Summary
Hepatocyte-like cells (HLCs) can be generated through directed differentiation or transdifferentiation. Employing two strategies, we generated induced pluripotent stem cell (iPSC)-HLCs and hiHeps from the same donor cell line. Both types of HLCs clustered distinctly from each other during gene expression profiling. In particular, differences existed in gene expression for phase II drug metabolism and lipid accumulation, underpinned by H3K27 acetylation status in iPSC-HLCs and hiHeps. While distinct phenotypes were achieved in vitro, both types of HLCs demonstrated similar phenotypes following transplantation into Fah-deficient mice. In conclusion, functional HLCs can be obtained from the same donor using two strategies. Global gene expression defined the differences between those populations in vitro. Importantly, both HLCs displayed partial but markedly improved hepatic function following transplantation in vivo, demonstrating plasticity and the potential for cell-based modeling in the dish and cell-based therapy in the future.
Keywords: transdifferentiation, directed differentiation, hepatocyte-like cells, gene expression pattern
Graphical Abstract
Highlights
-
•
hiHeps and iPSC-HLCs from the same donor are compared
-
•
hiHeps and iPSC-HLCs show distinct expression patterns and hepatic functions
-
•
Different expressions in hiHeps and iPSC-HLCs are partially attributed to H3K27ac
-
•
Both HLCs are further matured in the in vivo microenvironment of livers
In this article, Hui and colleagues show that hiHeps and iPSC-HLCs generated from the same donor display different gene expression patterns that correlate with their hepatic functions. Distinct H3K27ac modifications partially explain the functional differences between the two types of HLCs. Importantly, both HLCs show improved hepatic gene expression after repopulation in murine livers.
Introduction
In order to overcome the limitations of using human hepatocytes from donor liver organs, methods to derive hepatocyte-like cells (HLCs) from other cells have been studied intensively (Forbes et al., 2015, Rezvani et al., 2016). To date, many different types of HLCs have been successfully generated from human pluripotent stem cells (hPSCs), with some derivative HLCs exhibiting respectable human drug metabolism and liver repopulation in vivo (Rezvani et al., 2016). hPSC-HLCs are typically obtained from hPSCs in a stage-wise process (Gerbal-Chaloin et al., 2014, Hay et al., 2008, Si-Tayeb et al., 2010, Sullivan et al., 2010, Touboul et al., 2010), whereas hiHeps are obtained by directing cellular transdifferentiation from human fibroblasts, or other cell types, by the forced expression of specific hepatocyte transcription factors (Du et al., 2014, Huang et al., 2014). To understand the advantages of both systems, a systemic comparison between induced pluripotent stem cell (iPSC)-HLCs and hiHeps is necessary to realize their translational value and understand the basic mechanisms that underpin hepatic differentiation and liver organogenesis (Forbes et al., 2015). While studies have been performed in PSCs, derived from the inner cell mass of nuclear transfer embryos, and iPSCs (Ma et al., 2014), a systematic study comparing iPSC-HLCs and hiHeps from the same donor has not been performed.
PSC-HLCs generated by different protocols were compared in a recent study (Godoy et al., 2015). On the basis of gene expression, gene networks were established to predict for successful or failed hepatocyte differentiation. In these studies, HNF1, FXR, and PXR were highlighted as key transcription factors required to improve HLC differentiation. In a similar approach, we have performed direct comparison of iPSC-HLC and hiHep gene expression and function in vitro and in vivo. This is of the utmost importance as iPSC-HLCs and hiHeps demonstrate significant potential in the quest to accurately model human disease and develop immune-matched cell-based therapies for the clinic (Bhatia et al., 2014, Fox et al., 2014). More specifically, our studies provide important information on the mechanisms that underlie cell identity changes during the process of liver differentiation and regeneration (Szkolnicka and Hay, 2016). Umbilical cord-derived fibroblasts (UCFs) were used in these studies. They were isolated from the same donor prior to reprogramming and directed differentiation or transdifferentiation, thereby removing the influence of genetic variation observed in the population (Kajiwara et al., 2012). In summary, our studies focused on global gene expression and epigenetic remodeling to better understand the circuitry that underpins successful hepatic specification and cell function in vitro and in vivo.
Results
Generation of Functional iPSC-HLCs and hiHeps from the Same Donor
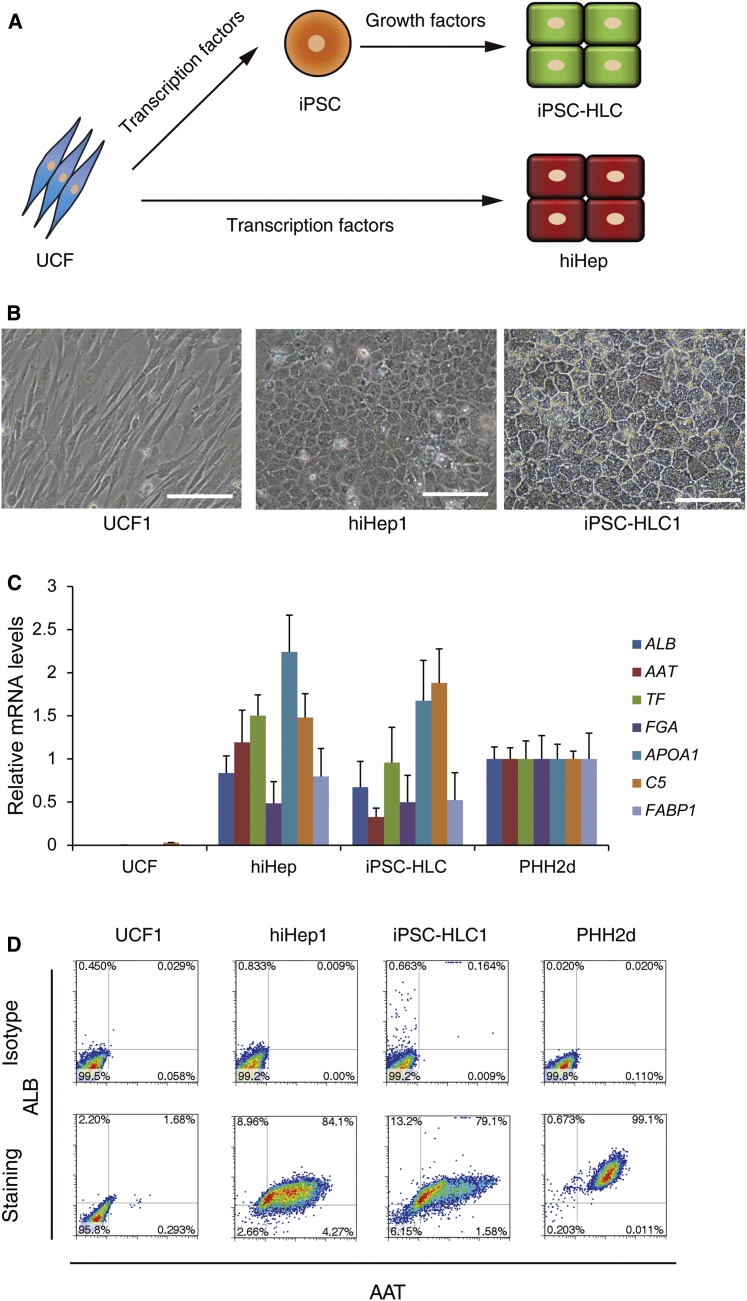
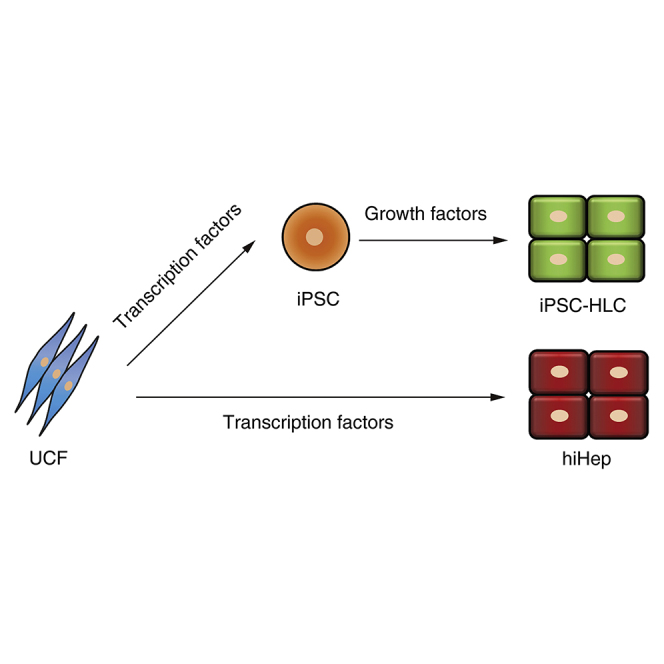
Human umbilical cord fibroblasts, UCF1 and UCF2, were generated from two individuals (Figures S1A and S1B). The UCF donor cells were reprogrammed to iPSCs using standard methodology (Takahashi et al., 2007). The derivative iPSC lines were termed iPSC1 (derived from UCF1) and iPSC2 (derived from UCF2). Both lines expressed pluripotent stem cell markers (NANOG and SSEA4) and were positive for alkaline phosphatase (Figures S2A–S2D). In association with increased OCT4 expression, the promoter of OCT4 was demethylated (Figure S2E). After transplantation into the immune-deficient mice, both iPSC lines formed teratomas comprising tissues derived from the three germ layers (Figure S2F). Taken together, these results confirm that we produced two bona fide iPSC lines that could be maintained with normal karyotype for more than 40 passages (Figure S2G).
Both iPSC cells were differentiated into HLCs following a published protocol (Szkolnicka et al., 2014). We also transdifferentiated UCF1 and UCF2 into hiHep using FOXA3, HNF1A, and HNF4A as previously published (Huang et al., 2014) (Figure 1A). To confirm cell identity, hiHeps and iPSC-HLCs were validated to be genetically identical with the parental lines by short tandem repeat typing (Table S1). Morphologically, both hiHeps and iPSC-HLCs displayed typical epithelial phenotype, forming tight junctions, and canaliculi monolayers became confluent (Figure 1B). Interestingly, the diameter of the iPSC-HLCs was approximately 25% larger than that of hiHeps (12.6 μm in hiHeps versus 15.8 μm in iPSC-HLCs). A more detailed analysis demonstrated that the expression levels of typical hepatic markers were comparable between hiHeps and iPSC-HLCs, and those approached the levels detected in primary human hepatocytes (PHHs) as determined by qPCR (Figure 1C). Hepatocellular specification was also monitored by flow cytometry, and around 80% hiHeps and iPSC-HLCs co-expressed ALBUMIN and α-1-antitrypsin (AAT) (Figure 1D). The expression and secretion of ALBUMIN and AAT were further confirmed by ELISA, using supernatants from iPSC-HLCs and hiHeps. Of note, both proteins were detected at levels comparable with that in PHH cultures (Figure S3A). These data together indicate that iPSC-HLC and hiHep cells were homogeneous populations displaying typical hepatocyte features.
Figure 1.
Generation of Hepatocyte-like Cells (HLCs) by Different Strategies
(A) Schematic diagram of the generation of HLCs by different strategies.
(B) Typical morphology of UCF, hiHep, and iPSC-HLC. hiHep1 and iPSC-HLC1 were derived from UCF1. Scale bar, 100 μm.
(C) Hepatic gene expression levels of HLCs were measured by qPCR. UCF included two independent replicates, UCF1 and UCF2; hiHep included four replicates from independent experiments (hiHep1, hiHep2, hiHep3, and hiHep4); iPSC-HLC included four replicates from independent experiments (iPSC-HLC1, iPSC-HLC2, iPSC-HLC3, and iPSC-HLC4); PHH included two independent replicates that were cultured for 2 days.
(D) Both hiHeps and iPSC-HLCs displayed a high percentage of ALB and AAT double-positive cells, as measured by flow cytometry. UCFs were used as negative control and PHHs cultured for 2 days were used as positive control.
See also Figures S1 and S2 and Table S1.
Differential Hepatocyte Gene Expressions in iPSC-HLCs and hiHeps
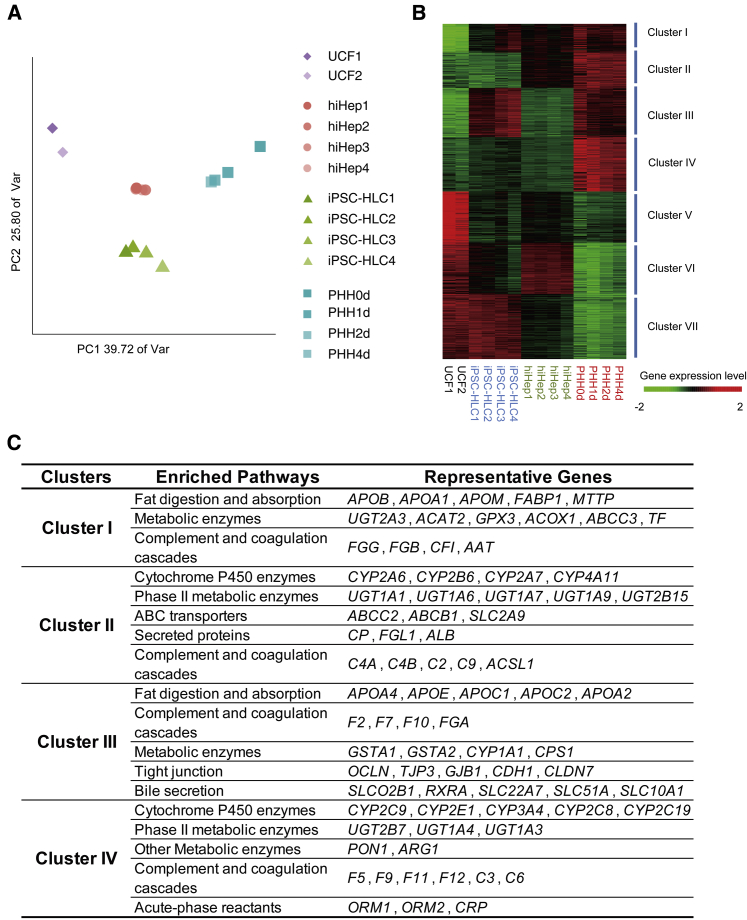
Following our initial characterization, we preformed genome-wide profiling of iPSC-HLCs and hiHeps and compared their gene expression (Table S2) with UCFs and PHHs controls. The top 4,000 most variably expressed genes between UCFs and PHHs that cultured for 1, 2, and 4 days were selected for further analysis. Whole-genome analysis using principal component analysis (PCA) confirmed that iPSC-HLCs, hiHeps, UCFs, and PHHs were clustered into distinct groups (Figure 2A).
Figure 2.
Transcriptome Analysis of hiHeps and iPSC-HLCs
(A) Principal component analysis (PCA) of four cell types using 4,000 genes with highest variance in UCFs and PHHs cultured for 1, 2, and 4 days. The percentages on the axes represent the variance explained by the respective axes. hiHep1 and hiHep2 were derived from UCF1, hiHep3 and hiHep4 were derived from UCF2; iPSC-HLC1 and iPSC-HLC2 were derived from iPSC1, iPSC-HLC3 and iPSC-HLC4 were derived from iPSC2. PHHs were fresh, or cultured for 1, 2, and 4 days.
(B) Hierarchical clustering of UCFs, hiHeps, iPSC-HLCs, and PHHs using 4,000 genes with highest variance in UCFs and PHHs cultured for 1, 2, and 4 days. The samples are the same as (A).
(C) Enriched pathways and representative genes in different cluster groups are summarized.
Following this, we focused our analyses on differential gene expression between iPSC-HLCs and hiHeps. Unsupervised clustering of hepatic genes was performed, following with gene ontology enrichment analysis (Figures 2B and 2C). Based on the different gene expression patterns of hiHeps, iPSC-HLCs, and PHHs, the highly expressed hepatic genes could be divided into seven clusters. Genes involving fat digestion and absorption (e.g., FABP1, APOB) and metabolism enzymes (e.g., GPX3 and ACOX1) were enriched in cluster I; their expression was induced in both hiHeps and iPSC-HLCs. There were some hepatic genes showing low expression in both iPSC-HLCs and hiHeps (cluster IV), including cytochrome P450-based metabolism genes (e.g., CYP2C9, CYP2E1, and CYP3A4) and coagulation complements (e.g., F5, F9, and F11).
Genes in cluster II were highly expressed in hiHeps but not in iPSC-HLCs. For example, the phase II metabolic enzymes (e.g., UGT1A1 and UGT1A6) and ABC transporters (e.g., ABCC2 and ABCB1) were highly enriched in cluster II genes. The expression of genes in cluster III was induced in iPSC-HLCs, and included fat digestion and absorption genes (e.g., APOA2 and FASN) and bile secretion genes (e.g., OATPB and NTCP) (Figure 2C). Moreover, in clusters V, VI, and VII, the typical fibrotic genes and pathways were downregulated in HLCs (Figure S4A). Taken together, these studies highlighted differential gene expression between iPSC-HLCs and hiHeps.
Comparison of hiHep and iPSC-HLC Function In Vitro
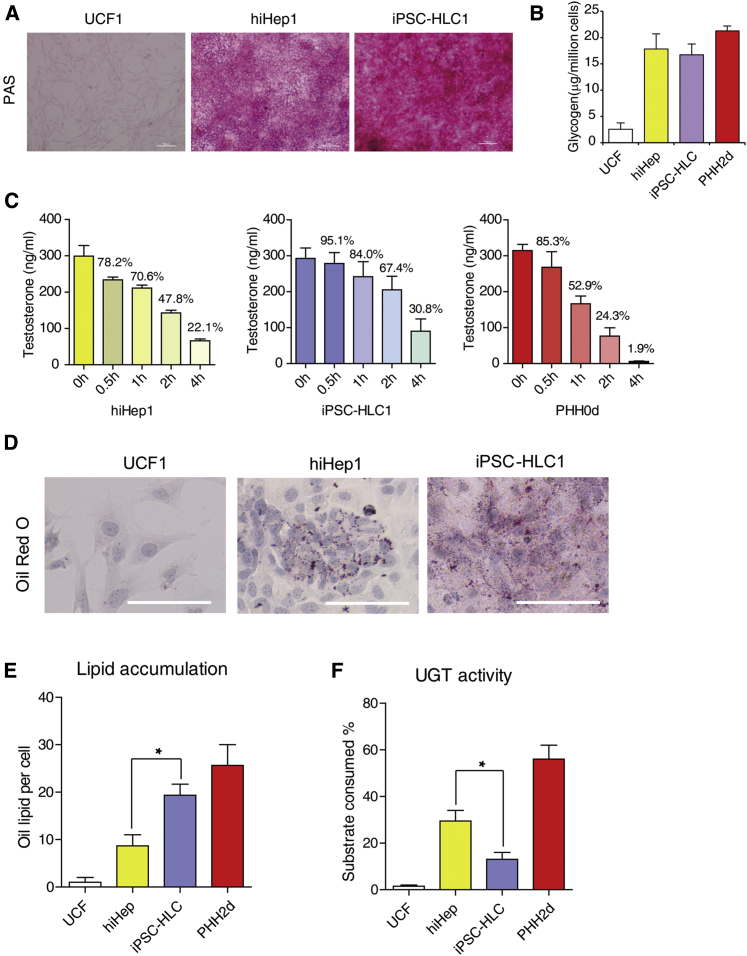
Whole-genome analyses of expression profiles elucidated differences between iPSC-HLCs and hiHeps. To understand its functional implications, we measured several key hepatic functions in both cell types. As highlighted from gene expression analyses, both cells carried comparable levels of glycogen storage abilities as determined by periodic acid-Schiff (PAS) staining and colorimetric assay (Figures 3A and 3B).
Figure 3.
Functional Characterizations of hiHeps and iPSC-HLCs
(A) Glycogen storage in different HLCs was measured by periodic acid-Schiff (PAS) staining. Scale bar, 100 μm.
(B) Glycogen storage in HLCs was determined quantitatively by colorimetric measurement (Abnova). UCF included two independent replicates, UCF1 and UCF2; hiHep included four replicates from independent experiments (hiHep1, hiHep2, hiHep3, and hiHep4); iPSC-HLC included four replicates from independent experiments (iPSC-HLC1, iPSC-HLC2, iPSC-HLC3, and iPSC-HLC4). PHH included two replicates cultured for 2 days from independent experiments.
(C) HLCs both eliminated testosterone as efficiently as PHHs. Concentrations of testosterone were determined by liquid chromatography-tandem mass spectrometry. Each time point had three replicates from independent experiments.
(D) iPSC-HLCs showed more lipid accumulation than hiHep as measured by oil red O staining. Scale bar, 100 μm.
(E) Lipid accumulation was quantified by oil lipid numbers per cell. UCF had two independent replicates, UCF1 and UCF2; hiHep had four replicates from independent experiments (hiHep1, hiHep2, hiHep3, and hiHep4); iPSC-HLC had four replicates from independent experiments (iPSC-HLC1, iPSC-HLC2, iPSC-HLC3, and iPSC-HLC4). PHH included two replicates cultured for 2 days from independent experiments. There was a significant difference between hiHep and iPSC-HLC. ∗p < 0.05.
(F) UGT activities of hiHeps and iPSC-HLCs were determined by the luminescence of remaining substrates. The combination of replicates is the same as in (E). There was a significant difference between hiHep and iPSC-HLC. ∗p < 0.05.
See also Figure S3.
To investigate phase I cytochrome P450 (CYP) induction, we treated hiHeps and iPSC-HLCs with 3-methylcholanthrene, phenobarbital, and rifampicin. hiHeps and iPSC-HLCs also possessed remarkable capacities to respond to these chemicals (Figures S3B and S3C). We specifically measured testosterone elimination as an indication of CYP3A function in iPSC-HLCs and hiHeps. Both cells could eliminate testosterone to ∼20%–30% of the original level within 4 hr (Figure 3C). This was approximately 10- to 15-fold less than PHHs. We next analyzed the biliary excretion capabilities. Both hiHeps and iPSC-HLCs expressed biliary excretion-related transporters (Figure S3D) and showed a high biliary excretion index (BEI), effluxing substrates cholyl-lysyl-fluorescein (CLF), D8-taurocholic acid (D8-TCA), and rosuvastatin at similar levels to PHH (Figure S3E). These results suggested that hiHeps and iPSC-HLCs possessed multiple hepatic functions at comparable levels.
Despite the above similarity between iPSC-HLCs and hiHeps, expression profile analysis predicted that iPSC-HLCs had faster lipid metabolism than hiHeps, whereas hiHeps might perform better in phase II metabolism. Indeed, lipid formation and accumulation in iPSC-HLCs was similar to that in PHHs and almost double than that in hiHeps (Figures 3D and 3E). In accordance with the transcriptomic datasets, we observed that hiHeps possessed greater UGT activity than iPSC-HLCs (Figure 3F).
Understanding iPSC-HLCs and hiHeps Cell Identity In Vitro
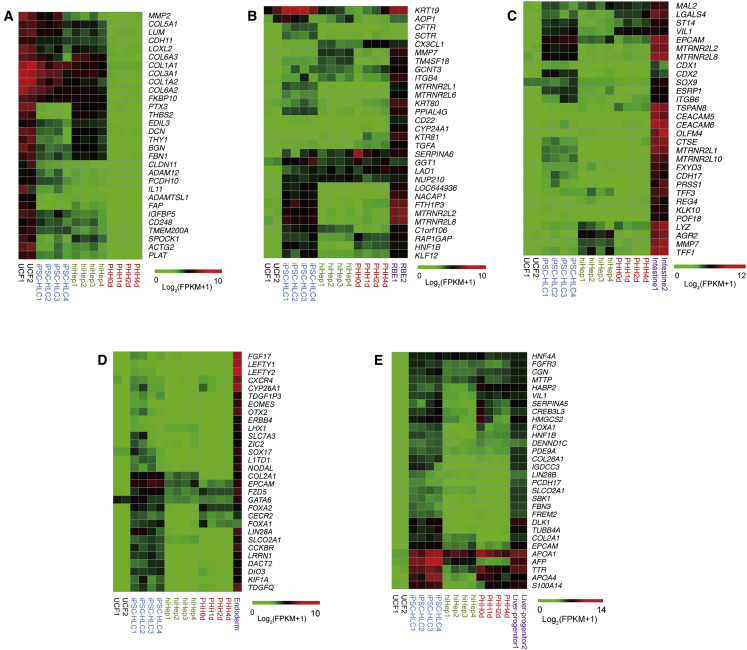
Next, we asked whether iPSC-HLCs and hiHeps retain molecular traces of their induction processes and whether those molecular traces are important components of hiHep and iPSC-HLC cell identity. We first characterized the elimination of fibroblast-specific gene expression in iPSC-HLCs and hiHeps. Markedly, fibroblast-related genes, selected according to published data (Buganim et al., 2012, Huang et al., 2014), were significantly extinguished in both iPSC-HLCs and hiHeps. Those included COL1A1, COL1A2, MMP14, and LOXL2 (Figures 4A, S4B, and S4C). However, some fibrotic genes were expressed at low levels in both iPSC-HLCs and hiHep cells (Figure 4A). These data suggest that the original fibroblast identity was efficiently but not fully erased in both hiHeps and iPSC-HLCs.
Figure 4.
Different Strategies Generate HLCs with Different Gene Expression Patterns
(A) A small amount of fibrotic genes remained after hiHep direct transdifferentiation from fibroblasts. hiHep1 and hiHep2 were derived from UCF1, hiHep3 and hiHep4 were derived from UCF2; iPSC-HLC1 and iPSC-HLC2 were derived from iPSC1, iPSC-HLC3 and iPSC-HLC4 were derived from iPSC2. PHHs were fresh, or cultured for 1, 2, and 4 days.
(B and C) hiHeps did not express cholangiocyte (B) and intestine (C) lineage-specific genes. The combination of replicates is the same as in (A).
(D and E) iPSC-HLCs remained definitive in endoderm (D) and liver progenitor (E) specific gene expression. The combination of replicates is the same as (A).
See also Figures S4 and S5 and Table S2.
We next investigated whether iPSC-HLCs and hiHeps retain any molecular traces related to transdifferentiation and differentiation. We first analyzed marker genes of bile duct cells, which share common progenitors with hepatocytes. Expression pattern analysis did not show significant bile duct gene expression in both hiHeps and iPSC-HLCs (Figure 4B). Following this, we analyzed for the expression of colon-specific genes (Forster et al., 2014). Notably, the expression of colon-specific genes was undetectable in hiHeps, indicating the specificity of the applied hepatic transdifferentiation protocol (Figure 4C). Interestingly, CDX2 was detected at low levels in iPSC-HLCs, indicating that low-level colon signatures persist in vitro (Figure S5C).
Following these studies, we characterized whether iPSC-HLCs retain molecular traces of endoderm progenitors (Cheng et al., 2012, Loh et al., 2014) and hepatoblasts (Yu et al., 2013). Intriguingly, iPSC-HLCs expressed several marker genes for endoderm progenitors (including FOXA2 and GATA6), and hepatoblasts (including AFP and EPCAM) (Figures 4D, 4E, and S4D). In contrast, these progenitor marker genes were undetectable in hiHeps, while the iPSC-HLCs expressed less ALBUMIN and TAT mRNA than the hiHep as shown in RNA-seq analysis (Figure S4D). The expression of progenitor marker genes was validated by q-PCR in iPSC-HLCs (Figure S5A). We performed co-staining for ALB (a marker for mature hepatocytes) and AFP (a marker for immature hepatocytes) in HLCs (Figure S5B). It was striking that almost all iPSC-HLCs were double positive for ALB and AFP, whereas no AFP-positive cells were detectable in hiHeps, suggesting that iPSC-HLCs retained some progenitor traces during the differentiation process.
Active Histone Modifications Are Detected during hiHeps and iPSC-HLCs
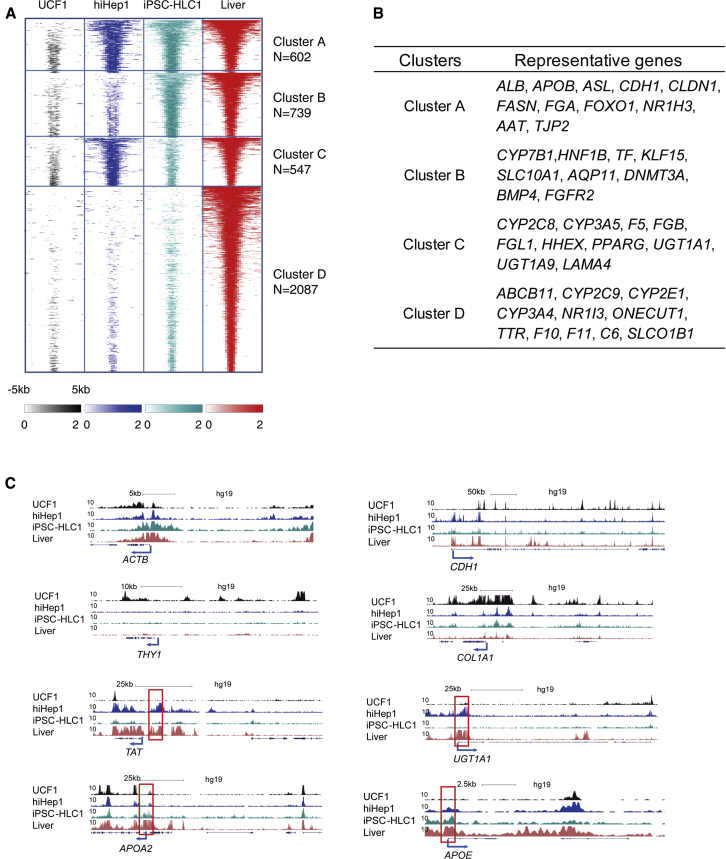
The characteristics of a differentiated cell are stably maintained by histone modifications (Holmberg and Perlmann, 2012). In our experiments, we chose to analyze an active pattern of histone modification, H3K27 acetylation (H3K27ac) at active enhancer and promoter regions in both iPSC-HLCs and hiHeps. Furthermore, UCFs and fresh human adult liver were used as negative and positive controls (Bonn et al., 2012, Creyghton et al., 2010) (Table S2). The genome-wide analysis of H3K27ac occupancy exhibited different patterns between the samples (Figures 5A and S6A) and was divided into eight different clusters. Cluster A included genes that have similar H3K27ac modifications in both HLCs and liver, including ALB, AAT, APOB, and CDH1 (Figure 5B). In cluster H, most H3K27ac modifications identified on fibrotic genes were erased in HLCs and liver (Figures S6A and S6B). Cluster D indicated hepatic genes that were not fully modified by H3K27ac in HLCs (Figure 5B). Specifically, the enhancers and promoters near THY1 and COL1A1 were enriched with H3K27ac in UCFs but erased in hiHeps and iPSC-HLCs (Figure 5C). In contrast, enhancer regions near CDH1, TAT, UGT1A1, APOA2, and APOE were enriched in H3K27ac in hiHep and iPSC-HLCs when compared with UCFs (Figure 5C). This finding not only demonstrated that both hiHeps and iPSC-HLCs gained stable epigenetic modifications but also proved that hiHeps and iPSC-HLCs had active hepatic gene expression. In accordance with previous analysis, there were differences between iPSC-HLCs and hiHeps with differential acetylation observed at enhancers near TAT and UGT1A1 in hiHep, whereas in iPSC-HLCs, enhancers near APOA2 and APOE were more active (Figure 5C).
Figure 5.
Characterization of H3K27ac Occupancy in Different Cell Types
(A) Distribution of the H3K27ac signals in the four samples. Genes that have H3K27ac in liver but not in UCF1 are shown.
(B) Gene Ontology analysis for the genes associated with H3K27ac peaks in different cluster groups in (A).
(C) Representative tracks of H3K27ac occupancy at housekeeping gene (ACTB), fibroblast-specific genes (THY1 and COL1A1), hepatic specific genes (CDH1, TAT, UGT1A1, APOA2, APOE) across the four cell types. Red boxes mark the different H3K27ac peaks between hiHeps and iPSC-HLCs.
Improvements in Hepatocyte Phenotype Were Detected Following hiHep and iPSC-HLC Transplant In Vivo
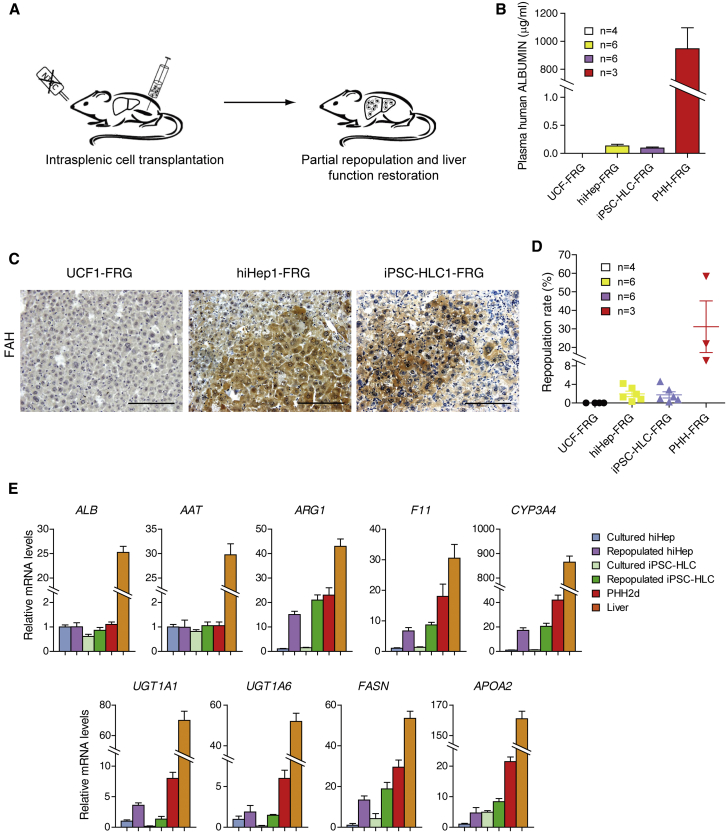
The analysis of histone active markers partially explained the similarities and differences between iPSC-HLCs and hiHeps. Given the plasticity of histone modifications, it is possible to further improve the hepatic features of iPSC-HLCs and hiHeps if a niche is supplied. We decided to transplant these cells into the liver, which may represent the best microenvironment for hepatocytes. hiHeps and iPSC-HLCs were transplanted into Fah−/−Rag2−/−Il2rg−/− (FRG) mice via splenic injection. FRG mice were kept alive with a supply of 2-(2-nitro-4-trifluoro-methylbenzyol)-1,3-cyclohexanedione (NTBC). After NTBC withdrawal, FRG mice usually die of metabolic liver disease within 4–6 weeks (Azuma et al., 2007). We transplanted five million hiHeps and iPSC-HLCs into FRG mice and analyzed liver repopulation after transplantation (Figure 6A). FRG mice transplanted with UCF and PHH were used as controls. After 2 months, human ALBUMIN levels were at comparable levels in the sera of FRG mice transplanted with hiHeps (153 ± 42 ng/mL) and iPSC-HLCs (114 ± 50 ng/mL) but were low compared with PHHs (0.91 ± 0.16 mg/mL) (Figure 6B). Immunohistochemical staining for human FAH showed that repopulation efficiencies were similar for hiHeps and iPSC-HLCs with contributions ranging from 0.3% to 4.2% (1.95% ± 1.49%) and 0.2%–4.6% (1.75% ± 1.65%) respectively, which were much lower than that of PHHs (Figures 6C, 6D, and S6C). Even though HLCs could repopulate into the livers of FRG mice, they did not secret human ALBUMIN as efficiently as PHHs, which suggested that they were not mature enough for hepatic function.
Figure 6.
In Vivo Characterization and Maturation of hiHeps and iPSC-HLCs after Repopulation
(A) Schematic outline of HLC transplantation into the livers of Fah−/−/Rag2−/−/Il2rg−/− mice (FRG). Each mouse was transplanted intrasplenically with 5 million HLCs.
(B) Human ALBUMIN levels in the sera of surviving mice from independent experiments were determined by ELISA. UCF-FRG included four mice transplanted with two UCF1 and two UCF2; hiHep-FRG included six mice transplanted with two hiHep1, one hiHep2, two hiHep3, and one hiHep4; iPSC-HLC-FRG included six mice transplanted with two iPSC-HLC1, one iPSC-HLC2, two iPSC-HLC3, and one iPSC-HLC4; PHH-FRG included three mice transplanted with fresh PHHs.
(C) The repopulation of HLCs in FRG mouse livers was determined by immunostaining for FAH. Scale bar, 100 μm.
(D) Repopulation rates of HLCs in the liver of FRG mice were calculated. The combination of replicates is the same as in (B).
(E) FAH-positive hiHeps and iPSC-HLCs were collected by microdissection from serial liver sections. The mRNA levels of indicated genes were measured in repopulated hiHeps and iPSC-HLCs, cultured hiHeps and iPSC-HLCs, PHH and liver by qPCR. Data are normalized to cultured hiHeps. Cultured hiHeps included four replicates from independent experiments (hiHep1, hiHep2, hiHep3. and hiHep4). Repopulated hiHeps included two independent replicates from hiHep1-FRG and two independent replicates from hiHep3-FRG. Cultured iPSC-HLCs included four replicates from independent experiments (iPSC-HLC1, iPSC-HLC2, iPSC-HLC3, and iPSC-HLC4). Repopulated iPSC-HLCs included two independent replicates from iPSC-HLC1-FRG and two independent replicates from iPSC-HLC3-FRG.
See also Figure S6.
To evaluate whether hiHeps and iPSC-HLCs were further matured following exposure to the in vivo niche, we micro-dissected repopulated nodules from liver sections. Expression of hepatocyte genes was determined in hiHeps and iPSC-HLCs by qPCR using human-specific primers. Compared with cultured hiHeps and iPSC-HLCs, the expression levels of several genes were significantly increased after repopulation, including CYP3A4 and F11 (Figure 6E). In addition, we characterized the mRNA levels of lipid metabolism and phase II metabolism-related genes. Markedly, the expression differences discovered in cultured HLCs were normalized in repopulated hiHeps and iPSC-HLCs, suggesting that the in vivo microenvironment improved hepatic gene expression. Taken together, our results demonstrate that iPSC-HLCs and hiHeps have the potential to be further matured if the niche they are maintained in is supportive.
Discussion
To rule out donor variability, iPSC-HLCs and hiHeps were generated from the same parental cells (Kajiwara et al., 2012, Ma et al., 2014). In general, both types of HLCs were comparable, with gene expression patterns similar to PHHs, but with obvious differences. In vitro functional analysis demonstrated that iPSC-HLCs and hiHeps were comparable, with HLCs demonstrating glycogen storage, ALBUMIN, alpha-1-antitrypsin, and biliary excretion. Notably, when iPSC-HLCs and hiHeps were transplanted into mouse livers, they repopulated the liver at comparable levels and demonstrated comparable gene expression. Taken together, these findings demonstrate that both directed differentiation and transdifferentiation successfully instruct hepatic differentiation, cell engraftment in vivo, and similar maturation.
While similarities were observed, differences between hiHeps and iPSC-HLCs existed in vitro. Comparison of global gene expression profiles highlighted that iPSC-HLCs express several endoderm progenitor- and hepatoblast-related marker genes as previously reported (Carpentier et al., 2014, Godoy et al., 2015). The induction of iPSC-HLCs was achieved using an affordable and facile three-staged protocol mimicking certain aspects of human hepatocyte development. The in vitro differentiation duration is significantly shorter than human development in utero, therefore residual molecular traces left during iPSC-HLC formation could have restricted cell maturation. In addition, the cell niche, and in particular the extracellular matrix, has been shown to be an important driver of HLC differentiation, and therefore these areas require further investigation (Cameron et al., 2015).
It was previously reported that mouse iHep cells induced by Foxa3 and Hnf4a express the colon epithelium-specific marker gene Cdx2, suggesting that transdifferentiation might detour cells into unwanted lineages if not properly controlled (Morris et al., 2014). However, we did not observe such colon-specific gene expression in our hiHeps. In our studies, the functional assays further validated those findings from gene expression analyses. These data suggest that both iPSC-HLCs and hiHeps have advantages in modeling different aspects of human liver biology “in a dish.” Specifically, iPSC-HLCs may be a better model to study fatty liver diseases, whereas hiHeps could be the choice for in vitro phase II drug metabolism. HLCs were compared with PHHs with and without culture in this study. While cultured hepatocytes do not completely reflect the nature of primary hepatocytes in vivo, they might be sufficient for these types of comparisons if one wishes to normalize the effect caused by in vitro culture.
Although functional differences were observed between the HLCs, cell repopulation of immune-deficient Fah−/− mice using iPSC-HLCs or hiHeps was comparable. Importantly, this translated into improvements in liver physiology and murine survival rates providing promise for the clinic in the future. Importantly, iPSC-HLC or hiHep plasticity was observed in vivo, indicating that HLCs responded to their environment, leading to improvements in cell phenotype. While these studies demonstrate significant advances, it should be noted that iPSC-HLCs and hiHeps were still not as efficient at engrafting as PHHs in FRG mice. This highlights the need to improve hepatic maturation and/or liver preconditioning, prior to cell transplantation in the future (Fisher and Strom, 2006, Grompe and Strom, 2013, Yang et al., 2017).
Our findings not only suggest potential applications for iPSC-HLCs and hiHeps but also provide insight about the process of differentiation and transdifferentiation. The identification of differentiated cells is mainly determined by regulation of tissue-specific transcription factors and epigenetic modifications (Holmberg and Perlmann, 2012). In support of this, we demonstrate that HLCs generated from both transdifferentiation and directed differentiation had stable epigenetic modifications, which had an impact on their function in vitro. These studies also confirm that both hiHeps and iPSC-HLCs are partially differentiated in 2D culture in vitro. Going forward, it will be necessary to identify new potential combinations of transcription and growth factors to improve transdifferentiation and directed differentiation. In addition, new strategies such as 3D differentiation (Rashidi et al., 2016, Takebe et al., 2013) and alternative substrates may improve the maturation of both types of HLCs (Cameron et al., 2015, Lim et al., 2016).
Despite the divergences between HLCs and PHHs, HLCs have been already proved useful for in vitro disease modeling and cell-based therapies (Forbes et al., 2015, Shi et al., 2016). Going forward, it is critical to unveil the differences that exist between HLCs and PHHs to improve cell phenotype further. This will lead to improved models for the lab and cells for the clinic in the future (Nicolas et al., 2017).
Experimental Procedures
Cell Culture and Generation of Hepatocyte-like Cells
UCFs were cultured in human fibroblast medium. hiHeps were generated from immortalized UCFs with transduction of FOXA3, HNF1A, and HNF4A, and cultured in hepatocyte-maintaining medium as previously reported (Huang et al., 2014). Human iPSCs were generated from UCFs by transduction of retrovirus (OCT4, SOX2, KLF4, and C-MYC). iPSC-HLCs were generated from iPSCs through a three-step method as previously reported (Szkolnicka et al., 2014).
Primary Hepatocyte Culture
PHHs from three individuals were purchased from Celsis In Vitro Technologies. PHHs were pooled together in the same number and plated at a density of 1.25 × 105/cm2. For the testosterone clearance assay, we used freshly thawed PHHs. For the other assays, the days of culture of PHHs are indicated. Institutional ethical committees approved the collection and use of human samples.
Mice
FRG mice were maintained with NTBC water at a concentration of 16 mg/L. NTBC water was withdrawn 1 week before the transplantation of HLCs. hiHeps and iPSC-HLCs (5 × 106 cells/animal) were injected into the spleens of the mice. All mouse experiments were approved by the Institutional Animal Care and Use Committee of the Institute of Biochemistry and Cell Biology and performed in accordance with institutional guidelines.
RNA-Seq Analysis
Total RNA was isolated from UCFs, hiHeps, iPSC-HLCs, and PHHs by Trizol. RNA-seq libraries were prepared with the Illumina TruSeq RNA Sample Preparation Kit. The fragmented and randomly primed 100 bp paired-end libraries were sequenced on an Illumina HiSeq 2000 sequencing system.
ChIP-Seq Analysis
Chromatin immunoprecipitation (ChIP) was performed using antibodies for acetylation of histone H3 lysine 27 acetylation (H3K27ac, Abcam Ab4729). DNA libraries of matched input and ChIP samples were prepared using the Illumina Truseq kit, indexed for multiplexed runs of four libraries per lane, and sequenced on an Illumina HiSeq2000 instrument.
Statistical Analysis
Data were subjected to Student’s t test. p < 0.05 was considered statistically significant. Data are presented as means ± SD.
Author Contributions
L.H. and G.W. conceived and supervised the study. Y.G. performed most of the experiments. X.Z. analyzed the RNA-seq and ChIP-seq data. L.Z. and J.C. performed the in vivo characterization of hiHeps and iPSC-HLCs. X.N., X.L., and G.P. characterized and analyzed the CYP metabolism and drug toxicity. Y.L. performed the H3K27ac ChIP experiment. D.C.H. assisted in the generation and characterization of iPSC-HLCs and manuscript writing. D.L., C.Y., Z.Z., Y.S., X. Chen, and X.Cheng helped with the preparation of UCF and characterization of human iPSCs. Y.G., X.Z., G.W., and L.H. analyzed the data and wrote the paper with suggestions from the other authors.
Acknowledgments
We are thankful to Dr. Yunwen Zheng (University of Tsukuba, Japan) for his suggestions on the experiments. This study is supported by CAS- CSIRO (153D31KYSB20160247), CAS-NWO Programme (153D31KYSB20150085), the National Natural Science Foundation of China (NSFC; 31630044, 31601186, 81471948, 31771431, 31271354, and 31401263), the Ministry of Science and Technology of China (MOST; 2013CB967103), the Science and Technology Commission of Shanghai Municipality (STCSM; 16JC1400202 and 15JC1400200), and Shanghai Zhangjiang National Independent Innovation Demonstration Zone Stem Cells Translational Medicine Industry Base Project (ZJ2014-ZD-002). D.C.H. was supported by the UK Regenerative Medicine Platform award (MR/L022974/1).
Published: November 22, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.10.019.
Contributor Information
Gang Wei, Email: weigang@picb.ac.cn.
Lijian Hui, Email: huilab@sibcb.ac.cn.
Accession Numbers
Original data are available in the NCBI Gene Expression Omnibus under accession number GEO: GSE103078.
Supplemental Information
References
- Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E., Strom S., Kay M.A., Finegold M., Grompe M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat. Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.N., Underhill G.H., Zaret K.S., Fox I.J. Cell and tissue engineering for liver disease. Sci. Transl. Med. 2014;6:245sr2. doi: 10.1126/scitranslmed.3005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczynski B., Riddell A., Furlong E.E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Buganim Y., Itskovich E., Hu Y.C., Cheng A.W., Ganz K., Sarkar S., Fu D.D., Welstead G.G., Page D.C., Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron K., Tan R., Schmidt-Heck W., Campos G., Lyall M.J., Wang Y., Lucendo-Villarin B., Szkolnicka D., Bates N., Kimber S.J. Recombinant laminins drive the differentiation and self-organization of hESC-derived hepatocytes. Stem Cell Reports. 2015;5:1250–1262. doi: 10.1016/j.stemcr.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier A., Tesfaye A., Chu V., Nimgaonkar I., Zhang F., Lee S.B., Thorgeirsson S.S., Feinstone S.M., Liang T.J. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J. Clin. Invest. 2014;124:4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Ying L., Lu L., Galvao A.M., Mills J.A., Lin H.C., Kotton D.N., Shen S.S., Nostro M.C., Choi J.K. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell. 2012;10:371–384. doi: 10.1016/j.stem.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Fisher R.A., Strom S.C. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82:441–449. doi: 10.1097/01.tp.0000231689.44266.ac. [DOI] [PubMed] [Google Scholar]
- Forbes S.J., Gupta S., Dhawan A. Cell therapy for liver disease: from liver transplantation to cell factory. J. Hepatol. 2015;62:S157–S169. doi: 10.1016/j.jhep.2015.02.040. [DOI] [PubMed] [Google Scholar]
- Forster R., Chiba K., Schaeffer L., Regalado S.G., Lai C.S., Gao Q., Kiani S., Farin H.F., Clevers H., Cost G.J. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports. 2014;2:838–852. doi: 10.1016/j.stemcr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox I.J., Daley G.Q., Goldman S.A., Huard J., Kamp T.J., Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345:1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbal-Chaloin S., Funakoshi N., Caillaud A., Gondeau C., Champon B., Si-Tayeb K. Human induced pluripotent stem cells in hepatology: beyond the proof of concept. Am. J. Pathol. 2014;184:332–347. doi: 10.1016/j.ajpath.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Godoy P., Schmidt-Heck W., Natarajan K., Lucendo-Villarin B., Szkolnicka D., Asplund A., Bjorquist P., Widera A., Stober R., Campos G. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 2015;63:934–942. doi: 10.1016/j.jhep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M., Strom S. Mice with human livers. Gastroenterology. 2013;145:1209–1214. doi: 10.1053/j.gastro.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Hay D.C., Fletcher J., Payne C., Terrace J.D., Gallagher R.C., Snoeys J., Black J.R., Wojtacha D., Samuel K., Hannoun Z. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Natl. Acad. Sci. USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J., Perlmann T. Maintaining differentiated cellular identity. Nat. Rev. Genet. 2012;13:429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- Huang P., Zhang L., Gao Y., He Z., Yao D., Wu Z., Cen J., Chen X., Liu C., Hu Y. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.T., Lee S.C., Gao Y., Kim K.P., Song G., An S.Y., Adachi K., Jang Y.J., Kim J., Oh K.J. Small molecules facilitate single factor-mediated hepatic reprogramming. Cell Rep. 2016;15:814–829. doi: 10.1016/j.celrep.2016.03.071. [DOI] [PubMed] [Google Scholar]
- Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y., Nichane M. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Morey R., O'Neil R.C., He Y., Daughtry B., Schultz M.D., Hariharan M., Nery J.R., Castanon R., Sabatini K. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A., Cahan P., Li H., Zhao A.M., San Roman A.K., Shivdasani R.A., Collins J.J., Daley G.Q. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas C.T., Hickey R.D., Chen H.S., Mao S.A., Lopera Higuita M., Wang Y., Nyberg S.L. Concise review: liver regenerative medicine: from hepatocyte transplantation to bioartificial livers and bioengineered grafts. Stem Cells. 2017;35:42–50. doi: 10.1002/stem.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidi H., Alhaque S., Szkolnicka D., Flint O., Hay D.C. Fluid shear stress modulation of hepatocyte-like cell function. Arch. Toxicol. 2016;90:1757–1761. doi: 10.1007/s00204-016-1689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani M., Grimm A.A., Willenbring H. Assessing the therapeutic potential of lab-made hepatocytes. Hepatology. 2016;64:287–294. doi: 10.1002/hep.28569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.L., Gao Y., Yan Y., Ma H., Sun L., Huang P., Ni X., Zhang L., Zhao X., Ren H. Improved survival of porcine acute liver failure by a bioartificial liver device implanted with induced human functional hepatocytes. Cell Res. 2016;26:206–216. doi: 10.1038/cr.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan G.J., Hay D.C., Park I.H., Fletcher J., Hannoun Z., Payne C.M., Dalgetty D., Black J.R., Ross J.A., Samuel K. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkolnicka D., Farnworth S.L., Lucendo-Villarin B., Hay D.C. Deriving functional hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2014;30 doi: 10.1002/9780470151808.sc01g05s30. 1G.5.1–1G.5.12. [DOI] [PubMed] [Google Scholar]
- Szkolnicka D., Hay D.C. Concise review: advances in generating hepatocytes from pluripotent stem cells for translational medicine. Stem Cells. 2016;34:1421–1426. doi: 10.1002/stem.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Touboul T., Hannan N.R., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Di Santo J. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- Yang J., Wang Y., Zhou T., Wong L.Y., Tian X.Y., Hong X., Lai W.H., Au K.W., Wei R., Liu Y. Generation of human liver chimeric mice with hepatocytes from familial hypercholesterolemia induced pluripotent stem cells. Stem Cell Reports. 2017;8:605–618. doi: 10.1016/j.stemcr.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., He Z.Y., You P., Han Q.W., Xiang D., Chen F., Wang M.J., Liu C.C., Lin X.W., Borjigin U. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.