Abstract
Distinct dynamin superfamily GTPases catalyze the constant fission and fusion of the elaborate mitochondrial networks that navigate the eukaryotic cytoplasm. Long believed to be the singular handiwork of dynamin-related protein 1 (Drp1), a cytosolic family member that transiently localizes to the mitochondrial surface, the execution of mitochondrial fission is now arguably believed to entail membrane remodeling events that are initiated upstream of Drp1 by ER-associated cytoskeletal networks and completed downstream by the prototypical dynamin, dynamin 2 (Dyn2). Recent developments in the field have also placed a sharp focus on the membrane microenvironment around the division apparatus and the potential facilitatory role of specific lipids in mitochondrial fission. Here, I will review current progress, as well as highlight the most visible gaps in knowledge, in elucidating the varied functions of the dynamin superfamily in the coordinated events of mitochondrial fission and fusion. The essential roles of protein and lipid cofactors are also highlighted.
Keywords: Mitochondrial dynamics, mitochondrial fission, mitochondrial fusion, dynamin-related proteins, cardiolipin, actin dynamics
Introduction
Originated as an endosymbiont from ancient bacteria approximately 2 billion years ago, the modern day mitochondrion is seldom present as a discrete entity in the eukaryotic cell (1,2). Instead, the many mitochondria that inhabit the cytoplasmic space form extensive, branched, and dynamic networks that are remodeled continuously by the coupled and compensatory actions of fission and fusion (3). The fusion, fission and transport of mitochondria, all of which occur over cytoskeletal tracks that traverse the extranuclear volume, are collectively referred to as ‘mitochondrial dynamics’. Many excellent review articles have articulated on the phenomenology of mitochondrial dynamics, and their implications in physiology and disease (4–8). In this review article, I will focus on the structural and functional mechanics of mitochondrial fission and fusion, and the putative roles and molecular mechanisms of various dynamin superfamily GTPases as ‘catalysts’ and/or ‘effectors’ of mitochondrial remodeling.
1. Mitochondrial remodeling: the setting. What, why, where, and how?
What?
Mitochondria, whether present as short, discrete units or organized in long, dynamic networks, can be approximated as ‘spherocylinders’ with two hemispherical ends capping a cylindrical (tubular) center that may vary greatly in length as well as in girth (diameter). Indeed, the mitochondria of most mammalian cells typically vary between approximately 200 nm to 1 micron in external diameter (9,10). Mitochondrial fission or fragmentation therefore entails a massive, local constriction of the bounding mitochondrial outer membrane (MOM) from a significantly large external diameter down to less than 4 nm in internal diameter to reach the theoretical limit for spontaneous membrane fission (11). Correspondingly, mitochondrial fusion initially involves the creation of a localized and narrow fusion pore between opposing MOM bilayers that then expands substantially to reach the large overall diameter of the tubular mitochondrion (12). Adding further complexity to this membrane-remodeling challenge is also the need for a concerted constriction, or homotypic fusion, of the MOM-encapsulated mitochondrial inner membrane (MIM) that protrudes repeatedly into the mitochondrial matrix as tubular folds of ‘cristae’.
Why?
Mitochondria, unlike intracellular transport vesicles, are not created de novo, i.e. from scratch from other donor membrane compartments. Instead, the existing mitochondria expand in size (length and girth), coordinately replicate their genome (mtDNA), and undergo fragmentation in order to be effectively partitioned into daughter cells during cell division (4). Apart from this obvious need, mitochondrial fission also serves many essential functions in a postmitotic cell. These include (i) enabling the transport of appropriately sized mitochondria along cytoskeletal tracks to various parts of the cell, e.g. down narrow axons to the ATP-demanding nerve termini (13), (ii) the excision of irreparably damaged, dysfunctional mitochondrial fragments toward ‘mitophagy’ (autophagy of mitochondria) (14), and (iiii) facilitating the release of intermembrane space (IMS)-localized pro-apoptotic cytochrome c into the cytosol during programmed cell death (15). Mitochondrial fusion, on the other hand, ensures organellar content homogeneity via the transcomplementation of mtDNA, proteins and lipids, and occurs presumably to combat the propagation of excessive reactive oxygen species (ROS)-induced mtDNA damage and dysfunction (4,16–18). The rates of mitochondrial fission and fusion in any given cell type are counterbalanced, tightly controlled, and are periodically altered in response to various physiological cues and a changing intracellular environment. All of the above physiological aspects are reviewed extensively in refs. (4–8,18–21).
Where?
Even though mitochondria are distributed indiscriminately throughout the cytosol, mitochondrial fission and fusion both occur almost exclusively at regions of ER-mitochondria-cytoskeleton contact (22–24). Extensively branched and narrow ER membrane tubules that emanate from the sheet-like peripheral ER of the perinuclear region intersect and radially enwrap the elongated mitochondria to mark sites of imminent mitochondrial division (25). Various bridging protein molecules, including those that ultimately catalyze mitochondrial fusion, facilitate the tethering of the tubular ER to the MOM (25–27). Importantly, these confined ER-mitochondria interfacial regions constitute focal points for the collaborative interaction of membrane-anchored actin-nucleating and actin-polymerizing factors that create localized, confined zones of directed actin polymerization (23,24,28). Either actin polymerization directed against the MOM surface from the circumnavigating ER membrane tubule and/or the contraction of cross-bridged actin-myosin II motor filaments networked radially around the mitochondrial tubule is believed to induce the greater membrane curvature (smaller diameter) characteristic of these fission-predisposed MOM sites. The other component of the cellular cytoskeleton, the microtubules, to which the mitochondria are anchored to, facilitate the transportation and distribution of mitochondria in mammalian cells (13,29–31). Incidentally, ER-mitochondrial contacts also occur on microtubules. Microtubule anchoring may provide the longitudinal membrane tension necessary for mitochondrial fission and for the effective separation of the newly formed mitochondrial poles (32,33).
How?
Although the ER-mitochondria-actin nexus induces the initial membrane curvature on mitochondria to mark sites of future fission, GTPases of the dynamin superfamily termed ‘dynamin-related proteins (DRPs)’ catalyze the ultimate constriction of mitochondria toward fission, or conversely, the close apposition of opposing mitochondrial membranes toward fusion (4,34). Key to the act of mitochondrial fission are dynamin-related protein 1 (Drp1), and arguably, dynamin 2 (Dyn2), both of which are recruited from their predominantly cytosolic locations transiently to mitochondrial fission sites (Fig. 1). A current model depicts mitochondrial fission in three hierarchical steps (10,35). The ER-actin network initially constricts mitochondria (step 1). Subsequent Drp1 helical self-assembly at ER-preconstricted fission sites and GTP hydrolysis-driven conformational rearrangements constrict the mitochondria further, although not to the point of fission (step 2). Finally, a more effective mechanoenzymatic constriction activity from Dyn2 self-assembly at Drp1-superconstricted regions ultimately catalyzes mitochondrial fission (step 3). Where, whether and how one step ends and the other one begins is unclear. How these events are coordinated in time and space also remains a mystery. Based on emergent data, we will consider an equally plausible non-hierarchical model for mitochondrial fission in the following sections. Likewise, key to the act of mitochondrial fusion are two dynamin family GTPases, mitofusins (Mfn1/2) and optic atrophy 1 (OPA1), which catalyze the independent fusion of the MOM and MIM, respectively. Unlike Drp1 and dynamin 2, Mfn1/2 and OPA1 are each anchored to their respective membrane via transmembrane (TM) segments (Fig. 2). Modeled on SNARES, the mechanisms of mitochondrial fusion has been divided similarly into tethering, docking and fusion steps (12). Using the recently available crystal structures of a minimal Mfn construct and their structural and functional resemblance to bacterial dynamin-like protein (BDLP) and mammalian atlastin (36–39) as a basis, we will speculate as to how GTP hydrolysis in Mfn1/2 and OPA1 may be mechanoenzymatically coupled to the distinct fusion of the mitochondrial double membrane.
Figure 1.
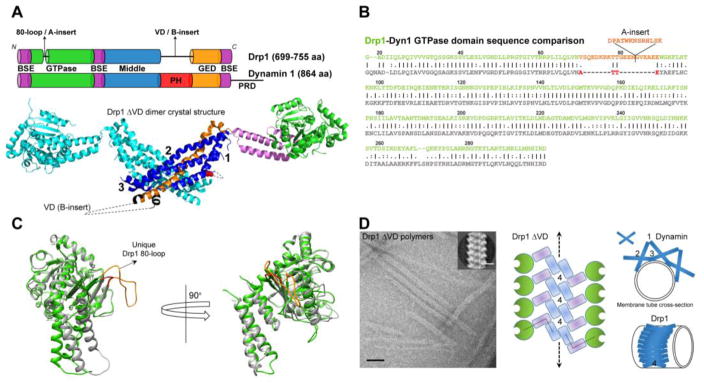
Fission DRPs. (A) (Top) Domain arrangements of Drp1 and dynamin. BSE, bundle signaling element; PH, pleckstrin homology domain; GED, GTPase effector domain; PRD, proline-rich domain. (Bottom) Colour-coded representation of the Drp1ΔVD dimer crystal structure (PDB 1D: 4BEJ). The location of interface 2 that mediates Drp1/dynamin crisscross dimerization and of interfaces 1 and 3 that mediate dimer-dimer interactions during self-assembly are shown. The location of the membrane-interacting VD is represented by a dotted loop at the base of the molecule. (B) Sequence alignment of the GTPase domains of Drp1 (green) and dynamin 1 (Dyn1; gray). Residue numbering corresponds to Drp1. The location and sequence of the 80-loop A-insert present in select Drp1 splice variants is indicated. Region in Dyn1 corresponding to the Drp1 80-loop is highlighted in red. (C) An overlay of the Drp1 (green; PDB ID:4H1U) and Dyn1 (gray; PDB ID: 2X2E) GTPase-GED fusion crystal structures clearly distinguishes the Drp1-specific 80-loop as a conspicuous structural feature that regulates Drp1 activity. (D) (left) Negative-stain EM images of Drp1ΔVD polymers. Scale bar, 50 nm. Inset shows 2D class average projection. Inset scale bar, 10 nm. (middle) A top-down view of interface 4-mediated linear filament formation in Drp1ΔVD. The putative locations of the domains are colour-coded as in panel A. (right) Model of the distinct modes of stalk-mediated dynamin and Drp1 polymerization on membrane tubes. Assembly interfaces are numbered adjacent to their locations.
Figure 2.
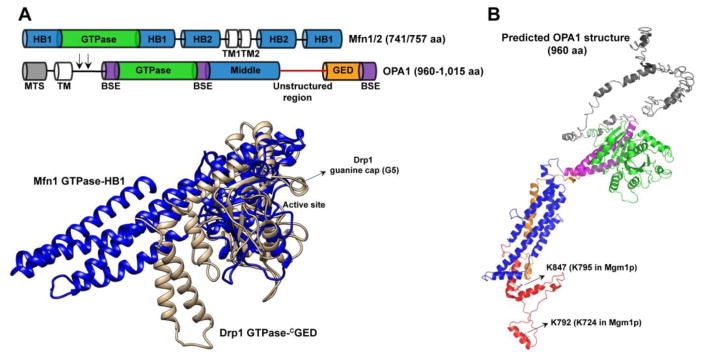
Fusion DRPs. (A) (Top) Domain arrangement of Mfn1/2 versus OPA1. HB1/2, helical bundle 1/2; TM, transmembrane region; MTS, mitochondrial targeting sequence. Arrows point to the locations of proteolytic cut sites in OPA1 that convert TM-anchored 1-OPA1 into soluble s-OPA1. (Bottom) Structural overlay of the Drp1 GTPase-GED fusion construct (brown) with the Mfn1 GTPase-HB1 fusion construct (blue; PDB ID: 5GOM). The guanine cap (G5) that facilitates GTP binding in Drp1 is conspicuously absent in Mfn1. (B) An I-TASSER (151) predicted structure of OPA1 based on homology to Drp1. The domains are colour-coded in correspondence with panel A. Residues 1-245 that include the MTS, TM, proteolytic cut sites and coiled-coil (CC) regions are shaded gray. Residues 246-277, 560-584 and 931-960 form the BSE. Residues 278-559 constitute the GTPase domain core, whereas residues 585-754 and 861-930 constitute the middle (MD) and GED regions. The largely unstructured region at the base of the molecule correspond to residues 755-860. The positions of conserved Lys (K) in OPA1 that in Mgm1p participate in phospholipid binding are shown.
2. The main cast: the DRPs of fission and fusion
In light of the very recent and exhaustive review articles written on the varied roles of dynamin superfamily members in mitochondrial dynamics (18,34), I will restrict my discussion here to the salient structural and functional features of the dynamin superfamily that delineate their specific roles in mitochondrial fission or fusion.
2. 1. Collaring Drp1 and Dyn2 in mitochondrial fission
Although classical dynamins (Dyn1, 2 and 3) are the prototypical (founding) dynamin-related proteins (DRPs) based on which others are described, Drp1 is the archetypal DRP (the perfect example) (40). Drp1 is more ancient in evolutionary origin and considered ancestral to the classical dynamins (41).
The notion that Drp1 functions as a ‘mechanochemical noose’ to constrict and divide mitochondria is however modeled largely on dynamin. Disruption of Drp1 activity either in yeast or in mammalian cells results in defective mitochondrial fission and a hyperfused mitochondrial network, an effect comparable in principle to the accumulation of trapped endocytic pits in nerve cells expressing disrupted dynamin activity (42–47). Electron-dense bands of Dnm1p (the yeast Drp1 homolog) observed at mitochondrial division sites (44) were likened to the electron-dense collars of mutant dynamin wrapped around the base of trapped endocytic pits at the presynaptic termini of temperature-sensitive shibire flies (46). Similar to dynamin, purified mammalian Drp1 and yeast Dnm1p were also shown to assemble into helical polymers in vitro (albeit with distinct geometries), and constrict tubular membrane templates over which they are assembled (albeit to different extents) upon GTP hydrolysis (48–52).
DRPs are large, modular GTPases that self-assemble into organized arrays, helical or otherwise, for function (Fig. 1A). Fission DRPs (e.g. classical dynamins and Drp1), which constitute polymers of a helical geometry over membranes are structurally and functionally distinct from fusion DRPs (e.g. Mfn1/2 and OPA1) that largely form organized, flat lattices as discussed below (53–56).
Fission DRPs dynamin and Drp1 contain a highly conserved N-terminal GTPase domain that characteristically homodimerizes in a nucleotide-dependent manner to cooperatively hydrolyze GTP (57,58) (Fig. 1A, top). GTPase domain dimerization occurs between adjacent rungs of the helical dynamin/Drp1 polymer and couples cooperative GTP hydrolysis to mechanoenzymatic membrane tube constriction (59). A four-helical bundle or ‘stalk’ composed of discontinuous middle (MD) and GTPase effector (GED) regions participates in lateral DRP-DRP self-interactions and establishes the helical geometry of dynamin/Drp1 self-assembly (60–63). The stalk connects the GTPase domain that is located farthest from the membrane surface to membrane-proximal lipid-binding regions that interact peripherally with the membrane bilayer (Fig. 1A, bottom). The stalk communicates with the GTPase domain via another discontinuous but short three-helical bundle called the ‘bundle signaling element or BSE’ (Fig. 1A, top). The BSE undergoes a dramatic conformational rearrangement relative to the GTPase domain in a so-called ‘powerstroke’ (59), which affects the geometry of stalk-mediated self-assembly and presumably allows for underlying membrane tube constriction toward fission (64–69). The GTPase domain, BSE and stalk thus constitute a conserved ‘mechanochemical core’ in both dynamin and Drp1. Whether and how such conformational rearrangements in fission DRPs drive membrane remodeling and fission are heavily debated and remain areas of intense investigation (70).
Regardless, key structural and functional differences distinguish Drp1 from classical dynamins. Despite a highly conserved active site (~75% similarity with dynamin) (Fig. 1B), Drp1 exhibits a ~10-fold-lower rate of cooperative GTP hydrolysis compared to Dyn1 (~200 min−1 for dynamin (71) versus ~20 min−1 for Drp1 (72)) upon helical self-assembly on target membranes. These kinetics likely reflect a ~10-fold greater KM (Michaelis constant) of Drp1 GTP hydrolysis (~200 μM) relative to Dyn1 (~20 μM) (73–75). A structural comparison of the Drp1 and Dyn1 GTPase domains provides a plausible explanation for this disparity. An overlay of the primary and 3D structures of the respective GTPase domains reveals the presence of a unique stretch of residues in Drp1 called the ‘80-loop’ (Fig. 1B, 1C). Absent in classical dynamins, 80-loop counterparts only exist in the DRPs of lower eukaryotes. These include Dnm1p and Vps1p from Saccharomyces cerevisiae and Dyn A from Dictyostelium discoideum (76). Connecting two β-strands of the central β-sheet, this loop is pronouncedly longer in Drp1 compared to dynamin and is positioned at the rim of the GTPase domain (Fig. 1C). The Drp1-specific 80-loop is also subject to alternative splicing and may include 13 additional residues called the ‘A-insert’ in select Drp1 splice variants (Fig. 1B) (73,77–79). A biochemical and biophysical comparison of the short (without A-insert) and long (with A-insert) ‘80-loop’ splice variants has further revealed a dramatically reduced cooperativity of GTP hydrolysis for the long variant compared to the short variant (by greater than 5-fold) (73). The collective data therefore suggest the Drp1-specific 80-loop functions to regulate the nucleotide-dependent, inter-rung dimerization of the GTPase domain (73). The 80-loop, identified as a potential binding site for either protein partners (76) or divalent cations (58), might function thus as a ‘kinetic timer’ to increase the residence time of the helical Drp1 polymer over mitochondrial fission sites by favoring Drp1 self-assembly over rapid, GTP hydrolysis-driven polymer disassembly (80). The above Drp1-specific structural and kinetic properties could be pertinent to the observed longer time-scale Drp1-mediated mechanoenzymatic constriction of mitochondria relative to dynamin (10). Moreover, the presence of 80-loop-to-80-loop dimerization contacts in the crystal packing of minimal Drp1 GTPase domain dimers suggests that these interactions might constitute an additional or an alternate interface for Drp1 self-assembly (58). Owing to its proximity to the BSE and a high density of negative charges at its tip, the 80-loop is also proposed to regulate BSE movement during the Drp1 ‘power stroke’ (58). The role and mechanism of the Drp1-specific 80-loop awaits further structural and functional elucidation.
Likewise, a structural and functional comparison of the Drp1 and dynamin stalks has revealed distinct alternate and preferential modes of stalk-mediated self-assembly. Crystal structures of nearly full-length dynamin and Drp1 have revealed that the stalks interact via distinct interfaces to establish the geometry of helical self-assembly (60–63). A largely hydrophobic ‘interface 2’ common to both Drp1 and dynamin positions two protein monomers in a crisscross fashion at the stalk midbody to create the dimeric building blocks of self-assembly (Fig. 1A, bottom). In dynamin, interfaces 1 and 3, located at the top and bottom of the stalk (Fig. 1A), proximal to the BSE and the membrane-binding region, respectively, participate in ‘end-to-end’ dimer-dimer interactions in a manner that registers a tilt between the dimers and favors uniform helical self-assembly. Consistently, interface 3-disrupting mutations in Dyn1 give rise to either linear dimeric arrays, as seen in the crystal lattice of interface 3-mutated Dyn1 (61), or helical polymers of a non-uniform geometry as observed on membrane templates (71,72). By contrast, an interface 3-perturbing mutation in Drp1 (C505A) remarkably does not disrupt Drp1 helical polymerization on membranes, suggesting an alternative and preferential Drp1-specific mode of stalk-mediated self-assembly. A unique assembly interface described for Drp1, interface 4, that permits a ‘back-to-back’ interaction of the dimeric stalks, instead of a ‘side-by-side’ one (Fig. 1D) (53,62), likely mediates this alternative self-assembly mode.
The membrane-interacting pleckstrin homology (PH) domain of dynamin and the intrinsically disordered variable domain (VD) of Drp1, in their unliganded states (i.e. non-membrane-bound states), function to auto-inhibit the premature higher-order self-assembly of dynamin/Drp1 in solution via the steric occlusion of one or more self-assembly interfaces (60–63,80). Deletion of the dynamin PH domain and the Drp1 VD, which mimics the autoinhibition-relieved, membrane-bound state of dynamin/Drp1 in solution, has clearly revealed the differential preferred interfaces and geometries of Drp1 and Dyn1 self-assembly (63,80). Whereas dynaminΔPH more efficiently forms helical polymers in solution relative to WT in keeping with the essential role of the now-exposed interface 3 in promoting helical dynamin self-assembly (63), Drp1ΔVD, by contrast, forms curvilinear polymers composed seemingly of interface 4-assembled dimeric subunits (Fig. 1D) (80). The intrinsically disordered VD thus appears to occlude interface 4 instead of, or in addition to, interface 3 in full-length Drp1. The inherent flexibility of the interface 4-assembled Drp1 polymer, in contrast to the interface 3-assembled curvature-restricted dynamin polymer, may be relevant for the curvature-adaptability of Drp1 self-assembly over the mitochondrial surface as well as over the progressively constricting mitochondrial fission sites in vivo (10,72,81). Nevertheless, the relative contributions and roles of these distinct interfaces in Drp1 function remain unresolved and awaits further mechanistic dissection.
The Drp1 VD also referred to as ‘insert B’ or ‘B-insert’ occurs in place of the dynamin PH domain. The VD is however absent in the Drp1 crystal structure, and is not structurally resolved in the current cryo-EM structure of membrane-bound yeast Dnm1p, leaving an apparent 3-to-4-nm gap (empty space) between the protein density and the membrane surface (52). This has given rise to speculation that Drp1/Dnm1p may not interact directly with the membrane bilayer, and may necessitate partner protein interactions (discussed later) for membrane anchoring (53). In support of this notion, a direct interaction between the yeast Dnm1p VD and its cognate mitochondrial membrane adaptor, Mdv1p, has been previously established (82). Regardless, recent studies have indicated that the Drp1 VD, which shares only 9% identity with the yeast Dnm1p VD (82), binds membrane phospholipids directly with a preference for the mitochondria-specific phospholipid, cardiolipin (CL) (72,83,84). Whereas the nature and functional consequence of Drp1 VD-membrane interactions are poorly understood, at least four Lys (K) residues located within the VD have been implicated in specific CL binding(83). Whether VD interactions with membranes and adaptors are mutually exclusive is unclear.
The VD, similar to the GTPase domain 80-loop, is subject to alternative splicing and varies in length between 111–148 amino acid residues among Drp1 splice variants. Intriguingly, recent studies have shown that the Drp1 VD splice variants differ significantly in their preferred geometry of helical self-assembly over membranes, with a longer VD correlating with a narrower helical diameter (73). These data suggest that the unstructured VD by nature of its variable hydrodynamic bulk functions as a ‘fulcrum’ or a ‘pivot’ to direct Drp1 self-assembly in defined helical geometries. The VD is also the site of multiple post-translational modifications (e.g. sumoylation) (79,83), which may further regulate the geometry of Drp1 self-assembly (85). The roles and mechanisms of the VD in regulating Drp1 function awaits further structural and functional examination.
Despite the relative wealth of information available for the structured and globular dynamin PH domain (~125 amino acid residues) and its role in dynamin-catalyzed endocytic vesicle scission (86–89), its apparent essential role in Dyn2-catalyzed mitochondrial fission remains unclear (10). The dynamin PH domain binds to negatively charged phospholipids, and exhibits specificity for phosphatidylinositol-4,5-biphosphate (PIP2), an essential lipid present typically at the plasma membrane. Although there are sparse studies implicating the involvement of PIP2 and its phosphatase, synaptojanin 2 in mitochondrial membrane fission (90–92), this has not been further investigated. Whether the Dyn2 PH domain binds other negatively charged phospholipids present on the mitochondrial surface, including cardiolipin (CL) and phosphatidic acid (PA), to anchor Dyn2 for mechanoenzymatic membrane constriction and fission is unknown. Incidentally, Dyn1 has been previously shown to selectively and deeply insert into PA-containing lipid monolayers (93). PH domain-membrane insertion is critical for dynamin-catalyzed membrane fission during endocytosis (88).
Classical dynamins including Dyn2, but not Drp1, contain a C-terminal unstructured proline-rich domain (PRD) (Fig. 1A, top). Dyn2 is initially recruited to endocytic fission sites at the plasma membrane via PRD interactions with the Src-homology 3 (SH3) domains of protein partners (94,95). Subsequent interactions of the PH domain with PIP2 on the highly curved membrane necks of invaginated endocytic pits promotes helical dynamin self-assembly for fission (96). Although the PRD is essential for Dyn2’s purported role in mitochondrial fission (10), the identity of its recruitment partners at mitochondrial fission sites remains elusive. A few likely candidates include SH3- and BAR (Bin/Amphiphysin/Rvs) domain-containing proteins such as sorting nexin 9 (SNX9) (97) and endophilin B1 (98) that localize to the mitochondria. However, their roles in mitochondrial fission are not firmly established.
Two binding partners common to Drp1 and Dyn2 are the actin cytoskeleton and microtubules, both of which are intimately associated with the mitochondria (79,81,99,100). Drp1 and Dyn2 both directly bind filamentous actin via their stalk regions, and robustly induce actin bundling and dynamics (100,101). The potential roles of these cytoskeletal elements in the localization and dynamics of Drp1 and Dyn2 at mitochondrial fission sites, as well as in force generation and membrane fission remain unexplored (102).
2. 2. TM-anchored DRPs of mitochondrial fusion
Unlike dynamin and Drp1 that share a common structural framework and a nearly identical domain arrangement, fusion DRPs Mfn1/2 and OPA1, remarkably, are architecturally dissimilar to each other (Fig. 2A, top). This likely reflects the unique physicochemical environments of the MOM versus the MIM as well as the additional roles of these DRPs in maintaining the distinct morphology and interactions of these two disparate membranes (4,6,103). Mfn1 is found exclusively at the MOM surface where it participates in homotypic or heterotypic interactions with Mfn1 or Mfn2 on opposing mitochondria (6,104). On the other hand, Mfn2 is also present at the ER membrane, and may be essential for ER-mitochondria membrane tethering at contact sites (26). OPA1 likewise also plays a critical role in maintaining the tubular morphology of the numerous ‘cristae’ that involute the MIM. IMS-facing OPA1 participates in membrane tethering interactions across the length of the cristae lumen and maintains cristae morphology (6). Full-length Mfn1/2 and OPA1 are both anchored via conventional TM segments to their respective membranes even though the locations of these TM segment(s) relative to the GTPase domains are topologically reversed between the two (Fig. 2A, top) (4).
Phylogenetically related more closely to BDLP than to dynamin or Drp1, Mfn1/2 adopts the recurrent ‘U-shaped’ polypeptide configuration with the N-terminal GTPase domain and the extreme C-terminal helical region present in close proximity, exposing the bulk of Mfn1/2 thus to the cytosol (36,37,105). The solitary four-helical bundle ‘stalk’ of dynamin/Drp1 (Fig. 1A, bottom) is effectively replaced in Mfn1/2 by a bipartite four-helical bundle composed of helical bundles 1 (Fig. 2A, bottom) and 2 (HB1 and HB2; termed ‘neck’ and ‘trunk’, respectively, in BDLP) (36–38). Loop segments that connect the helices of HB1 and HB2, by analogy to BDLP, function seemingly as a collective, flexible ‘hinge’ to effect dramatic ‘swivel-like’ nucleotide-dependent conformational rearrangements that displace HB1 and HB2 relative to each other (36–38,106). Replacing the ‘paddle’ segments located at the base of the BDLP trunk that peripherally interact with membranes are two closely spaced TM segments of inverse topology at the base of HB2 in Mfn1/2 that permanently integrate these DRPs permanently into the MOM bilayer (105).
Although the functional requirement of the TM anchoring of Mfn1/2, as opposed to the peripheral association of BDLP, in relation to membrane tethering and fusion is yet unclear, a mechanism based partly on models for BDLP in bacterial membrane fusion and for atlastin (39,107) in ER membrane fusion has been proposed for Mfn1/2. The recently solved 3D structures of a minimal Mfn1 construct containing only the GTPase domain and HB1 are virtually identical to that of the GTPase and neck domains of full-length BDLP (36–38). Based on this observation and the dramatic conformational rearrangements that ensue in full-length BDLP upon GTP hydrolysis, as observed under different nucleotide-bound structural states (106), a similar order of events is envisaged for Mfn1/2 (105,108). In this model, Mfn1/2 present in a GTP-bound ‘open’ conformation, which positions HB1 and HB2 in an ‘end-on’ arrangement, mediates the in trans tethering of opposing MOM bilayers via the transition state-dependent dimerization of GTPase domains across the two membranes. Subsequent GTP hydrolysis and inorganic phosphate (Pi) release, in yet another ‘power stroke’, effect a dramatic buckling of this elongated structure to a ‘closed’ conformation which forces HB1 and HB2 to adopt a more ‘side-by-side’ arrangement, presumably thus pulling the two tethered membrane bilayers closer together for fusion. Consistent with this model, recent cryo-electron tomography (cryo-ET) studies of isolated yeast mitochondria trapped at various stages of the fusion process reveal three distinct steps in the mechanism of Mfn1/2 (Fzo1p in yeast)-mediated MOM fusion (12). The first step involves ‘membrane tethering’ wherein the MOM tips of opposing mitochondria are bridged at an intermembrane distance of ~7 nm by an organized, dense lattice of Mfn1/2 molecules emanating from both partner membranes. This likely corresponds to the open, GTP-bound (nonhydrolyzable GMP-PNP-stabilized) conformation of Mfn1/2 paired across the two membrane bilayers via the transition state-dependent in trans dimerization of the GTPase domains. The second step involves ‘membrane docking’, wherein the bridging Mfn1/2 lattice is reorganized upon GTP hydrolysis and/or GDP/Pi release into a ‘docking ring’ that outlines the periphery of the area of contact between the now more closely juxtaposed MOM bilayers with an intermembrane distance of ~1–3 nm (down from ~7 nm in the GTP-bound state). Although no protein density is observed at the regions of closest approach between the membranes, it is nevertheless posited that a transition to the closed, GDP-bound conformation, coupled to Mfn1/2 lattice disassembly and reorganization, facilitates this transition. Interestingly however, the Mfn1/2 molecules that constitute the docking ring at the edges of these contact areas still maintain an intermembrane distance of ~7 nm, which might reflect a reversal to the GTP-bound open conformation post-membrane docking and -lattice reorganization. The third and final step involves ‘membrane fusion’ wherein a partial disassembly of the docking ring catalyzes the formation of a toroidal membrane pore of ~40-nm diameter between the docked membrane bilayers that may possibly expand further to complete MOM fusion. What maintains membrane docking in the absence of protein density, or how the Mfn1/2 lattice-to-docking-ring transition is accomplished, or whether the Mfn1/2 molecules that constitute the docking ring undergo yet another round of nucleotide-dependent structural transition to catalyze pore formation, remain unknown. How the fusion pore is expanded further around the periphery of membrane contact in the face of docking ring disassembly is still a mystery. Regardless, together these studies have provided unprecedented insight into the plausible mechanisms of MOM fusion, and have offered several starting points for further mechanistic exploration.
In contrast to Mfn1/2, mammalian OPA1 contains a single TM segment at its extreme N-terminus that connects to the more centrally located, IMS-facing GTPase domain via an intervening coiled-coil (CC) region (Fig. 2A, top). Remarkably, however, C-terminal to the GTPase domain, OPA1 is more similar to dynamin/Drp1 than to Mfn1/2 or BDLP (4). Like dynamin and Drp1, OPA1 contains well-defined MD and GED regions that are connected by a putatively unstructured polypeptide region akin to the Drp1 VD. Although no high-resolution structures are yet available for OPA1 and no detailed mechanisms proposed, such domain arrangement would essentially place the GTPase domain in close proximity to the membrane with the stalk and the unstructured region positioned at a distance away from the membrane surface, a topography essentially opposite to that of dynamin/Drp1 or Mfn1/2. Interestingly however, TM-anchored full-length OPA1 (referred to as long OPA1 or l-OPA1) can also be proteolytically cleaved by proteases YME1L and OMA1 to generate a soluble form of OPA1 (referred to as s-OPA1) that resides in the IMS (109), and which could peripherally interact with membranes in a more typical dynamin/Drp1-like topography. This plausible alternate topography or ‘orientation-switch’ of s-OPA1 may be relevant to its purported role in mitochondrial fission as suggested previously (109) and discussed further below. Moreover, unlike in the case of Mfn1/2, OPA1 is not required on both opposing membranes for MIM fusion suggesting that OPA1-OPA1 trans-interactions are not required for membrane tethering (110). This observation raises the possibility that TM-anchored OPA1 may interact directly with specific lipids on the opposing MIM, likely through the unstructured polypeptide region located at the base of its stalk (Fig. 2B), in order to tether and fuse membranes. A very recently published study indeed corroborates this notion (111).
Notwithstanding the paucity of information on mammalian OPA1, much structural and functional insight have been gleaned from studies of its yeast ortholog, Mgm1p. Mgm1p, which likewise undergoes proteolytic processing to yield l-Mgm1p and s-Mgm1p variants, however does not exhibit any basal GTPase activity unless the long and short variants are paired in a heterotypic manner via their GTPase domains (54). Moreover, whereas both l-Mgm1p and s-Mgm1p can participate in homotypic interactions across opposing MIM, their heterotypic pairing occurs only on the same MIM (54). These data, by analogy to Mfn1/2, suggest that the transition between membrane tethering and the GTP hydrolysis-dependent membrane docking and fusion steps for OPA1 is not progressive (or automatic) and relies critically on the balance (ratio) of long and short variants at MIM fusion sites. Regardless, an l-OPA1 variant incapable of being proteolytically cleaved to generate s-OPA1 was shown to be sufficient for both mitochondrial fusion and cristae maintenance in mammalian cells (109). How or whether l-OPA1 exhibits GTPase activity in a more physiological setting is unknown. s-OPA1, incompetent for membrane fusion in the absence of l-OPA1 either in vitro or in vivo, is however sufficient for the maintenance of cristae morphology (112). Furthermore, s-OPA1 expression has been shown to promote mitochondrial fission instead of fusion indicating that s-OPA1 activity at the MIM maybe somehow coupled to Drp1 activity at the MOM for a concerted fission of the mitochondrial double membrane (109). Indeed, very recent data suggest that a MOM-independent, OPA1-regulated constriction of the MIM is a priming event for mitochondrial division (113). In contrast to s-OPA1, s-Mgm1p is sufficient for liposome fusion in vitro (56).
A standout enzymatic characteristic of the mitochondrial fusion DRPs compared to the fission DRPs is the marked absence of a robust GTPase activity. The minimal Mfn1 GTPase domain-HB1 construct exhibits a maximal basal turnover rate of between ~0.3–3.0 per minute depending on the experimental conditions used (36,37), whereas OPA1 or Mgm1p, which exhibits discernible basal GTPase activity only in the short, soluble form, turns GTP over at a maximal rate of ~1.0 per minute (54,55,114). A structural comparison of the Mfn1 GTPase domain with that of Drp1 reveals the molecular bases of their disparate enzymatic activities (Fig. 2A, bottom). In contrast to Drp1, the active site of Mfn1 is exposed with a flat guanine-nucleotide binding interface. A signature motif of the fission DRPs termed the ‘guanine cap’ or ‘G5’, which arches over the binding pocket and tightly wraps the nucleotide is completely missing in Mfn1 explaining the relatively poor affinity for guanine nucleotides (36,37). In addition, the Mfn1 nucleotide-binding pocket is shielded by a bulky Trp residue (W239), conserved only in BDLP, that requires dislocation by incoming GTP for the proper docking of the nucleotide (36). Furthermore, the GTPase domain dimerization interface stabilized in the presence of transition state analogs, GDP/AlF4− or GDP/BeF3−, is substantially smaller in Mfn1 in comparison to dynamin (~995 Å2 vs. 2,500 Å2), and is stabilized primarily by electrostatic interactions versus exclusive hydrophobic, in trans interactions in dynamin (36). The reduced GTP hydrolysis rates of the fusion DRPs, by disfavoring lattice disassembly, may facilitate the formation of an appropriate area of initial intermembrane contact between opposing mitochondria to achieve stable membrane tethering. Subsequent cooperative GTP hydrolysis-driven lattice reorganization and membrane docking accomplishes membrane fusion. How Mfn1/2 GTPase activity is spatiotemporally coordinated toward productive membrane fusion warrants further investigation.
In contrast to dynamin and Drp1 that utilize defined stalk interfaces to direct helical self-assembly over tubular membrane templates, Mfn1/2 and OPA1 (Mgm1p) primarily constitute organized and extended ‘flat lattices’ on membrane surfaces (54–56). Although not much is known ultrastructurally about MOM-anchored Mfn1/2 in this regard, dimeric building blocks of MIM-anchored s-Mgm1p (OPA1 ortholog) have been shown to self-organize in a triangular, trimer-of-dimers arrangement (54). These hexamers (trimer-of-dimer subunits) ultimately form the higher-order repeating unit of the extended protein lattice. Hexamer-hexamer stacking interactions between opposing membranes presumably achieve membrane tethering with an intermembrane distance of ~15 nm (55,56). GTP binding has been shown to reorganize and spatially constrict this lattice to apparently promote membrane docking, whereas GTP hydrolysis has been shown to drive membrane fusion (56). Contradicting results have been obtained for Mfn1 in that the deletion of the TM segment has been shown to either ablate (37) or have no tangible effect (36) on the nucleotide-independent formation of Mfn1 dimers. Regardless, in the presence of either GTP or transition-state GTP analogs, Mfn1 dimers form higher-order complexes specifically via GTPase domain dimerization suggesting a similar mechanism to s-Mgm1p (36). A role for HB2, previously implicated in membrane tethering (115), has now been suggested in nucleotide-independent Mfn1 dimerization (36). Detailed high-resolution structural studies are in order to unravel the distinct mechanisms of fusion DRP self-assembly as well as identify their discrete nucleotide-dependent conformational transitions during membrane fusion.
Despite the lack of high-resolution structural data, specific ‘K’ residues within the putative unstructured region of s-Mgm1p (the structural equivalent of Drp1 VD) have been implicated in the binding of fusogenic membrane phospholipids, CL and PA (55). PA is a minor component of the MIM, whereas CL is highly enriched, and constitutes nearly 20% of the MIM phospholipids (116,117). Interestingly, K724 and K795 in s-Mgm1p (55), whose alanine substitutions abrogate lipid binding both in vitro and in vivo, are highly conserved in mammalian OPA1 (K792 and K847 in OPA1, respectively) (Fig. 2B) and likely perform similar functions. Similar to s-Mgm1p, s-OPA1 interacts with CL-enriched liposomes, which likewise stimulates OPA1 GTPase activity through the promotion of membrane tethering (114). Likewise, TM-anchored l-Mgm1p also preferentially partitions into CL-containing membranes (54). Whether this association also involves the unstructured region is not known as no correlated stimulation of GTPase activity is observed. Of interest, both s-Mgm1p and s-OPA1 can interconvert between flat lattices and protein-decorated membrane tubes (a la Drp1) suggesting alternate modes of membrane interactions (56,114). Whether this entails a toggle-like mechanism involving reversible unstructured region-membrane interactions remains unexplored. In addition, s-Mgm1p has also been shown to induce GTP-stimulated local membrane bending (ruffling) of liposomes suggesting an intrinsic capacity for fusogenic membrane remodeling (118). In the case of Mfn1/2, the local production of PA by enzymatic CL hydrolysis facilitates Mfn1/2-mediated MOM fusion (119), although whether Mfn1/2 interacts directly with PA is unknown. In the case of ER atlastin, it has been shown that the extreme C-terminal helix inserts into the membrane bilayer and may contribute to membrane bending (120). Whether a functional equivalent exists in Mfn1/2 is yet unclear.
3. The functional mechanics of mitochondrial fission
In this section, I will discuss the current hierarchical model for mitochondrial membrane remodeling and the emergent data that outline a different scenario. I will posit alternative mechanisms at play that may be involved in mitochondrial membrane fission.
3. 1. The ER-cytoskeleton-mitochondrion nexus: the first cut is the deepest
Recent work indicates that the ER orchestrates mitochondrial division, both spatially and temporally, by organizing and regulating curvature-inducing and force-generating protein molecules at fission sites (22–24,28). The initial finding that mitochondrial division in both yeast and mammalian cells occur at sites of ER tubule circumscription of mitochondria led to the subsequent discovery that the mitochondria, which approximate ~500 nm in overall diameter, are preconstricted to a significantly smaller diameter (<200 nm) exclusively at ER-mitochondria contact sites (22). This occurs independently of Drp1 (Dnm1p), but is dependent on ER-associated actin (23). In yeast, these preconstrictions range between ~150–200 nm in diameter, which closely approximates the diameter of the Dnm1p helices formed over negatively charged membranes in vitro (52), albeit in the absence of GTP hydrolysis. What this size-correspondence would be in mammalian cells is still not clear since mammalian Drp1 with its multiple splice variants displays a much more dynamic range of helical diameters, ranging from ~50-nm to ~150 nm in diameter even in the absence of GTP hydrolysis (73). Regardless, the ER-actin-mediated constriction of mitochondria is hypothesized to generate a cylindrical membrane template that is well suited for Drp1/Dnm1p helical self-assembly (22) (Fig. 3). Actin filament formation at the ER-mitochondria interface is nucleated and elongated cooperatively by ER-anchored inverted formin 2 (INF2) and MOM-associated Spire1C (24,28). Actin filament elongation in the radial direction of the mitochondria is proposed to exert pressure on the MOM to drive mitochondrial constriction (Fig. 3). What restricts this constriction to a specific diameter, or whether actin polymerization comes to an arrest at this stage, as postulated, is however unclear. It may be that the turgor pressure of the mitochondria coupled with the mechanical resistance of the mitochondrial double membrane to bending counteracts any further actin-mediated constriction; that is until Drp1 self-assembly facilitates further remodeling utilizing unconventional mechanisms as described below. ER tubule-mitochondria contact also depend on microtubules (32). A SNARE, syntaxin 17 (Syn17), which localizes to ER-mitochondria contact sites, directly binds both microtubules and Drp1 and may function to colocalize ER tubules and Drp1 at future fission sites (121). Syn17-mediated bridging of ER tubule-associated microtubules to specifically GTP- and adaptor-bound Drp1 at ER-mitochondria contact sites (121) could further contribute mechanical forces for localized mitochondrial constriction and fission.
Figure 3.
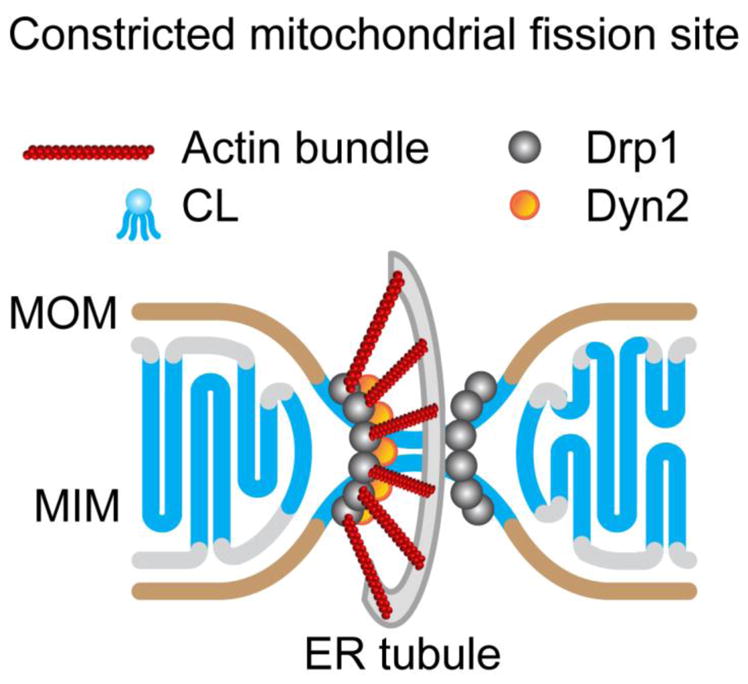
Molecular organization at a mitochondrial fission site. Shown is a cartoon illustration of an ER tubule-constricted mitochondrial division site accentuating the putative, relative spatial arrangement of the key molecular players involved in the mechanics of mitochondrial fission.
3. 2. Constriction by Drp1: so near yet so far?
Yeast Dnm1p helices in the absence of essential protein and lipid partners constrict negatively charged tubular membrane templates to an outer diameter of only about 70 nm implying that Dnm1p/Drp1 does not have the dynamic range to constrict membranes to the point of fission (52). Yet, subsequent studies with mammalian Drp1 on either artificial or physiologically relevant membrane templates have shown that Drp1 can indeed constrict membrane tubes down to ~30 nm or less upon GTP hydrolysis (49,122). The dynamic range of Drp1 is therefore comparable to that of prototypical Dyn1, which upon GTP binding superconstricts membrane tubes from ~50 nm outer diameter down to ~37 nm (~3.7 nm luminal diameter; the theoretical limit for spontaneous fission) (66). For reasons yet unclear however, Drp1, unlike dynamin, does not mediate the complete fission of membrane tubules (122). On membrane tubes containing a physiologically relevant mixture of phospholipids, including CL and PE relevant to mitochondrial morphology and fission (117), Drp1 self-assembly in the constant presence of GTP has been shown create similarly narrow but localized ‘superconstrictions’ (<30 nm diameter) that are primed for fission (122). Interestingly, the generation of localized Drp1-mediated superconstrictions entail the sequestration, clustering and non-bilayer phase transition of negative curvature-promoting cone-shaped CL, alongside PE, by the Drp1 VD (122). It is therefore plausible that the sequestration of cone-shaped lipids at ER-mitochondria contact sites upon Drp1 self-assembly partially counteracts the bending resistance of the mitochondria and augments the high negative membrane curvature required for a deeper ER-actin-mediated constriction (123)(Fig. 3). Thus, ER-actin and Drp1 may function cooperatively, and not in a strictly hierarchical manner, to create and stabilize mitochondrial superconstrictions. Interestingly, actin filaments directly bind Drp1 and promote Drp1 GTPase activity, which likely enhances the cooperative membrane remodeling (81,101).
3. 3. Actin’ like dynamin in mitochondrial fission
Yeast do not possess a classical dynamin like Dyn2, yet exhibit mitochondrial fission. Therefore, the role of Dyn2 in mammalian mitochondrial fission is likely tailored for a regulatory purpose instead of for direct membrane remodeling. In the current hierarchical model for mitochondrial fission, Dyn2 is proposed to superconstrict membrane tubules that are preconstricted by Drp1(10). This however may create an ‘area conflict’ for Drp1 and Dyn2 helical self-assembly. In vivo imaging data of Dyn2-depleted mitochondria reveal transient splitting of Drp1 puncta, which apparently then bridge a naked, superconstricted membrane tube of ~70 nm diameter awaiting Dyn2 arrival for further progression toward fission (10). How such an energetically unfavorable tubular membrane configuration is stabilized in the absence of a protein coat is nevertheless a mystery. Actin dynamics at fission sites (124), essential for mitochondrial division, likely mediates this splitting of the dynamic Drp1 scaffold and applies tensile forces to stabilize these tubular membrane intermediates. Furthermore, actin has been shown to stimulate Drp1 GTPase activity in collaboration with MOM-anchored Drp1 partner proteins (e.g. Mff, see below) and may facilitate Drp1 disassembly (81). Interestingly, the only recognized binding partner of Dyn2 that exists at mitochondrial fission sites is actin itself (100). Dyn2, via mechanisms yet unexplored, likely regulates actin dynamics at these sites to resolve Drp1-superconstricted tubular membrane intermediates toward fission (Fig. 3). Actin directly binds to the dynamin middle domain whose accessibility in turn is regulated by the PH domain (60,61). The dynamin PRD, even in the absence of binding partner interactions, regulates dynamin GTPase activity (125). Thus, all of these regulatory Dyn2 activities are found essential for mitochondrial fission (10). The proposed hierarchical mechanochemical role of Dyn2 in mitochondrial fission may therefore not be a foregone conclusion but demands further scientific verification. Furthermore, it is not clear whether Dyn2 recruitment to fission sites necessitates helical Drp1 self-assembly (10). Why this Dyn2-dependent mechanism operates in mammals but not in yeast is an unresolved mystery.
4. The supporting cast: the essential protein and lipid cofactors of mitochondrial fission
Although Drp1 can directly bind negatively charged membrane phospholipids including CL under favorable conditions in vitro (72,83,84), recruitment of cytosolic Drp1 to mitochondria in vivo relies critically on its interactions with various MOM-anchored transmembrane protein adaptors (receptors), namely mitochondrial fission factor (Mff), mitochondrial dynamics proteins of 49 and 51 kDa (MiD49/51), and fission factor 1 (Fis1) (18). Among these, Mff and MiD49/51 play a more prominent role in mammalian cells (126). Mff and MiD49/51 are found exclusively in metazoans (MiD49/51 only in chordate metazoans) and do not have orthologs in yeast (127,128). Moreover, MiD49/51 strictly localizes to the mitochondria, whereas Mff and Fis1 are also present in peroxisomes, which in turn depend on Drp1 for fission (129). MiD49 and MiD51 are largely redundant in structure and function, although a controversial and unresolved regulation of MiD51 activity, but not of MiD49, by cytosolic ADP has been proposed (126,127,129–132). The role of Fis1 in mammals remains controversial. Although Fis1 is essential for Dnm1p recruitment to mitochondria in yeast (133), it is largely dispensable for Drp1 recruitment and mitochondrial fission in mammalian cells (126,134,135). Nevertheless, the absence of Fis1 in mammalian cells perturbs mitophagy and apoptosis through mechanisms that are yet unclear (126,136–139).
Mff and MiD49/51 can independently recruit Drp1 to mitochondria in vivo to promote fission (129,135). Mff and MiD49/51 also bind Drp1 independently in vitro and can promote Drp1 self-assembly even in the absence of membranes (75,130,131,140). However, in vivo, knockdown of each adaptor substantially reduces Drp1 mitochondrial recruitment, and the combined ablation of Mff and MiD49/51 completely abrogates Drp1 mitochondrial localization (141). These data suggest MOM CL (present at <10 % of total phospholipids) cannot by itself recruit Drp1, and that Drp1-CL interactions occur either coincidentally with, or downstream of, Drp1 recruitment by protein adaptors. Interestingly, Mff has also been shown to preferentially partition into CL-enriched membranes in vitro (73) and may facilitate such an interaction. Whether this holds true also for MiD49/51 remains to be determined.
Advanced imaging techniques have revealed that Mff and MiD51 colocalize at ER-mitochondria contact sites to direct Drp1-mediated mitochondrial fission (22,142). Furthermore, recent studies have indicated that MiD51 (a.k.a. MIEF1) and Mff interact with each other to constitute a ternary complex with Drp1 (126). These data suggest that Mff and MiD51 function cooperatively with Drp1 to mediate mitochondrial fission. Paradoxically however, MiD51 and Mff exert opposite effects on Drp1 GTPase activity (126). On membranes, Mff copolymerizes with Drp1 to stimulate Drp1 GTPase activity, whereas Drp1 copolymerization with MiD51 suppresses Drp1 GTPase activity (126). In a ternary mixture containing Drp1, Mff and MiD51, MiD51 effectively reduces the Mff-stimulation of Drp1 GTPase activity (126). Based on the observation that the presence of the VD inhibits stable Drp1-Mff interactions in vitro (140,143), it has been proposed that a displacement of the VD via upstream CL interactions promotes subsequent Drp1-Mff co-assembly. Yet, in other studies, Mff has been shown to also bind full-length Drp1 and stimulate Drp1 GTPase activity independent of CL, suggesting that Drp1 VD-CL interactions conversely occur downstream of Drp1-Mff interactions (73,81). MiD51 suppression of Mff-stimulated GTPase activity also occurs independent on CL (126), indicating that Drp1-phospholipid interactions likely occur post-adaptor recruitment. This could explain why only a subset of the dynamic Drp1 puncta found on the mitochondrial surface, where Drp1 is necessarily associated with both protein adaptors and phospholipids, progress toward fission (81). The reduced rate of GTP hydrolysis effected by MiD51 may stabilize Drp1-Mff complexes on the mitochondrial surface until an appropriate membrane microenvironment, e.g. ER-mitochondrial contact site, is encountered to effect Drp1 VD-CL interactions and progress toward productive fission. MiD49/51 may thus function as a ‘kinetic timer’ of mitochondrial fission. How these adaptors cooperatively regulate Drp1 function, both spatially and temporally, and link their differential activities toward mitochondrial fission remains to be determined.
CL is the signature phospholipid of the mitochondria. Enriched in the mitochondrial inner membrane (MIM) where it constitutes nearly 1 out of 5 lipids, CL is also found in relatively minor but significant quantities (3–10%) in the mitochondrial outer membrane (MOM) (18,116). Furthermore, CL content can approach 25% at MIM-MOM contact sites that speckle the mitochondrial surface (144). CL promotes the helical self-assembly and enzymatic stimulation of Drp1 over membranes independent of protein adaptors in vitro (72,122,145). In mammalian cells, MOM surface-exposed CL likewise appears to play a critical role in regulating mitochondrial fission. Degradation of CL to phosphatidic acid (PA) by Drp1-associated, MOM-localized mitochondrial phospholipase D (mitoPLD) inhibits Drp1 polymers in fission, firmly establishing a role for mitochondrial surface-localized CL and CL metabolism in mitochondrial fission (145). Degradation of PA to cone-shaped, but uncharged, diacylglycerol (DAG) by the enzyme ‘lipin’ relieves this inhibition and instead promotes mitochondrial fission, presumably through the DAG-mediated stabilization of negative membrane curvature (117,145). Yeast devoid of CL still exhibit mitochondrial fission (146,147). CL-deficient yeast however accumulate phosphatidylglycerol (PG), which is a CL precursor and a minor anionic lipid under normal conditions (148,149). Similar to CL in various biophysical properties, PG likely compensates for the lack of CL and rescues mitochondrial fission. A similar scenario may also play out in CL-deficient mammalian cells, which also do not exhibit an overt mitochondrial phenotype (150). Regardless, the mechanism by which CL fulfills its role in mitochondrial fission warrants further exploration, given that the existing paradigm does not invoke a role for specific Drp1-phospholipid interactions (53). The nature, molecular identity, specificity, hierarchy and functional consequences of Drp1 VD-phospholipid interactions in mitochondrial fission also remain largely unknown.
5. The CL link to coupled mitochondrial fusion
Conversion of CL to fusogenic PA at the MOM surface, while inhibiting Drp1-mediated mitochondrial fission (145), remarkably promotes Mfn1/2-mediated mitochondrial fusion (119). Thus, CL metabolism at the MOM functions a bi-functional ‘on/off toggle’ to govern the balance of mitochondrial fission and fusion. Whether PA-producing mitoPLD, which physically associates with Drp1 (145), also interacts with Mfn1/2, or whether Drp1 and Mfn1/2 colocalize (or possibly even associate) in the same microenvironment to respond oppositely to metabolic cues is unknown. CL, owing to its greater shape polymorphism (bilayer and non-bilayer orientations) could either be fusogenic or pro-fission (117). MIM CL organization could regulate OPA1 topography (orientation) at the MIM by promoting either fusogenic lattice formation (via l-OPA1 N-terminal TM-anchoring) or fission-prone MIM tubulation (via s-OPA1 unstructured region peripheral membrane interactions). Thus, CL organization at the MIM can function as a ‘topographical switch’ for OPA1 and coordinately balance MIM fission and fusion. In yeast, Ugo1p, a MOM-associated protein functions to coordinate MOM and MIM fusion by physically bridging MOM Fzolp (Mfn1/2 ortholog) to MIM Mgm1p (OPA1 ortholog) and additionally participates in lipid mixing at fusion sites (4). A similar molecule is however yet to be discovered in mammals. Regardless, whether such a collusion exists among DRPs in executing the finely tuned balance of fission and fusion in mitochondrial dynamics is yet to be thoroughly investigated.
Concluding remarks
The last decade has witnessed the evolution of the field of mitochondrial dynamics from mostly phenomenology and physiology (or pathophysiology) to precise structures and mechanisms. The advent of high-resolution 3D structures (snapshots) for most, if not all, of the molecular players involved have provided ideal vantage points for an elaborate investigation of their intricate biochemical and biophysical mechanisms. Yet as outlined above, much is unknown about how these protein activities are coordinated in space and time. For instance, despite the availability of the Drp1 (ΔVD) structure, much remains unknown about how Drp1 interacts specifically with membrane phospholipids, a critical facet of its function. Furthermore, the structural transitions in Drp1 that precede or follow interactions with either adaptor proteins or phospholipids, as well as the order of molecular events required for productive mitochondrial fission remain unclear. How adaptor proteins and phospholipids cooperate with Drp1, mechanistically, to mediate mitochondrial fission remains unresolved, and poses a fundamental gap in our understanding of mitochondrial dynamics. The newly established roles of Dyn2 and ER-actin dynamics in mitochondrial fission also warrant a deeper mechanistic investigation. How fission and fusion DRPs assemble and function disparately, yet coordinate their activities to sustain the mitochondrial fission-fusion balance still remains to be deciphered. With the ongoing revolution in high-resolution electron cryo-microscopy (cryo-EM), and the application of highly sophisticated biophysical approaches to investigate protein and membrane dynamics, greater mechanistic insight might be just around the corner.
Acknowledgments
I wish to thank Jason Mears and Ryan Clinton (CWRU) for their assistance in the preparation of figures and in the EM analysis of Drp1ΔVD polymers. This work is supported by a US National Institutes of Health (NIH) grant 1R21NS101554-01 awarded to RR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kowald A, Kirkwood TB. Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proc Natl Acad Sci U S A. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 4.Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 5.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pernas L, Scorrano L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu Rev Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 7.Wai T, Langer T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol Metab. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 9.Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt T, Cavellini L, Kuhlbrandt W, Cohen MM. A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. Elife. 2016:5. doi: 10.7554/eLife.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125:2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter V, Singh AP, Kvansakul M, Ryan MT, Osellame LD. Splitting up the powerhouse: structural insights into the mechanism of mitochondrial fission. Cell Mol Life Sci. 2015;72:3695–3707. doi: 10.1007/s00018-015-1950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy M, Reddy PH, Iijima M, Sesaki H. Mitochondrial division and fusion in metabolism. Curr Opin Cell Biol. 2015;33:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naon D, Zaninello M, Giacomello M, Varanita T, Grespi F, Lakshminaranayan S, Serafini A, Semenzato M, Herkenne S, Hernandez-Alvarez MI, Zorzano A, De Stefani D, Dorn GW, 2nd, Scorrano L. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc Natl Acad Sci U S A. 2016;113:11249–11254. doi: 10.1073/pnas.1606786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naon D, Scorrano L. At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim Biophys Acta. 2014;1843:2184–2194. doi: 10.1016/j.bbamcr.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015:4. doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horbay R, Bilyy R. Mitochondrial dynamics during cell cycling. Apoptosis. 2016;21:1327–1335. doi: 10.1007/s10495-016-1295-5. [DOI] [PubMed] [Google Scholar]
- 30.Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Kanfer G, Kornmann B. Dynamics of the mitochondrial network during mitosis. Biochem Soc Trans. 2016;44:510–516. doi: 10.1042/BST20150274. [DOI] [PubMed] [Google Scholar]
- 32.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varadi A, Johnson-Cadwell LI, Cirulli V, Yoon Y, Allan VJ, Rutter GA. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J Cell Sci. 2004;117:4389–4400. doi: 10.1242/jcs.01299. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, Yoon Y. Mitochondrial fission and fusion. Biochem Soc Trans. 2016;44:1725–1735. doi: 10.1042/BST20160129. [DOI] [PubMed] [Google Scholar]
- 35.McBride HM, Frost A. Cell biology: Double agents for mitochondrial division. Nature. 2016;540:43–44. doi: 10.1038/nature20482. [DOI] [PubMed] [Google Scholar]
- 36.Cao YL, Meng S, Chen Y, Feng JX, Gu DD, Yu B, Li YJ, Yang JY, Liao S, Chan DC, Gao S. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature. 2017;542:372–376. doi: 10.1038/nature21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi Y, Yan L, Yu C, Guo X, Zhou X, Hu X, Huang X, Rao Z, Lou Z, Hu J. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol. 2016;215:621–629. doi: 10.1083/jcb.201609019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 39.Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci U S A. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 41.Purkanti R, Thattai M. Ancient dynamin segments capture early stages of host-mitochondrial integration. Proc Natl Acad Sci U S A. 2015;112:2800–2805. doi: 10.1073/pnas.1407163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koenig JH, Ikeda K. Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 48.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 49.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 52.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol. 2013;23:R891–899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rujiviphat J, Meglei G, Rubinstein JL, McQuibban GA. Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J Biol Chem. 2009;284:28682–28686. doi: 10.1074/jbc.M109.044933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abutbul-Ionita I, Rujiviphat J, Nir I, McQuibban GA, Danino D. Membrane tethering and nucleotide-dependent conformational changes drive mitochondrial genome maintenance (Mgm1) protein-mediated membrane fusion. J Biol Chem. 2012;287:36634–36638. doi: 10.1074/jbc.C112.406769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wenger J, Klinglmayr E, Frohlich C, Eibl C, Gimeno A, Hessenberger M, Puehringer S, Daumke O, Goettig P. Functional mapping of human dynamin-1-like GTPase domain based on x-ray structure analyses. PLoS One. 2013;8:e71835. doi: 10.1371/journal.pone.0071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chappie JS, Mears JA, Fang S, Leonard M, Schmid SL, Milligan RA, Hinshaw JE, Dyda F. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 2011;147:209–222. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 61.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reubold TF, Faelber K, Plattner N, Posor Y, Ketel K, Curth U, Schlegel J, Anand R, Manstein DJ, Noe F, Haucke V, Daumke O, Eschenburg S. Crystal structure of the dynamin tetramer. Nature. 2015;525:404–408. doi: 10.1038/nature14880. [DOI] [PubMed] [Google Scholar]
- 64.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 65.Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- 66.Sundborger AC, Fang S, Heymann JA, Ray P, Chappie JS, Hinshaw JE. A dynamin mutant defines a superconstricted prefission state. Cell Rep. 2014;8:734–742. doi: 10.1016/j.celrep.2014.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivasan S, Dharmarajan V, Reed DK, Griffin PR, Schmid SL. Identification and function of conformational dynamics in the multidomain GTPase dynamin. EMBO J. 2016;35:443–457. doi: 10.15252/embj.201593477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mattila JP, Shnyrova AV, Sundborger AC, Hortelano ER, Fuhrmans M, Neumann S, Muller M, Hinshaw JE, Schmid SL, Frolov VA. A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature. 2015;524:109–113. doi: 10.1038/nature14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, Kirchhausen T, Kozlov MM, Lenz M, Low HH, McMahon H, Merrifield C, Pollard TD, Robinson PJ, Roux A, Schmid S. Membrane fission by dynamin: what we know and what we need to know. EMBO J. 2016;35:2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macdonald PJ, Francy CA, Stepanyants N, Lehman L, Baglio A, Mears JA, Qi X, Ramachandran R. Distinct Splice Variants of Dynamin-related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity. J Biol Chem. 2016;291:493–507. doi: 10.1074/jbc.M115.680181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stowell MH, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- 75.Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci U S A. 2013;110:E1342–1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. Embo J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uo T, Dworzak J, Kinoshita C, Inman DM, Kinoshita Y, Horner PJ, Morrison RS. Drp1 levels constitutively regulate mitochondrial dynamics and cell survival in cortical neurons. Exp Neurol. 2009;218:274–285. doi: 10.1016/j.expneurol.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon Y, Pitts KR, Dahan S, McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol. 1998;140:779–793. doi: 10.1083/jcb.140.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strack S, Wilson TJ, Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol. 2013;201:1037–1051. doi: 10.1083/jcb.201210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Francy CA, Alvarez FJ, Zhou L, Ramachandran R, Mears JA. The Mechanoenzymatic Core of Dynamin-related Protein 1 Comprises the Minimal Machinery Required for Membrane Constriction. J Biol Chem. 2015;290:11692–11703. doi: 10.1074/jbc.M114.610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015:4. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bui HT, Karren MA, Bhar D, Shaw JM. A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment. J Cell Biol. 2012;199:613–622. doi: 10.1083/jcb.201207079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ugarte-Uribe B, Muller HM, Otsuki M, Nickel W, Garcia-Saez AJ. Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J Biol Chem. 2014;289:30645–30656. doi: 10.1074/jbc.M114.575779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Figueroa-Romero C, Iniguez-Lluhi JA, Stadler J, Chang CR, Arnoult D, Keller PJ, Hong Y, Blackstone C, Feldman EL. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23:3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- 88.Ramachandran R, Pucadyil TJ, Liu YW, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mehrotra N, Nichols J, Ramachandran R. Alternate pleckstrin homology domain orientations regulate dynamin-catalyzed membrane fission. Mol Biol Cell. 2014;25:879–890. doi: 10.1091/mbc.E13-09-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rosivatz E, Woscholski R. Removal or masking of phosphatidylinositol(4,5)bisphosphate from the outer mitochondrial membrane causes mitochondrial fragmentation. Cell Signal. 2011;23:478–486. doi: 10.1016/j.cellsig.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nemoto Y, Arribas M, Haffner C, DeCamilli P. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J Biol Chem. 1997;272:30817–30821. doi: 10.1074/jbc.272.49.30817. [DOI] [PubMed] [Google Scholar]
- 92.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burger KN, Demel RA, Schmid SL, de Kruijff B. Dynamin is membrane-active: lipid insertion is induced by phosphoinositides and phosphatidic acid. Biochemistry. 2000;39:12485–12493. doi: 10.1021/bi000971r. [DOI] [PubMed] [Google Scholar]
- 94.Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 95.Okamoto PM, Herskovits JS, Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 96.Bethoney KA, King MC, Hinshaw JE, Ostap EM, Lemmon MA. A possible effector role for the pleckstrin homology (PH) domain of dynamin. Proc Natl Acad Sci U S A. 2009;106:13359–13364. doi: 10.1073/pnas.0906945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, Fazel A, Bergeron JJ, Trudeau LE, Burelle Y, Gagnon E, McBride HM, Desjardins M. Parkinson’s Disease-Related Proteins PINK1 and Parkin Repress Mitochondrial Antigen Presentation. Cell. 2016;166:314–327. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 98.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanabe K, Takei K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-Tooth mutant. J Cell Biol. 2009;185:939–948. doi: 10.1083/jcb.200803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu C, Yaddanapudi S, Weins A, Osborn T, Reiser J, Pollak M, Hartwig J, Sever S. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hatch AL, Ji WK, Merrill RA, Strack S, Higgs HN. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell. 2016;27:3109–3121. doi: 10.1091/mbc.E16-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hatch AL, Gurel PS, Higgs HN. Novel roles for actin in mitochondrial fission. J Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ikon N, Ryan RO. Cardiolipin and mitochondrial cristae organization. Biochim Biophys Acta. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schrepfer E, Scorrano L. Mitofusins, from Mitochondria to Metabolism. Mol Cell. 2016;61:683–694. doi: 10.1016/j.molcel.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 105.Formosa LE, Ryan MT. Mitochondrial fusion: Reaching the end of mitofusin’s tether. J Cell Biol. 2016;215:597–598. doi: 10.1083/jcb.201611048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Low HH, Sachse C, Amos LA, Lowe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Byrnes LJ, Singh A, Szeto K, Benvin NM, O’Donnell JP, Zipfel WR, Sondermann H. Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 2013;32:369–384. doi: 10.1038/emboj.2012.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Daumke O, Roux A. Mitochondrial Homeostasis: How Do Dimers of Mitofusins Mediate Mitochondrial Fusion? Curr Biol. 2017;27:R353–R356. doi: 10.1016/j.cub.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 109.Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ban T, Ishihara T, Kohno H, Saita S, Ichimura A, Maenaka K, Oka T, Mihara K, Ishihara N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol. 2017;19:856–863. doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 112.Lee H, Smith SB, Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J Biol Chem. 2017;292:7115–7130. doi: 10.1074/jbc.M116.762567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cho B, Cho HM, Jo Y, Kim HD, Song M, Moon C, Kim H, Kim K, Sesaki H, Rhyu IJ, Sun W. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat Commun. 2017;8:15754. doi: 10.1038/ncomms15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet. 2010;19:2113–2122. doi: 10.1093/hmg/ddq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 116.Ha EE, Frohman MA. Regulation of mitochondrial morphology by lipids. Biofactors. 2014;40:419–424. doi: 10.1002/biof.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Frohman MA. Role of mitochondrial lipids in guiding fission and fusion. J Mol Med (Berl) 2015;93:263–269. doi: 10.1007/s00109-014-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rujiviphat J, Wong MK, Won A, Shih YL, Yip CM, McQuibban GA. Mitochondrial Genome Maintenance 1 (Mgm1) Protein Alters Membrane Topology and Promotes Local Membrane Bending. J Mol Biol. 2015;427:2599–2609. doi: 10.1016/j.jmb.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 119.Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 120.Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci U S A. 2012;109:E2146–2154. doi: 10.1073/pnas.1208385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arasaki K, Shimizu H, Mogari H, Nishida N, Hirota N, Furuno A, Kudo Y, Baba M, Baba N, Cheng J, Fujimoto T, Ishihara N, Ortiz-Sandoval C, Barlow LD, Raturi A, Dohmae N, Wakana Y, Inoue H, Tani K, Dacks JB, Simmen T, Tagaya M. A Role for the Ancient SNARE Syntaxin 17 in Regulating Mitochondrial Division. Dev Cell. 2015;32:304–317. doi: 10.1016/j.devcel.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 122.Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell. 2015;26:3104–3116. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boyd KJ, Alder NN, May ER. Buckling Under Pressure: Curvature-Based Lipid Segregation and Stability Modulation in Cardiolipin-Containing Bilayers. Langmuir. 2017;33:6937–6946. doi: 10.1021/acs.langmuir.7b01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moore AS, Wong YC, Simpson CL, Holzbaur EL. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat Commun. 2016;7:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barylko B, Wang L, Binns DD, Ross JA, Tassin TC, Collins KA, Jameson DM, Albanesi JP. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry. 2010;49:10592–10594. doi: 10.1021/bi101343p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Osellame LD, Singh AP, Stroud DA, Palmer CS, Stojanovski D, Ramachandran R, Ryan MT. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem. 2013;288:27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Loson OC, Liu R, Rome ME, Meng S, Kaiser JT, Shan SO, Chan DC. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 2014;22:367–377. doi: 10.1016/j.str.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Richter V, Palmer CS, Osellame LD, Singh AP, Elgass K, Stroud DA, Sesaki H, Kvansakul M, Ryan MT. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J Cell Biol. 2014;204:477–486. doi: 10.1083/jcb.201311014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loson OC, Meng S, Ngo H, Liu R, Kaiser JT, Chan DC. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 2015;24:386–394. doi: 10.1002/pro.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 139.Wang K, Long B, Jiao JQ, Wang JX, Liu JP, Li Q, Li PF. miR-484 regulates mitochondrial network through targeting Fis1. Nat Commun. 2012;3:781. doi: 10.1038/ncomms1770. [DOI] [PubMed] [Google Scholar]
- 140.Clinton RW, Francy CA, Ramachandran R, Qi X, Mears JA. Dynamin-related Protein 1 Oligomerization in Solution Impairs Functional Interactions with Membrane-anchored Mitochondrial Fission Factor. J Biol Chem. 2016;291:478–492. doi: 10.1074/jbc.M115.680025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Otera H, Miyata N, Kuge O, Mihara K. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J Cell Biol. 2016;212:531–544. doi: 10.1083/jcb.201508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Elgass KD, Smith EA, LeGros MA, Larabell CA, Ryan MT. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J Cell Sci. 2015;128:2795–2804. doi: 10.1242/jcs.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu R, Chan DC. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol Biol Cell. 2015;26:4466–4477. doi: 10.1091/mbc.E15-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem. 1990;265:18797–18802. [PubMed] [Google Scholar]
- 145.Adachi Y, Itoh K, Yamada T, Cerveny KL, Suzuki TL, Macdonald P, Frohman MA, Ramachandran R, Iijima M, Sesaki H. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol Cell. 2016;63:1034–1043. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gohil VM, Thompson MN, Greenberg ML. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J Biol Chem. 2005;280:35410–35416. doi: 10.1074/jbc.M505478200. [DOI] [PubMed] [Google Scholar]
- 147.Joshi AS, Thompson MN, Fei N, Huttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem. 2012;287:17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Khalifat N, Rahimi M, Bitbol AF, Seigneuret M, Fournier JB, Puff N, Arroyo M, Angelova MI. Interplay of packing and flip-flop in local bilayer deformation. How phosphatidylglycerol could rescue mitochondrial function in a cardiolipin-deficient yeast mutant. Biophys J. 2014;107:879–890. doi: 10.1016/j.bpj.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pokorna L, Cermakova P, Horvath A, Baile MG, Claypool SM, Griac P, Malinsky J, Balazova M. Specific degradation of phosphatidylglycerol is necessary for proper mitochondrial morphology and function. Biochim Biophys Acta. 2016;1857:34–45. doi: 10.1016/j.bbabio.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Raemy E, Montessuit S, Pierredon S, van Kampen AH, Vaz FM, Martinou JC. Cardiolipin or MTCH2 can serve as tBID receptors during apoptosis. Cell Death Differ. 2016;23:1165–1174. doi: 10.1038/cdd.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]