation pressure in the water flea Daphnia magna. Ecology Letters. 2016;19:180–190. doi: 10.1111/ele.12551. [DOI] [PubMed] [Google Scholar]
- Tang S, Wu Y, Ryan CN, et al. Distinct expression profiles of stress defense and DNA repair genes in Daphnia pulex exposed to cadmium, zinc, and quantum dots. Chemosphere. 2015;120:92–99. doi: 10.1016/j.chemosphere.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Techer D, Milla S, Fontaine P, Viot S, Thomas M. Acute toxicity and sublethal effects of gallic and pelargonic acids on the zebrafish Danio rerio. Environmental Science Pollution Research International. 2015;22:5020–5029. doi: 10.1007/s11356-015-4098-2. [DOI] [PubMed] [Google Scholar]
- Vincenzi S. Extinction risk and eco-evolutionary dynamics in a variable environment with increasing frequency of extreme events. J R Soc Interface. 2014;11:20140441. doi: 10.1098/rsif.2014.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LY, Wang YS, Cheng H, Zhang JP, Yeok FS. Cloning of the Aegiceras corniculatum class I chitinase gene (AcCHI I) and the response of AcCHI I mRNA expression to cadmium stress. Ecotoxicology. 2015;24:1705–1713. doi: 10.1007/s10646-015-1502-0. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JC. Ecological processes and the ecology of stress: the impacts of abiotic environmental factors. Functional Ecology. 2013;27:37–44. [Google Scholar]
- Yampolsky LY, Zeng E, Lopez J, et al. Functional genomics of acclimation and adaptation in response to thermal stress in Daphnia. BMC Genomics. 2014;15:859–870. doi: 10.1186/1471-2164-15-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Tegenfeldt F, Kuznetsov D, et al. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Research. 2017;45:D744–D749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Huang BB, Zou M, et al. Genome-wide identification of lineage-specific genes within Caenorhabditis elegans. Genomics. 2015;106:242–248. doi: 10.1016/j.ygeno.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Zou HX, Pang QY, Zhang AQ, et al. Excess copper induced proteomic changes in the marine brown algae Sargassum fusiforme. Ecotoxicology Environmental Safety. 2015;111:271–280. doi: 10.1016/j.ecoenv.2014.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.