Abstract
Highly pathogenic avian influenza H5N1 viruses are circulating in many countries. We recently discovered that these viruses have been transmitted to pigs on multiple occasions in Indonesia. To investigate whether avian H5N1 influenza viruses adapted to mammals through their introduction into pigs, we examined the growth of avian and swine isolates in cell culture and compared their pathogenicity in mice. We found that swine isolates were less virulent to mice than avian isolates, suggesting that the viruses became attenuated during their replication in pigs. Continuous surveillance of H5N1 viruses among pigs is clearly warranted.
Keywords: influenza, H5N1, chicken, pig, virulence
The highly pathogenic H5N1 avian influenza A virus was isolated first from geese in Guangdong province, China in 1996 [1]. Since then, the viruses have reached across Asia, Europe, and Africa, giving rise to >380 human infections with a high mortality rate of >60% [2-4]. In Indonesia, the H5N1 virus outbreaks were first confirmed in poultry in December 2003. The virus spread rapidly and was identified in almost all the provinces of Indonesia by the end of 2006 [4, 5]. Human infection with an H5N1 virus was first confirmed in western Java in July 2005. As of December 16, 2008, Indonesia had more than 139 confirmed human cases with 113 deaths [4]. This is the highest case number and mortality rate (>80%) among all affected countries, increasing the risk of emergence of a pandemic virus.
Pigs have been considered an intermediate host for avian influenza viruses to adapt to humans [6]. Epithelial cells in pig trachea can be infected with avian and human influenza viruses and support the replication of both types of virus because they bear receptors for these viruses [7, 8]. These features raise two possibilities for the emergence of a human pandemic virus. One possibility is the emergence of a reassortant virus of avian and human viruses via the co-infection of both viruses in a pig [8, 9]. Alternatively, repeated replication in pigs may allow an avian virus to adapt to grow efficiently in pig epithelial cells and replicate efficiently in mammalian cells [8].
In the 20th century, we experienced three human pandemics caused by the introduction of avian influenza virus into the human population. Previous studies revealed that the Spanish pandemic was caused by an avian influenza virus that adapted to humans and the Asian and Hong Kong pandemics were caused by avian-human reassortant viruses [10-12], although it was not proven that these strains were generated in pigs.
Available evidence suggests that swine H1N1 influenza viruses are sporadically transmitted into humans [13, 14]. Moreover, swine workers have been shown to have increased levels of antibodies to swine influenza viruses, suggesting that humans are susceptible to swine-adapted influenza viruses [15, 16]. H5N1 infections in pigs have been reported rarely or gone unnoticed altogether because pigs infected with H5N1 influenza viruses show no influenza-like signs or symptoms [17, 18]. There have been no reports on whether adaptation to pigs makes avian viruses adapt to replicate in other mammalian hosts.
We previously investigated the prevalence of H5N1 influenza viruses among pigs in Indonesia during 2005-2007 and discovered that pigs were asymptomatically infected with H5N1 viruses every year [Nidom et al., unpublished]. While Indonesian H5N1 viruses were exclusively classified into three clades, i.e., clade 2.1.1-2.1.3, based on phylogenetic analysis of their HA genes [19], our phylogenetic analysis of the HA gene revealed that our Indonesian swine H5N1 influenza isolates were classified into three sub-lineages, i.e., clade 2.1.1, 2.1.3, and an undetermined clade (Fig.1). In addition, phylogenetic analyses of the remaining seven viral genes illustrated that the phylogenetic relationships established for the HA gene were maintained, and swine isolates were most closely related to isolates from chickens kept nearby, indicating transmission of avian H5N1 viruses into pigs via infected chickens on at least three independent occasions [Nidom et al., unpublished].
Fig.1. Phylogenetic relationship between the HA genes of H5N1 influenza A viruses from pigs and birds.
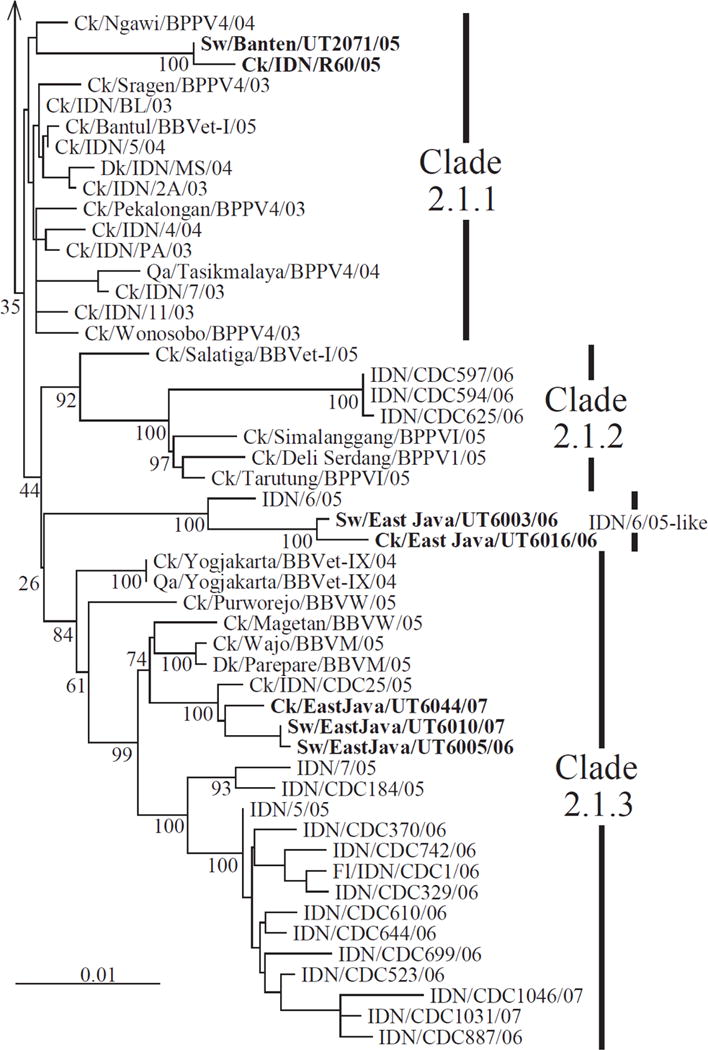
HA residues 281-1672 of Indonesian viruses were analyzed by neighbor-joining methods using Clustal W based on evolutionary validation. The tree was rooted to A/goose/Guangdong/1/96. Numbers below or above the branch nodes indicate neighbor-joining bootstrap values. Viruses isolated from pigs and their closest relatives isolated from domestic poultry are indicated by boldface. The sequence data used in this analysis have been submitted for publication and more detailed analyses were performed. Here we show an abbreviated phylogenetic tree to indicate the genetic relationship between the test viruses.
Here, to determine the pathogenicity of H5N1 pig isolates for mammalian hosts, we compared the virus growth property in mammalian cells and pathogenicity to mice of the swine isolates and the closest related H5N1 chicken isolates.
We analyzed two chicken and three swine isolates, i.e., A/chicken/East Java/UT6044/07, A/swine/East Java/UT6005/06, and A/swine/East Java/UT6010/07 for clade 2.1.3 and A/chicken/East Java/UT6016/06 and A/swine/East Java/UT6003/06 for the undetermined clade (A/Indonesia/6/05-like). Although A/chicken/Indonesia/R60/05 was most closely related to our swine isolate in clade 2.1.1, we were unable to obtain this virus and could, therefore, only analyze these five strains.
All of the H5N1 viruses were isolated in 10-day-old embryonated chicken eggs. All virus stocks were prepared from the allantoic fluid of infected eggs and stored at -80°C until use. The infectivity of stock viruses was determined in 10-day-old embryonated chicken eggs and Madin-Darby canine kidney (MDCK) cells according to standard procedures. The 50% egg infectious dose (EID50) and the 50% tissue culture infectious dose (TCID50) were calculated prior to mouse experiments by the Reed-Muench method [20]. To determine the 50% lethal dose for mice (MLD50), female five-week-old BALB/c mice (Japan SLC, Hamamatsu, Japan) were intranasally inoculated with 50μl of 10-fold serial dilutions containing doses ranging from 101 to 107 EID50 and monitored for survival over a 14-day period.
Growth properties in eggs and MDCK cells demonstrated that all viruses, whether from chickens or pigs, grew well in both eggs and MDCK cells, indicating that these swine H5N1 viruses grow as well as chicken viruses (Table 1). Among the avian influenza viruses isolated from humans, several have adapted to grow better in mammalian cells than avian isolates [21-23]. However, among the swine viruses isolated in our study, none adapted to grow substantially better in mammalian cells than chicken isolates.
Table 1.
Growth and pathogenicity comparison between avian and swine H5N1 viruses.
| Virus | HA classificationa | Virus growthb
|
MLD50 (log10EID50) |
|
|---|---|---|---|---|
| log10EID50/ml | log10TCID50/ml | |||
| A/chicken/East Java/UT6016/06 | A/Indonesia/6/05-like | 8.5 ± 0.5 | 8.8 ± 0.3 | 3.8 |
| A/swine/East Java/UT6003/06 | 9.6 ± 0.4 | 9.1 ± 0.3 | > 6.1 | |
|
| ||||
| A/chicken/East Java/UT6044/07 | 2.1.3 | 8.4 ± 0.3 | 8.6 ± 0.1 | 2.2 |
| A/swine/East Java/UT6005/06 | 9.4 ± 0.1 | 9.2 ± 0.4 | > 6.1 | |
| A/swine East Java/UT6010/07 | 9.5 ± 0.5 | 9.0 ± 0.4 | 3.5 | |
See the phylogenetic tree of Fig.1.
All data are the mean ± standard deviations from three independent experiments.
A comparison of pathogenicity in mice demonstrated that all of the swine H5N1 viruses were less virulent or attenuated to mice than the chicken viruses (Table 1); two swine strains (A/swine/East Java/UT6005/06 and A/swine/East Java/UT6003/06) were strongly attenuated in mice.
A genetic comparison between the chicken and swine strains identified several amino acid candidates potentially responsible for the attenuation in mice (Table 2). As for A/Indonesia/6/05-like viruses, there were twelve amino acid differences between the chicken and swine viruses. We were unable to determine which mutations were strongly correlated to low virulence in mice because these mutations are frequently found among avian and human H5N1 viruses. As for clade 2.1.3 viruses, A/swine/East Java/UT6005/06 had three amino acids that are thought to be responsible for attenuation in mice. Two amino acids in the PB2 protein, glutamine 13 and isoleucine 150, where mutations were found in Sw/UT6005, were highly conserved among the influenza A viruses, suggesting that these two mutations may be responsible for attenuation.
Table 2.
Amino acid differences between the proteins of swine and chicken strains.
| HA classification | A/Indonesia/6/05-like | HA classification | Clade 2.1.3 | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Virus | Ck UT6016 |
Sw UT6003 |
Virus | Ck UT6044 |
Sw UT6005 |
Sw UT6010 |
||
|
|
|
|||||||
| MLD50 (log10EID50)a | 3.8 | >6.1 | MLD50 (log10EID50) | 2.2 | >6.1 | 3.5 | ||
| Protein position | Protein position | |||||||
| PB2 | 526 | R | K | PB2 | 13c | Q | H | Q |
| PA | 231 | T | A | 150d | G | V | G | |
| 618 | A | T | 184 | T | A | A | ||
| 684 | G | E | 317 | L | M | M | ||
| HA1b | 36 | A | T | 365 | I | M | M | |
| 133 | A | S | 678 | G | D | D | ||
| NP | 50 | S | N | PB1-F2 | 4 | G | E | E |
| 318 | S | P | 47 | S | N | N | ||
| NA | 266 | S | G | PA | 367 | R | K | K |
| NS1 | 27 | M | L | 397 | K | E | E | |
| 74 | G | D | 547 | D | N | N | ||
| 180 | F | L | HA1 | 156 | A | T | T | |
| NA | 17 | I | V | V | ||||
| 41 | G | R | G | |||||
| 197 | K | R | R | |||||
| M2 | 23 | S | N | N | ||||
| 29 | A | V | V | |||||
| 68 | V | I | I | |||||
| NS1 | 124 | K | I | I | ||||
| 212 | N | S | S | |||||
| NS2 | 60 | I | V | V | ||||
| 92 | N | N | T | |||||
The same data as shown in Table 1.
H5 numbering
Gln at 13 position of the PB2 protein is widely conserved among H1N1, H3N2, and H5N1 viruses.
Gly-to-Val substitution at 150 position of the PB2 protein is a novel mutation among H1N1, H3N2 and H5N1 viruses.
Since our swine strains were isolated from pigs with no apparent influenza-like symptoms, the decrease of pathogenicity in mice suggests that the H5N1 viruses may have lost their pathogenicity in mammals during replication in pigs. Given that for the H5N1 viruses to cause a pandemic, they would likely become attenuated in humans, becoming attenuated in mammals may be a prelude toward the generation of a pandemic strain.
The fact that highly pathogenic H5N1 avian influenza viruses were transmitted to pigs on multiple occasions in Indonesia has afforded the opportunity for avian H5N1 viruses to adapt to growth in pigs asymptomatically. Pigs also serve as a source for the emergence of avian-human hybrid reassortant viruses [9, 24], increasing the risk of a pandemic strain surfacing. Given the gravity of this situation, it is clear from this study that the continuous surveillance and management of H5N1 viruses among pigs is a necessity.
Acknowledgments
We thank Susan Watson for editing the manuscript. This work was supported by a grant-in-aid for Specially Promoted Research and by a contract research fund for the Program of Funding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministries of Education, Culture, Sports, Science, and Technology and by grants-in-aid of Health, Labor, and Welfare of Japan, by ERATO (Japan Science and Technology Agency), and by the National Institute of Allergy and Infectious Diseases Public Health Service research grants.
Abbreviations
- HA
hemagglutinin
- MDCK
Madin-Darby canine kidney
- EID50
50% egg infective dose
- TCID50
50% Tissue culture infective dose
- MLD50
50% mouse lethal dose
- Ck
chicken
- Dk
duck
- Fl
feline
- Qa
quail
- IDN
Indonesia
- Sw
swine
- PB2
polymerase basic protein 2
References
- 1.Xu X, Subbarao K, Cox NJ, et al. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 2.Li KS, Guan Y, Wang J, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Smith GJD, Zhang SY, et al. H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Cumulative number of confirmed human cases of avian influenza A/ (H5N1) reported to WHO. 2008 Available at; http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_12_16/en/index.html. Accessed 24 Dec 2008.
- 5.OIE. Update on highly pathogenic avian influenza in animals (Type H5 and H7) 2006 Available at; http://www.oie.int/downld/AVIAN%20INFLUENZA/A2006_AI.php. Accessed 24 Dec 2008.
- 6.Scholtissek C, Burger H, Kistner O, et al. The nucleoprotein as a possible major factor in determing host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 7.Kida H, Ito T, Yasuda J, et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 8.Ito T, Couceiro JNSS, Kelm S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castrucci MR, Donatelli I, Sidoli L, et al. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 10.Glaser L, Stevens J, Zamarin D, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholtissek C, Rohde W, Von Hoyningen V, et al. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 13.Kendal AP, Goldfield M, Noble GR, et al. Identification and preliminary antigenic analysis of swine influenza-like viruses isolated during an influenza outbreak at Fort Dix, New Jersey. J Infect Dis. 1977;136:S381–S385. doi: 10.1093/infdis/136.supplement_3.s381. [DOI] [PubMed] [Google Scholar]
- 14.Rota PA, Rocha EP, Harmon MW, et al. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989;27:1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray GC, McCarthy T, Capuano AW, et al. Swine workers and swine influenza virus infections. Emerg Infect Dis. 2007;13:1871–1878. doi: 10.3201/eid1312.061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Reeth K. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet Res. 2007;38:243–260. doi: 10.1051/vetres:2006062. [DOI] [PubMed] [Google Scholar]
- 17.Choi YK, Nguyen TD, Ozaki H, et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol. 2005;79:10821–5. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipatov AS, Kwon YK, Sarmento LV, et al. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS Pathog. 2008;4:e1000102. doi: 10.1371/journal.ppat.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO/OIE/FAO H5N1 Evolution Working Group. Towards a unified nomenclature system for highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:e1. doi: 10.3201/eid1407.071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed LJ, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 21.Fiszon B, Hannoun C, Garcia-Sastre A, et al. Comparison of biological and physical properties of human and animal A (H1N1) influenza viruses. Res Virol. 1989;140:395–404. doi: 10.1016/s0923-2516(89)80118-9. [DOI] [PubMed] [Google Scholar]
- 22.Shinya K, Hamm S, Hatta M, et al. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 23.Hatta M, Hatta Y, Kim JH, et al. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claas EC, Kawaoka Y, de Jong JC, et al. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]


