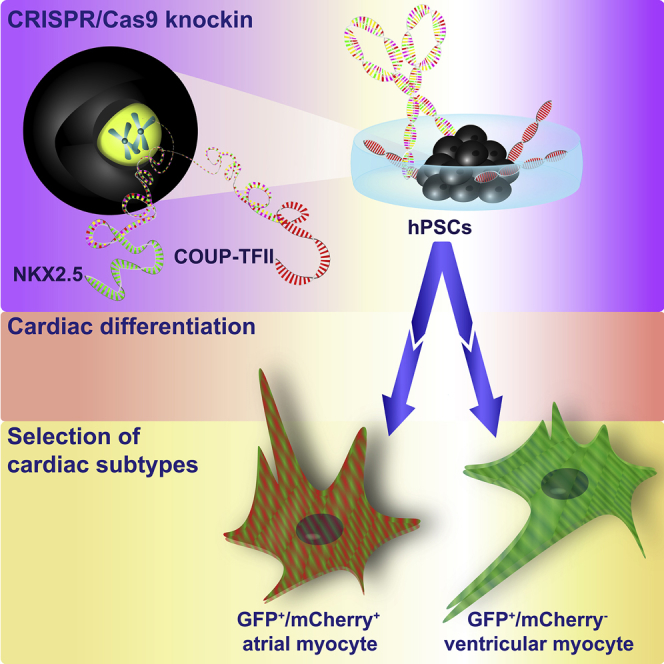
Summary
Reporter cell lines have already proven valuable in identifying, tracking, and purifying cardiac subtypes and progenitors during differentiation of human pluripotent stem cells (hPSCs). We previously showed that chick ovalbumin upstream promoter transcription factor II (COUP-TFII) is highly enriched in human atrial cardiomyocytes (CMs), but not ventricular. Here, we targeted mCherry to the COUP-TFII genomic locus in hPSCs expressing GFP from the NKX2.5 locus. This dual atrial NKX2.5EGFP/+-COUP-TFIImCherry/+ reporter line allowed identification and selection of GFP+ (G+)/mCherry+ (M+) CMs following cardiac differentiation. These cells exhibited transcriptional and functional properties of atrial CMs, whereas G+/M− CMs displayed ventricular characteristics. Via CRISPR/Cas9-mediated knockout, we demonstrated that COUP-TFII is not required for atrial specification in hPSCs. This new tool allowed selection of human atrial and ventricular CMs from mixed populations, of relevance for studying cardiac specification, developing human atrial disease models, and examining distinct effects of drugs on the atrium versus ventricle.
Keywords: human embryonic stem cells, cardiac differentiation, atrial specification, COUP-TFII-mCherry fluorescent stem cell reporter, CRISPR/Cas9, COUP-TFII-knockout
Graphical Abstract
Highlights
-
•
Dual NKX2.5EGFP/+-COUP-TFIImCherry/+ hPSCs to identify atrial cardiomyocytes
-
•
COUP-TFII is not required for atrial specification of hPSCs in vitro
In this article, Passier and colleagues developed an atrial fluorescent stem cell reporter by CRISPR/Cas9-mediated knockin of mCherry at the genomic locus of COUP-TFII in a cardiac NKX2.5EGFP/w reporter line. As validated at the transcriptional and functional level, the nuclear receptor COUP-TFII successfully marks atrial cardiomyocytes but is not required for atrial specification during in vitro differentiation of hPSCs.
Introduction
Human pluripotent stem cells (hPSCs) can differentiate to all cells of the human body. This has resulted in high expectations for applications in regenerative medicine, drug discovery, disease modeling in vitro, and developmental biology (Keller, 2005). Primary human cardiomyocytes (CMs) are extremely difficult to obtain from biopsies and to maintain in culture, so that hPSC-derived CMs (hPSC-CMs) have been rapidly implemented as an alternative not only in disease modeling in vitro but also in pre-clinical drug testing and safety pharmacology (Beqqali et al., 2009, Braam et al., 2010, Maddah et al., 2015, van Meer et al., 2016, Sala et al., 2016). Despite substantial improvements in the efficiency of hPSC differentiation to CMs during the last decade, the majority of directed cardiac differentiation protocols yield heterogeneous CM populations, largely composed of ventricular CMs (Mummery et al., 2012). Recently, we demonstrated efficient generation of atrial CMs from human embryonic stem cells (hESCs) (Devalla et al., 2015). These hESC-derived atrial CMs (hESC-AM), resemble human fetal atrial CMs at the molecular and functional level and have already proven to be a predictive and reliable pre-clinical model for atrial selective pharmacology (Devalla et al., 2015). Although hESC-AMs represented the majority of CMs (approximately 85%) in our directed differentiation protocol, other cardiac subtypes (mostly ventricular CMs with less than 1% of nodal cells) were also present.
To select hESC-AM populations and study their differentiation in vitro, we generated a fluorescent atrial hESC reporter line. Previously, we have shown that chick ovalbumin upstream promoter transcription factors I and II (COUP-TFI and II or NR2F1 and NR2F2) are highly expressed in retinoic acid (RA)-induced hESC-AMs, but not in ventricular CMs (hESC-VMs) (Devalla et al., 2015). In both the human fetal and adult heart, COUP-TFI and II are expressed in myocardial cells of the atria, but not the ventricles (Devalla et al., 2015). COUP-TFs belong to the steroid/thyroid hormone receptor superfamily, which shares high homology with retinoid and RA receptor subfamily members (Tsai and Tsai, 1997). Along with evolutionarily conserved protein-encoding sequences, the expression of COUP-TFII is also broadly identical in mouse, chick, zebrafish, frog, and Drosophila (Tsai and Tsai, 1997). During murine heart development, COUP-TFII is first detected in the visceral mesoderm and sinus venosus, then progresses to the common atrium, and becomes restricted to CMs of the atrial chambers at later stages of development (Pereira et al., 1999, Wu et al., 2013). This indicates that COUP-TFII is an important atrial-enriched transcription factor and prompted us to develop an atrial hESC reporter line using CRISPR/Cas9 genome-editing technology to insert sequences encoding the red-fluorescent protein mCherry into one allele of the genomic COUP-TFII locus. Since COUP-TFII expression is not confined to CMs, but is also expressed in other mesodermal cell types (for example, venous endothelial cells, skeletal muscle, and kidneys) (Lee et al., 2004, You et al., 2005, Yu et al., 2012), as well as endodermal (for example, liver and pancreas) (Zhang et al., 2002) and some ectodermal derivatives (cerebellum, eye, and ear) (Kim et al., 2009, Tang et al., 2010, Tang et al., 2005), we chose the well-established human cardiac NKX2.5EGFP/+ reporter (Elliott et al., 2011) to develop a unique dual reporter line that would allow identification and purification of hESC-AMs. Transcriptional and functional analysis of sorted GFP+ (G+)/mCherry+ (M+) double-positive CMs clearly demonstrated their atrial identity, whereas G+/M− CMs belonged to the ventricular lineage. In addition, we found that complete loss of COUP-TFII did not affect the differentiation toward AMs, based on both molecular and functional analysis. Purification of hESC-AMs will likely be important for optimization and standardization of assays in cardiac drug screening and modeling atrial diseases, such as atrial fibrillation, and understanding underlying molecular mechanisms for atrial specification and disease.
Results
Generation of a Fluorescent Dual Reporter by CRISPR/CAS9-Mediated Targeting of COUP-TFII in hESC-NKX2.5-GFP
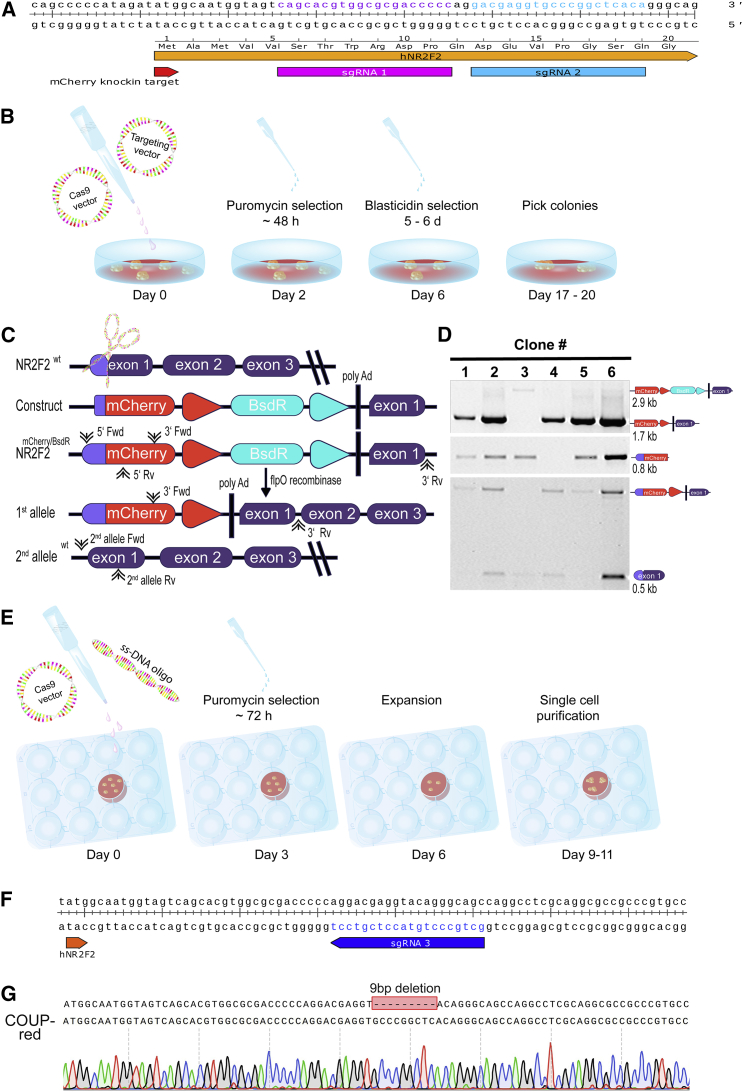
To generate an atrial hESC reporter line, we inserted sequences encoding the red fluorophore mCherry into the genomic locus of COUP-TFII (NR2F2) in the hESC NKX2.5EGFP/+ cardiac reporter line (NKX-GFP) (Elliott et al., 2011) using CRISPR/Cas9-mediated genome editing. Two different single-guide RNAs (sgRNAs) were designed to direct double-strand breaks within exon 1 of COUP-TFII (sgRNA 1 and 2) (Figure 1A). NKX-GFP hESCs were transfected with the COUP-TFII-mCherry targeting vector and one of the sgRNAs co-expressed from the Cas9 vector (Figures 1B and 1C). After antibiotic selection, the excision of the blasticidin-resistance gene was mediated using flippase site-specific recombination (Figure 1C). Correctly targeted clones displayed a 0.8 kb PCR product following screening of the 5′ end and a 2.9 kb product (1.7 kb after blasticidin excision) of the 3′ end (Figure 1D). Following clonal selection by fluorescence-activated cell sorting (FACS), correct targeting of the subclones as well as excision of the blasticidin-resistance cassette was reconfirmed by PCR. In addition, a PCR screen was performed to determine whether mCherry was inserted into one or both COUP-TFII alleles (Figure 1D). For subclones in which mCherry was monoallelic targeted, the genomic integrity of the wild-type (WT) COUP-TFII allele was confirmed by Sanger sequencing of the PCR product. Following sequencing, we identified that, in all mCherry monoallelic-targeted clones from sgRNA 1 and 2, the other allele possessed either insertions or deletions in the COUP-TFII coding sequence. Consequently, none of the resulting COUP-TFII-mCherry knockin clones were heterozygous for both mCherry and COUP-TFII.
Figure 1.
CRISPR/Cas9-Mediated Knockin of mCherry into COUP-TFII
(A) Location and sequence of single-guide RNAs (sgRNAs) 1 and 2 in the COUP-TFII locus.
(B) Schematic representation of the CRISPR/Cas9 plasmid-based targeting and dual-selection protocol.
(C) Schematic overview of wild-type (WT) COUP-TFII, COUP-TFII-mCherry targeting construct, and the resulting targeted (first allele) and WT COUP-TFII (second allele) alleles with forward (FWD) and reverse (RV) primer binding sites for screening.
(D) PCR screen to identify targeted clones: Upper panel: 3′ end screen and confirmation of excision of the blasticidin-resistance cassette in 1, 2, 4, 5, and 6, but not 3 (targeted, but blasticidin-resistance cassette still present). Middle panel: 5′ end screen shows that clones 1–3, and 5 and 6, were targeted at the 5′ end. Lower panel: screen to determine if heterozygous (clones 2, 3, 4, and 6) or homozygous (clones 1 and 5) mCherry knockin occurred.
(E) Schematic representation of the CRISPR/Cas9-mediated correction using a WT COUP-TFII single-stranded oligonucleotide (ssDNA oligo).
(F) Sequence corresponding to the 9 bp deletion in the COUP-TFII locus along with the annealing sites for sgRNA 3 designed to repair the deletion.
(G) Sanger sequencing confirming correction of the second COUP-TFII allele from the COUP-red clone after correction.
See also Figures S1 and S2.
To produce a COUP-TFIImCherry/+ hESC line, we designed a strategy to correct a 9 bp deletion in one of the subclones (Figure 1E). sgRNA 3 (Figure 1F) was designed to specifically anneal to the deleted region, while a single-stranded DNA (ssDNA) oligonucleotide (ssDNA oligo) containing part of the WT COUP-TFII genomic sequence (sequence available in the Supplemental Experimental Procedures) was used as the template to mediate gene repair through homologous recombination. The sgRNA, along with Cas9 and the ssDNA oligo were transfected into COUP-TFII-targeted hESCs that carried the 9 bp deletion before subcloning (Figures 1E and 1G). No off-target effects by the sgRNAs were detected at any of the top four analyzed in-silico-predicted genomic sites in all lines tested (Figure S1). Undifferentiated cells expressed the stem cell marker TRA-1-60 (Figure S2A) and karyotyping demonstrated no chromosomal abnormalities in the genetically modified hESCs (Figure S2B).
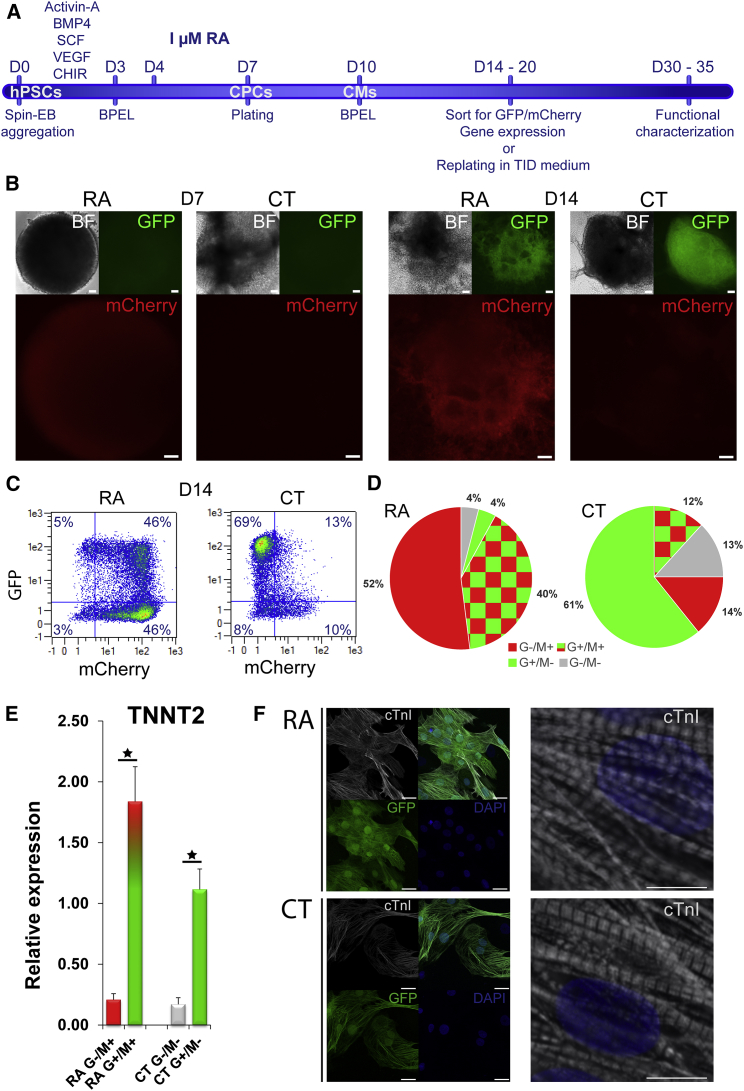
NKX2.5EGFP/+-COUP-TFIImCherry/+ hESCs Robustly Express mCherry upon Induction with RA
To initiate cardiac differentiation, hESCs from the corrected NKX2.5EGFP/+-COUP-TFIImCherry/+ dual reporter line, hereafter called “COUP-red”, were differentiated toward mesoderm by adding growth factors activin-A, BMP4, VEGF, SCF, and the GSK-3 inhibitor Chir99021 after the formation of aggregates by centrifugation (spin-embryoid bodies [spin-EBs]) (Figure 2A) as described previously (Devalla et al., 2015). For efficient directed cardiac differentiation towards the atrial fate, 1 μM RA was added to the EBs from day 4 to day 7 without additional medium changes (Devalla et al., 2015). In agreement with previous reports demonstrating that RA induces COUP-TFII expression (Kruse et al., 2008), only upon treatment with RA could mCherry fluorescence be detected in a subset of cells 36 hr (day 6) after RA addition; this was followed later at day 14 by a broader and higher mCherry fluorescence intensity (Figure 2B). As expected, only a small subpopulation of cells in control differentiation (CT) (no RA) displayed mCherry fluorescence, which did not overlap with GFP (Figures 2B and 2C). Only in the presence of RA did the majority of cells robustly express mCherry in combination with increased GFP fluorescence (Figure 2B), suggesting that these cells co-expressed transcription factors COUP-TFII and NKX2.5. In contrast, in CT cultures, the majority of M+ cells were negative for GFP (NKX2.5) (Figure 2B). We used flow cytometry to quantify the percentage of GFP- and mCherry-expressing cells. In agreement with our previous findings, more than 90% of the RA-treated cells were mCherry positive at day 14 (Figures 2C and 2D), and approximately half of these cells also expressed GFP. On the other hand, the majority of G+ cells were mCherry positive (∼90% based on G+/M+: 41% ± 2%, G−/M+: 52% ± 3%, G+/M−: 4% ± 1%, G−/M−: 4% ± 1%; n = 4; mean ± SEM) (Figures 2C and 2D). By contrast, cells from CT differentiation displayed a high percentage of G+/M− cells together with lower percentages of the other fractions (G+/M+: 12% ± 1%, G−/M+: 14% ± 2%, G+/M−: 61% ± 4%, G−/M−: 13% ± 2%; n = 4; mean ± SEM) (Figures 2C and 3D).
Figure 2.
Characterization of the Dual Human NKX2.5EGFP/+-COUP-TFIImCherry/+ Stem Cell Line (COUP-red) as Atrial Reporter
(A) Atrial-directed pin-embryoid body (spin-EB) protocol and treatment with retinoic acid (RA).
(B) mCherry expression together with GFP and bright-field (BF) images at D7 and D14 of differentiation in RA and control (CT) differentiations. Scale bar, 100 μm.
(C) Representative flow cytometry plots depicting the percentage of GFP-positive (G+) or mCherry (M+) cells at D14 of differentiation in RA or CT differentiations.
(D) Averaged G/M percentage calculated from four independent differentiations (n = 4): RA condition (G+/M+: 41% ± 2%, G−/M+: 52% ± 3%, G+/M−: 4% ± 1%, G−/M−: 4% ± 1%) versus CT (G+/M+: 12% ± 1%, G−/M+: 14% ± 2%, G+/M−: 61% ± 4%, G−/M−: 13% ± 2%). Data are displayed as means ± SEM
(E) Relative mRNA expression of cardiac troponin (TNNT2) in sorted G/M populations at day 20 of differentiation. Data are displayed as means ± SEM; ∗p < 0.05.
(F) Immunostaining of cardiac troponin (cTnI) together with endogenous GFP expression and DAPI as nuclear staining in unsorted cultures after dissociation and re-plating. Scale bars, 25 μm (left), 2.5 μm (right).
See also Figure S3.
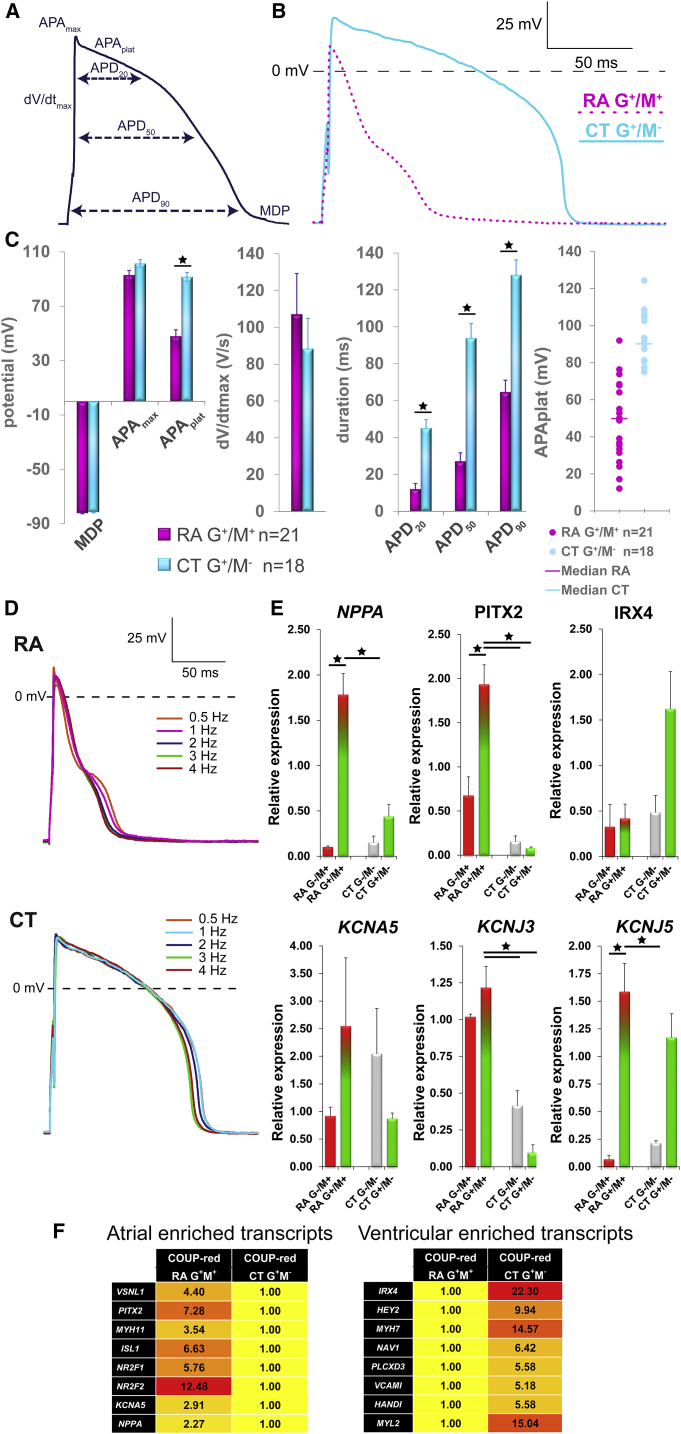
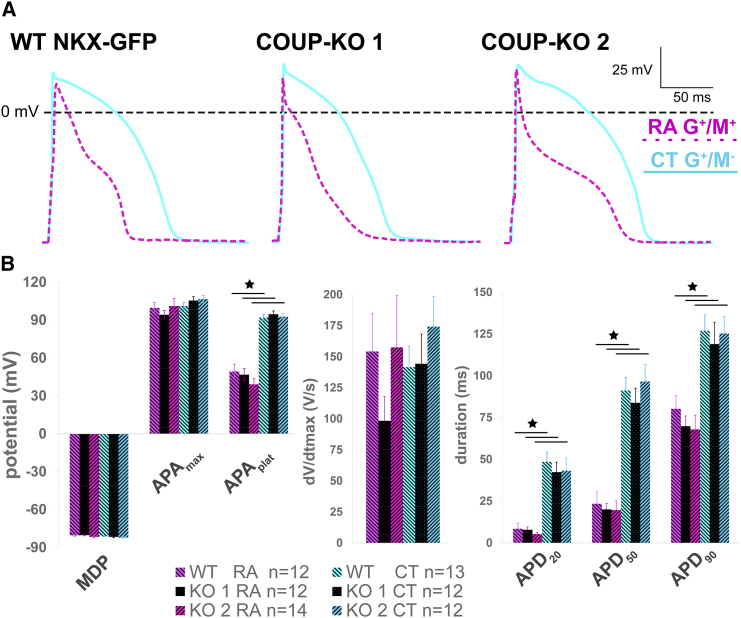
Figure 3.
M+ COUP-red CMs Exhibit Atrial Properties
(A) Analyzed AP parameters.
(B) Representative AP of G+/M+ CMs generated from RA-treated and G+/M− CMs from CT differentiations at 1 Hz stimulation.
(C) Averaged maximum diastolic potential (MDP), maximum AP amplitude (APAmax) and AP plateau amplitude (APAplat), maximum upstroke velocity (dV/dtmax), and AP duration at 20%, 50%, and 90% repolarization (APD20, APD50, and APD90). Scatterplot depicting the APAplat of single CMs and calculated median from RA and CT conditions. n = 21 cells for RA G+/M+ and n = 18 cells for CT G+/M−; from 3 independent differentiations. Data are displayed as means ± SEM; ∗p < 0.05.
(D) Typical AP traces stimulated at different frequencies.
(E) Transcriptional profiling of selected atrial or ventricular-specific genes by qPCR of G+/M+ and G−/M+ fractions from RA differentiations compared with G+/M− and G−/M− fractions from CT differentiations at D20 of differentiations (n = 4). Data are displayed as means ± SEM; ∗p < 0.05.
(F) Genome-wide transcriptional profiling by microarray. Heatmaps to display the fold difference of selected atrial- and ventricular-enriched transcripts in RA-treated G+/M+ and CT G+/M− from COUP-red at a threshold of 2-fold difference.
See also Figure S3.
COUP-TFII RNA and protein levels could be identified in RA-treated samples but not in CT. As expected, protein COUP-TFII levels in the heterozygous COUP-red reporter were reduced by half (Figure S3D). Expression of the COUP-TFII homolog COUP-TFI was not affected (Figure S3E).
The COUP-TFII Reporter Reliably Identifies Functional Human Atrial Cardiomyocytes
Both cardiac subpopulations expressed cardiac troponin (Figures 2E and 2F) and, after approximately 10 days of differentiation, spontaneously contracting cells appeared in RA and in CT differentiations of COUP-red (Movies S1 and S2).
Since G+/M+ double-positive cells were expected to have an atrial identity and G+/M− cells ventricular, we characterized the functional phenotype of sorted CMs by evaluating their action potentials (APs) by patch-clamp methodology (Figure 3A; raw data available in Table S1). In agreement with our previous study, typical AP traces stimulated at 1 Hz showed faster repolarization in RA-treated G+/M+ CMs compared with non-treated G+/M− CT CMs (Figure 3B). Whereas maximal diastolic potential, maximal AP amplitude (APAmax), and maximal AP upstroke velocity (dV/dtmax) did not differ between populations, G+/M+ CMs from RA-treated differentiations exhibited marked reduction in average AP plateau amplitude (APAplat) and AP duration at 20%, 50%, and 90% repolarization compared with G+/M− CMs (Figure 3C). Most importantly, despite some biological variation in APAplat of single CMs, none of the G+/M− CMs exhibited atrial AP properties, and, vice versa, none of G+/M+ CMs had a ventricular phenotype (Figure 3C). Also at higher pacing frequencies, cells maintained their characteristic AP properties (Figure 3D).
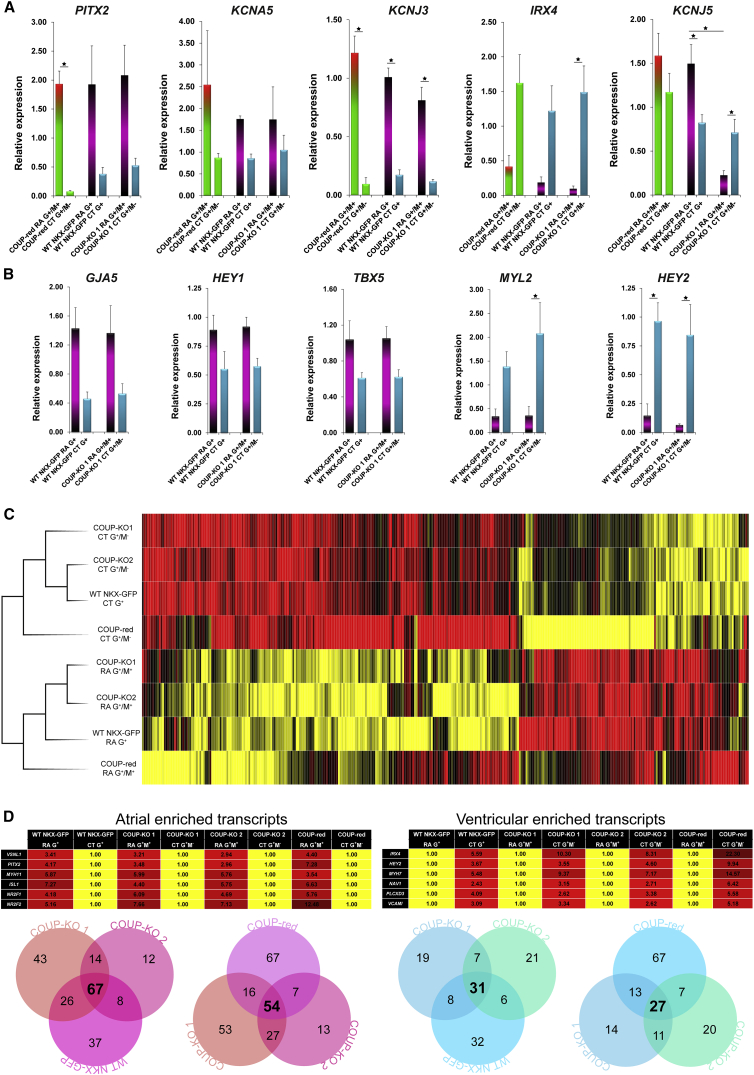
As expected, quantitative PCR (qPCR) of sorted G+/M+ CMs showed increased mRNA expression of the atrial markers NPPA and PITX2 and downregulation of the ventricular gene IRX4 compared with G+/M− CMs (Figure 3E). Moreover, G+/M+ CMs were characterized by enhanced expression of the atrial ion channel genes, potassium voltage-gated channel subfamily A member 5 (KCNA5), potassium voltage-gated channel subfamily J members 3 (KCNJ3), and 5 (KCNJ5) (Figure 3E). In agreement with the qPCR data, human whole-genome-wide transcriptional profiling of G+/M+ CMs from RA and G+/M− CMs from CT, demonstrated upregulation of atrial-enriched transcripts, including VSNL1, PITX2, MYH11, ISL1, NR2F1 and 2, KCNA5, and NPPA, in hESC-AMs (G+/M+ CMs), and enhanced expression of ventricular genes, such as IRX4, HEY2, MYH7, NAV1, PLCXD3, VCAM1, HAND1, and MYL2 in hESC-VMs (G+/M− CMs) (Figure 3F). Together, functional and transcriptional analysis confirmed atrial identity of G+/M+ CMs (hESC-AMs) and ventricular identity of G+/M− CMs (hESC-VMs).
Identity of mCherry-Positive Non-CMs
Besides efficient identification and selection of hESC-AMs and hESC-VMs, RA-treated cultures of the dual COUP-red reporter also clearly indicated a NKX2.5-negative, COUP-TFII-positive (G−/M+) (presumably non-CM) subpopulation. Besides in atrial CMs, COUP-TFII is also expressed in the epicardium (Lin et al., 2012), smooth muscle, and venous endothelial cells (You et al., 2005). Indeed, human whole-genome-wide transcriptional analysis by microarray showed more than 600 transcripts with 2-fold upregulation in G−/M+ compared with G+/M+ cells; gene ontology (GO) identified enrichment of several GO terms, including cardiovascular system and blood vessel development, that had the highest GO enrichment scores (Figure S3A), as well as terms related to the differentiation of epicardial cells (Figure S3B). Quantification by flow cytometry at day 13 of differentiation showed that 26% of G−/M+ cells expressed the endothelial cell, smooth muscle, and fibroblast marker CD90 (Thy-1), with 4% of cells double-positive for CD90 and the endothelial marker platelet endothelial cell adhesion molecule (PECAM or CD31) (Figure S3B). In agreement, a subpopulation (4%) of G−/M+ cells co-expressed the endothelial markers CD31 and VE-cadherin (CD144) together with a large (47%) intermediate population expressing either CD31 (10%) or CD144 (37%) (Figure S3B). However, at day 21, the majority of non-CMs (both M+ and M−) expressed SMA mRNA (encoded by smooth muscle ACTA2) and protein, while we did not observe PECAM protein expression, which suggested that the majority of G−/M+ subpopulation belong to the smooth muscle cell lineage at this stage (Figure S3C).
Since COUP-TFII is also expressed in many endodermal derivatives, we studied the endodermal marker α-fetoprotein (AFP). Only very few cells expressed AFP suggesting that cells with endodermal identity constituted a very minor subpopulation in both RA as well as CT cultures (Figure S3B).
CRISPR/Cas9-Mediated Knockout of COUP-TFII in hESCs
After establishing that the COUP-red dual reporter line is useful for the selection of atrial and ventricular CMs, we determined whether COUP-TFII is required for specification of the atrial lineage during in vitro differentiation. Although COUP-TFII has been shown as essential for atrial specification in two mouse models (Pereira et al., 1999, Wu et al., 2013), information on its role in atrial patterning of the human heart is limited. Following insertion of mCherry into the COUP-TFII genomic locus of one allele, we identified different disrupting genotypic modifications on the second COUP-TFII allele which introduced a premature stop codon, yielding two independent cell lines with complete knockout (hereafter called “COUP-KO 1” generated with sgRNA 1 and “COUP-KO 2” independently generated with sgRNA 2) (Figure S4).
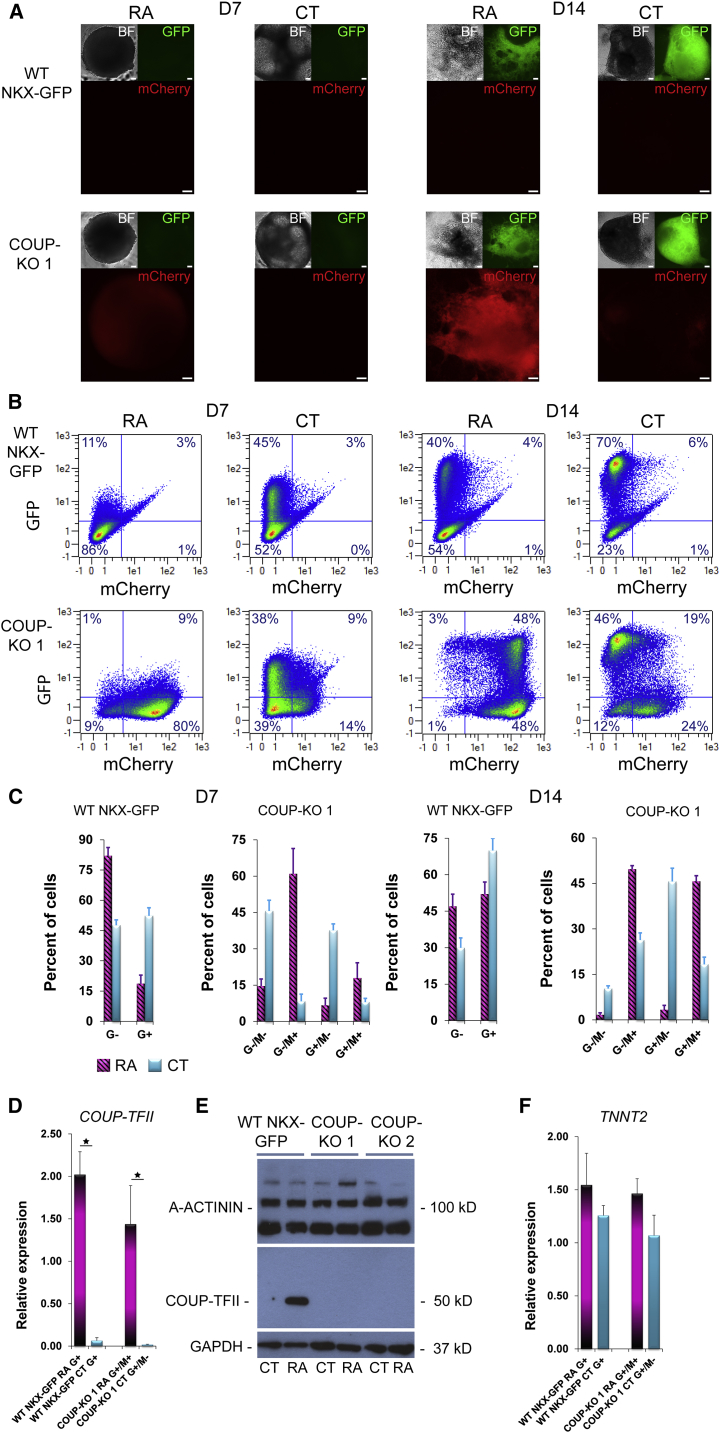
We then attempted to differentiate the COUP-KO cell lines to the atrial lineage by treatment with RA. As for RA and CT differentiation of the COUP-red reporter, both COUP-KO cell lines rapidly upregulated mCherry upon RA treatment starting at day 6 (Figure S5A), and robustly expressed mCherry in majority of cells by day 14 of differentiation, whereas CT differentiations yielded predominantly G+/M− populations (Figures 4A and S5B). In flow cytometry, both RA and CT differentiation of COUP-KO lines resulted in a similar distribution of G/M populations compared with the COUP-red reporter (Figures 4B, 4C, S5C, and S5D; Table 1). Importantly, we observed no obvious differences in onset or overall percentages of GFP in RA and CT differentiation from COUP-KO compared with WT NKX-GFP cells with two functional COUP-TFII alleles (Figures 4B, 4C, S5C, and S5D; Table 1). Complete knockout of COUP-TFII protein was confirmed by western blot in differentiated CMs from both COUP-KO lines, whereas there was prominent expression of COUP-TFII in RA-treated WT NKX-GFP cells (Figures 4D and 4E).
Figure 4.
Characterization of COUP-KO Lines
(A) mCherry overlapping with GFP and bright-field (BF) images at D7 and D14 of differentiation in RA or CT differentiations from WT NKX-GFP or COUP-KO cells from subclone COUP-KO 1. Scale bars, 100 μm.
(B) Representative flow cytometry plots depicting the percentage of GFP-positive (G+) and mCherry (M+) cells at D7 and D14 of differentiation in RA or CT samples from WT NKX-GFP or COUP-KO 1.
(C) Averaged G/M percentage calculated from three independent differentiations (n = 3) at day 7 of differentiation in RA (G+/M+, 18% ± 6%; G−/M+, 61% ± 10%; G+/M−, 7% ± 3%; G−/M−, 15% ± 3%) and CT (G+/M+, 8% ± 2%; G−/M+, 8% ± 3%; G+/M−, 38% ± 3%; G−/M−, 46% ± 4%) and at day 14 in RA (G+/M+, 46% ± 2%; G−/M+, 50% ± 1%; G+/M−, 4% ± 2%; G−/M−, 2% ± 1%) and CT (G+/M+, 18% ± 2%; G−/M+, 26% ± 2%; G+/M−, 46% ± 2%; G−/M−, 10% ± 1%), together with WT NKX-GFP cells (D7, CT G+/M+, 5% ± 3%; G+/M− 65% ± 3%; G−/M− 30% ± 3%; G−/M+, 1% ± 1%; RA G+/M+, 4% ± 1%; G+/M− 48% ± 4%; G−/M− 45% ± 4%; G−/M+, 2% ± 1%; D14, CT G+/M+, 3% ± 1%; G+/M− 49% ± 3%; G−/M− 48% ± 3%; G−/M+, 0% ± 0%; RA G+/M+, 4% ± 1%; G+/M− 15% ± 4%; G−/M− 81% ± 4%; G−/M+, 1% ± 1%). n = 3 independent differentiations; mean ± SEM.
(D) mRNA expression of COUP-TFII in COUP-KO 1 cells at day 14 of differentiation (n = 4 independent differentiations; mean ± SEM; ∗p < 0.05).
(E) Validation of complete knock out of COUP-TFII expression by western blot in unpurified differentiated CMs from RA and CT differentiations from COUP-KO cells compared with WT NKX-GFP cells.
(F) mRNA expression of cardiac troponin (TNNT2) in differentiated CMs from RA and CT differentiations from COUP-KO 1 and WT NKX-GFP cells (n = 4 independent differentiations).
See also Figures S4, S5, and S8.
Table 1.
Comparison of G/M Percentages in RA and CT Differentiation between Different Cell Lines at Day 14 of Differentiation
| Line | G+/M+ (%) | G+/M– (%) | G−/M+ (%) | G−/M– (%) |
|---|---|---|---|---|
| WT NKX-GFP RA (n = 3) | – | 52 ± 5 | – | 47 ± 5 |
| COUP-red RA (n = 4) | 41 ± 2 | 4 ± 1 | 52 ± 3 | 4 ± 1 |
| COUP-KO 1 RA (n = 3) | 46 ± 2 | 3 ± 2 | 50 ± 1 | 2 ± 1 |
| COUP-KO 2 RA (n = 2) | 37 ± 5 | 5 ± 2 | 56 ± 4 | 3 ± 2 |
| WT NKX-GFP CT (n = 3) | – | 70 ± 5 | – | 30 ± 4 |
| COUP-red CT (n = 4) | 12 ± 1 | 61 ± 4 | 14 ± 2 | 13 ± 2 |
| COUP-KO 1 CT (n = 3) | 18 ± 2 | 46 ± 4 | 26 ± 2 | 10 ± 1 |
| COUP-KO 2 CT (n = 2) | 18 ± 4 | 42 ± 4 | 27 ± 4 | 14 ± 3 |
Data are displayed as average percentages ± SEM from n = independent differentiations.
COUP-KO hESCs Differentiate to Functional Atrial Cardiomyocytes
Much like the directed cardiac differentiation of WT NKX-GFP and COUP-red hESCs, RA and CT cells of both COUP-KO lines expressed similar levels of cardiac troponin (TNNT2) (Figure 4F) and developed into functional contracting CMs within 10 days (Movies S3 and S4).
To establish the atrial identity of G+/M+ CMs, we analyzed sorted cells by perforated patch clamp and compared them with WT NKX-GFP CMs (Figure 5A; raw data available in Table S1). Average AP characteristics of RA-treated G+/M+ and CT G+/M− CMs from both COUP-KO lines were identical to their equivalents from WT NKX-GFP cells (Figure 5B). G+/M+ CMs from RA-treated COUP-KO differentiations showed a significant reduction in APAplat and AP duration compared with G+/M− CMs despite functional loss of COUP-TFII (Figure 5B). This was consistent with functional expression of atrial-specific potassium ion channels, which are responsible for the fast repolarization in atrial, but not ventricular CMs. Interestingly, RA-treated and CT G+/M+ CMs from COUP-KO 1 and COUP-red lines exhibited comparable atrial AP characteristics, suggesting that COUP-TFII deletion did not affect atrial differentiation in the RA, as well as in the CT condition (Figure S6A). Similarly, G+/M− CMs from CT and RA displayed typical ventricular characteristics (Figure S6B).
Figure 5.
COUP-KO-derived CMs Functionally Resemble their WT Counterparts
(A) Representative action potentials (APs) of G+/M+ CMs generated from RA-treated and G+/M− CMs from CT differentiations of WT NKX-GFP, COUP-KO1, and COUP-KO 2 cells.
(B) Averaged maximum diastolic potential (MDP), maximum AP amplitude (APAmax) and AP plateau amplitude (APAplat), maximum upstroke velocity (dV/dtmax), and AP duration at 20%, 50%, and 90% repolarization (APD20, APD50, and APD90). n = 12 cells for WT NKX-GFP RA G+, n = 12 cells for COUP-KO 1 RA G+/M+, and n = 14 cells for COUP-KO 2 RA G+/M+; n = 13 cells for WT NKX-GFP CT G+, n = 12 cells for COUP-KO 1 CT G+/M−, and n = 12 cells for COUP-KO 2 CT G+/M−; from three independent differentiations. Data are displayed as means ± SEM; ∗p < 0.05.
See also Figure S6.
Transcriptional Profiling of WT, COUP-KO, and COUP-red Cardiomyocytes
Since we did not observe functional differences between COUP-KO CMs and the original WT NKX-GFP CMs, we next studied their molecular profiles by qPCR on day 14 of differentiation. CMs were FACS selected from both COUP-KO lines (G+/M+ from RA-treated and G+/M− from CT samples) and compared with G+ WT NKX-GFP CMs from RA-treated and CT samples (approximately 85% atrial in RA and largely ventricular CMs in CT). Similar to CMs differentiated from the COUP-red reporter or WT NKX-GFP hESCs, RA-treated G+/M+ CMs from the COUP-KO lines exhibited enhanced expression of atrial-enriched PITX2 and atrial potassium ion channel genes KCNA5 and KCNJ3, along with simultaneous downregulation of the ventricular marker IRX4 (Figures 6A, S5E, and S5F). In contrast, KCNJ5 expression was downregulated in RA-treated G+/M+ CMs from COUP-KO hESCs compared with their corresponding RA-treated equivalents from WT NKX-GFP or COUP-red cells (Figures 6A and S5E). To determine whether there were additional transcriptional changes, we quantified the expression of genes previously reported to be targets of COUP-TFII. In comparison with G+/M− CMs (ventricular), G+/M+ COUP-KO CMs (atrial) exhibited increased expression of the atrial-enriched gap junction gene GJA5 and the atrial transcription factors HEY1 and TBX5, as well as decreased expression of ventricular-enriched genes, MYL2 and HEY2 (Figures 6B, S5E, and S5F). Importantly, these expression levels were comparable with levels of these genes in RA and CT differentiations of the WT NKX-GFP line.
Figure 6.
G+M+ CMs Lacking Functional COUP-TFII Exhibit Atrial Characteristics on Transcriptional Level
(A and B) qPCR analysis of selected atrial or ventricular transcripts of sorted COUP-KO 1 derived G+/M+ fractions from RA and G+/M− populations from CT compared with G+ populations of WT NKX-GFP RA and CT differentiations at D14 (n = 4 independent differentiations). Data are displayed as means ± SEM; ∗p < 0.05.
(C) Hierarchical clustering of G+/M+ RA fractions and G+/M− CT fractions of two independent subclones COUP-KO 1 and 2 revealed high similarity to their G+ WT NKX-GFP or G+/M+ and G+/M− COUP-red equivalents.
(D) Heatmaps: fold difference of atrial- and ventricular-enriched transcripts with Venn diagrams to display the overlap of differentially expressed genes between RA (atrial transcripts, left panel) and CT (ventricular transcripts, right panel) in COUP-KO 1, 2, and WT NKX-GFP or COUP-red at a threshold of 2-fold difference.
See also Figures S6 and S7 and Table S3.
Since knockout of COUP-TFII did not apparently result in major differences in expression of COUP-TFII target genes, we next carried out human whole-genome transcriptional profiling by microarray hybridization, for which sorted RA-treated and CT CMs from both COUP-KO and WT NKX-GFP lines in week 3 of differentiation were used. In agreement with our qPCR data, hierarchical clustering identified two clusters of RA-treated and CT CMs independent of the line (KO, COUP-red, or WT) from which they were derived (Figure 6C). As expected, we observed upregulation of atrial transcripts in RA-treated CMs (including VSNL1, PITX2, MYH11, ISL1, and NR2F1 and 2) and increased expression of ventricular transcripts in CT CMs (including IRX4, HEY2, MYH7, NAV1, PLCXD3, and VCAM1) from all three lines independent of their COUP-TFII genotype, suggesting that knockout of COUP-TFII had little effect on induction or repression of an atrial or ventricular-specific transcriptional program in hESC-CMs (Figure 6D; Table S3). The majority of affected genes which had a clear atrial (up in RA) or ventricular identity (up in CT) showed an overlap between two or three lines (Figure 6D). Since the atrial- and ventricular-enriched CM populations from the WT NKX-GFP line were not just one subtype, we further studied the overlap of atrial (14 transcripts) and ventricular transcripts (7 transcripts) in both COUP-KO lines based on a 2-fold change (Figure 6D). Based on GEO expression profiles, we found strong atrial and ventricular identities in respective groups, indicated by upregulation of ALDH1A2, TUBB2A, ZNF385B, SLC9A3R1, ZNF533, and ADD3 in RA, and LAMB1 and EMILIN2 in CT. These subtype-specific transcripts were differently expressed between the COUP-KO lines and the WT NKX-GFP line, but the majority (ZNF385B, SLC9A3R1, ZNF533, and ADD3 in RA, as well as LAMB1 and EMILIN2 in CT) overlapped with the COUP-red reporter, indicating that the COUP-TFII reporter line faithfully recapitulated atrial identity, which is not affected by the loss of COUP-TFII. In addition, other transcripts with an atrial identity could be identified in the individual COUP-KO lines, but were lost in the common upregulated pool since the 2-fold change cutoff was not met. For example, the atrial-specific ion channel-encoding gene KCNA5 exhibited a 2-fold enhanced expression in COUP-KO 2, and a 1.7-fold upregulation in COUP-KO 1.
Role of COUP-TFI and COUP-TFII Isoforms during Atrial Differentiation of WT and COUP-KO Lines
There is a high degree of homology and overlapping expression patterns between COUP-TFII and COUP-TFI. Moreover, we recently demonstrated that both COUP-TFs similarly regulate atrial-specific potassium channel genes, KCNA5 and KCNJ3 (Devalla et al., 2015). COUP-TFI gene expression and protein levels were similar in WT NKX-GFP and COUP-KO CMs, indicating that COUP-TFI expression does not compensate for the complete loss of COUP-TFII expression (Figures S7A and S7B).
To date, six different transcript variants of human COUP-TFII have already been identified (Table S2). Structural variant 1 (NR2F2-001, NM_021005) encodes full-length COUP-TFII protein (isoform a) with 414 amino acids (aa) and is composed of the typical elements of nuclear receptors: an N-terminal ligand-independent activation domain (activation function 1; aa: 1–79), followed by a highly conserved DNA-binding domain (DBD) (aa: 80–144), and a ligand-binding domain (LBD) (aa: 195–414) at the C-terminal end (Figure S7C) (Achatz et al., 1997, Yamazaki et al., 2013). Structural variants 2 (NR2F2-002, NM_001145155), 3 (NR2F2-201, NM_001145156), and 4 (NR2F2-003, NM_001145157) encode truncated protein isoforms (isoforms b and c), which share the same LBD, but lack the DBD domain (Figure S7C) (Achatz et al., 1997, Yamazaki et al., 2013). Similarly, structural variant 5 has a major truncation, but at the C-terminal. In contrast, non-coding short variant 6 is transcribed into a microRNA. Since the predominant transcript variant 1 of COUP-TFII is expressed predominantly in hESC-AMs and also because only variant 1 contains the transactivating DBD domain, sgRNAs were designed to specifically recognize this isoform for the CRISPR/Cas9 gene-editing strategy (Figure 1A). Since little is known about COUP-TFII isoform expression in hESC-AMs or hESC-VMs, we determined their expression by qPCR and western blot in RA and CT cultures from WT NKX-GFP and COUP-KO hESCs. Expression of all variants was strongly induced by RA. No differences in regulation of gene expression were observed between WT NKX-GFP and COUP-KO cells (Figure S7D). Correspondingly, western blotting confirmed the differential expression pattern of each COUP-TFII variant between RA and CT (Figure S7E). Interestingly, after overexposure of the blots, we detected enhanced expression of COUP-TFII isoform “c” primarily after induction with RA in WT NKX-GFP and also COUP-KO (Figure S7E). Two additional bands at 40 and 60 kDa were also evident, which may correspond to unknown isoforms of COUP-TFII (Figure S7E). Following increased exposure, we also identified low levels of COUP-TFII isoform a in CT condition from WT NKX-GFP, but not from COUP-KO hESCs (Figure S7E). Since truncated COUP-TFII variants b and c lack the DBD, it is unlikely that they directly affect expression of COUP-TFII target genes, although we cannot exclude these isoforms playing a role in indirect transcriptional regulation and thereby compensate for the loss of the dominant isoform a.
Discussion
Efficient procedures to differentiate defined cardiac subtypes from hPSCs are important for disease modeling and drug screening. Previously, we demonstrated robust generation of atrial CMs and their application for predictive pharmacology (Devalla et al., 2015). Although these cultures were enriched for atrial CMs (80%–90%), other subtypes were still present. Here, we therefore generated a fluorescent human atrial stem cell reporter based on the nuclear receptor COUP-TFII, which is predominantly expressed in human atrial, but not ventricular CMs (Devalla et al., 2015). Specifically, we generated a dual fluorescent NKX2.5EGFP/+-COUP-TFIImCherry/+ reporter line by targeting the human NKX-GFP hESC cardiac reporter (Elliott et al., 2011) via CRISPR/Cas9-mediated genome editing. RA treatment during differentiation of the COUP-red line resulted in rapid induction of mCherry. This is consistent with our previous data demonstrating that COUP-TFII expression is rapidly increased in differentiating cardiac progenitor cells within days of RA treatment (Devalla et al., 2015). As expected, only upon induction with RA does mCherry partially overlap with NKX2.5-GFP, indicating COUP-TFII-expressing CMs. COUP-red hESCs efficiently developed into contracting CMs with functional sarcomeres. Transcriptional and functional characterization of G+/M+ CMs established their atrial phenotype, demonstrating that this COUP-red dual reporter line reliably captures cells that are directed towards the atrial CM lineage.
Modulating RA signaling upon mesoderm induction has been demonstrated to guide differentiation towards epicardial cells, which subsequently undergo epithelial-to-mesenchymal transition to epicardial-derived smooth muscle cells (Iyer et al., 2015). Moreover, COUP-TFII is expressed in venous endothelial, as well as smooth muscle cells (You et al., 2005). In agreement with these findings, the majority of mCherry-positive non-CMs (G−/M+) displayed a smooth muscle-like profile, as indicated by transcriptional profiling, as well as the expression analysis of smooth muscle cell and endothelial markers.
In mice, homozygous loss of COUP-TFII resulted in lethal hemorrhages and edema formation in neural and myocardial tissues (Pereira et al., 1999). Homozygous deletion or myocardial specific deletion of COUP-TFII have been demonstrated to severely affect development of both atria during early embryonic development (Pereira et al., 1999, Wu et al., 2013). While primitive atrial chambers of COUP-TFII mouse mutants had a similar size compared with WT littermates during early phase of development, loss of COUP-TFII impeded rightward expansion of the common atrium (Pereira et al., 1999). Tissue-specific deletion of COUP-TFII forced CMs of the atrial chambers to acquire morphological and functional characteristics typical of ventricular cells (Wu et al., 2013). These findings suggest an important role for COUP-TFII in maintaining an atrial fate, while simultaneously suppressing a ventricular phenotype. In accordance, we had shown in differentiated hESC-AMs that COUP-TFII is able to bind to the promoters of atrial-specific ion channel genes KCNA5 and KCNJ3 and regulates their expression in hESC-derived CMs (Devalla et al., 2015). We investigated here the atrial differentiation potential of hESCs after CRISPR/Cas9-mediated inactivation of both COUP-TFII alleles. We successfully derived functionally contracting CMs from these COUP-KO hESCs. As indicated by similar percentages of NKX2.5-GFP-positive CMs, onset and overall efficiency of atrial differentiation were not affected by complete deletion of COUP-TFII. Morphological characterization of EBs from either RA or CT differentiations revealed high similarity to their equivalent WT NKX-GFP EBs throughout the whole differentiation procedure. Electrophysiological characterization somewhat surprisingly demonstrated clear atrial identity of G+/M+ CMs. In accordance, the global transcriptional profile of G+/M+ CMs derived from COUP-KO hESCs was very similar to that of hESC-AMs derived from WT NKX-GFP, as well as the heterozygous COUP-red atrial reporter, with upregulation of atrial-enriched genes and downregulation of ventricular-enriched genes. Only a very small group of genes were differentially expressed between CMs from both COUP-KO lines and WT NKX-GFP CMs. To confirm this, we determined expression levels of various genes that have previously been shown to be direct targets of COUP-TFII by qPCR. These atrial- or ventricular-specific candidates showed similar expression patterns in COUP-KO and WT NKX-GFP CMs. Of all of these COUP-TFII target genes, only KCNJ5, encoding the atrial-specific inward rectifier potassium channel 4 (Kir3.4), was decreased in the heterozygous COUP-red reporter and even further after complete KO of COUP-TFII, although this was not evident from whole-genome profiling by microarray. Kir3.4 subunits form functional heteromeric ion channels only after assembly with Kir3.1 subunits, which are encoded by KCNJ3. In correspondence to the transcriptional profile, AP properties of either G+/M+ or G+/M− CMs derived from COUP-KO cells were identical to those of their WT equivalents. Together these findings suggested that COUP-TFII might not be essential for RA-induced atrial CM differentiation or for suppression of the ventricular phenotype in hPSCs. Previously, we had shown that short hairpin RNA (shRNA)-mediated knockdown of COUP-TFs in hESC-derived CMs resulted in a change of gene expression of KCNA5, KCNJ3, and KCNJ5 (Devalla et al., 2015). The difference in COUP-TFII-dependent gene regulation with respect to KCNA5 and KCNJ3 between COUP-TFII KO and knockdown may reflect a higher non-specific off-target effect of shRNAs (Schulte-Merker and Stainier, 2014). Alternatively, differences in acute versus chronic gene disruption may contribute to the observed difference (Rossi et al., 2015). Whereas acute effects were studied following COUP-TFII knockdown in day 30 atrial CMs previously, in the present study we analyzed gene expression in sorted CMs, which had a complete loss of COUP-TFII already from the start of differentiation.
Besides a high degree of homology between COUP-TFII and COUP-TFI (NR2F1), these two nuclear receptors also have overlapping expression patterns in human fetal, as well adult hearts (Devalla et al., 2015). Knockout of COUP-TFII did not lead to increased levels of COUP-TFI, suggesting that it is not likely that COUP-TFI compensates for the loss of COUP-TFII. Nevertheless, we cannot exclude redundancy and the possibility that COUP-TFI levels may be sufficient to initiate and complete atrial differentiation in hPSCs. Interestingly, in addition to the predominant COUP-TFII isoform a, we detected isoform c predominantly in RA-treated and not in CT samples. Even though isoform a is predominant, it is possible that COUP-TFII variants b or c, which lack the DBD, may affect atrial differentiation via a mechanism independent of direct gene regulation. However, so far this mechanism of action has not been described for COUP-TFII. Interestingly, isoforms a and b, but not c, have identical orthologs in mouse.
Although various de novo or inherited mutations, including missense mutations of human COUP-TFII have been related to atrioventricular septal defects and other congenital heart defects, such as tetralogy of Fallot, aortic stenosis, coarctation of the aorta, hypoplastic left heart syndrome, and ventricular septal defects, loss-of-function mutations in COUP-TFII have not been implicated as contributing to atrial defects exclusively (Al Turki et al., 2014).
Even though hPSC ventricular CMs have been used successfully to recapitulate a variety of cardiac disease phenotypes and drug-induced toxicities, less attention has been paid to disease modeling using hPSC atrial CMs. Previously, a bacterial artificial chromosome reporter construct, in which expression of a fluorescence is driven by expression of the atrial-specific gene sarcolipin, has been described and used for identification of hPSC atrial CMs (Josowitz et al., 2014). However, enhanced Sarcolipin expression was also detected in CMs of ventricles with reduced systolic function (Zheng et al., 2014), and is also highly expressed in non-CMs, such as human skeletal muscle (Minamisawa et al., 2003). As mentioned previously, COUP-TFII is also expressed in other non-atrial cell types and tissues. These are important factors to consider for the proper use of lineage reporters.
Selection of hPSC-AMs is crucial for gaining mechanistic insights into processes underlying atrial specification of hPSCs during differentiation in vitro and will improve understanding of cardiac diseases specially affecting atrial CMs, as well as identifying atrial-specific surface markers. Because cardiac differentiation of different hPSC lines is not equally efficient, identification of atrial-specific surface markers would allow purification of atrial CMs generated, for example, from patient-specific induced pluripotent stem cells to model atrial-specific (genetic) disorders, such as atrial fibrillation in vitro. A precedent for this approach was the identification of SirpA and VCAM1 as surface markers for NKX2.5-expressing CMs using the NKX2.5EGFP/+ hESC line (Elliott et al., 2011).
In summary, we have generated a dual NKX2.5EGFP/+-COUP-TFIImCherry/+ hPSC line, which allows the identification and selection of functional atrial CMs. Furthermore, we showed that COUP-TFII is dispensable for acquiring an atrial phenotype during RA-directed differentiation of hESCs. Most importantly, this unique reporter line, which allows selection of hESC-AMs, will be a valuable tool for studying atrial differentiation, disease modeling, and early drug screening. Selected populations of hESC-AMs will impact optimization and standardization for hPSC-dependent assays, facilitate studies related to underlying mechanisms of atrial-specific diseases, and advance the development of new strategies to cure underlying causes of certain cardiac disease.
Experimental Procedures
Maintenance of hESCs
hESCs were maintained as undifferentiated colonies either in Essential 8 medium on vitronectin (Gibco, Thermo Fisher Scientific), or on irradiated mouse embryonic fibroblasts in hESC medium (Devalla et al., 2015).
Vector Construction
The sgRNAs (sgRNAs 1–3) were designed either with the web tool CRISPR Design (http://crispr.mit.edu) or Benchling (https://benchling.com/crispr). Oligos containing the sgRNA sequences (Integrated DNA Technologies) were cloned into the pSP-Cas9(BB)-2A-puro vector (PX459; Addgene) as described previously (Ran et al., 2013). The COUP-TFII-mCherry targeting vector was generated in three steps. See Supplemental Experimental Procedures for extended description and sequences.
hESC Transfection
A total of 0.25–1.0 × 106 hESCs was simultaneously transfected with 1.6–4 μg PX459-sgRNA vector and either 2 μg COUP-TFII-mCherry targeting vector or 8 pmol of the 110 bp COUP-TFII WT ss-oligo (synthesized as an Ultramer from IDT) using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Additional information, vector maps, and sequences are available in the Supplemental Experimental Procedures.
Genomic DNA Isolation, PCR Screening, and Sequencing
Genomic DNA was extracted from the transfected hESCs using the QuickExtract DNA Extraction Solution (EpiCentre). Correctly modified clones were identified by PCR-based screening strategies using Platinum High Fidelity Taq DNA polymerase (Gibco, Thermo Fisher Scientific) followed by Sanger sequencing of the PCR product (Macrogen). See the Supplemental Experimental Procedures for primer sequences.
Cardiac Differentiation of hESCs
Differentiation to hESC-derived CMs was performed using a previously described spin-EB protocol (Devalla et al., 2015).
Microscopy and Video Acquisition
Bright-field and fluorescent images or videos at 10× magnification were acquired with a Leica AF-6000LX microscope (Leica Microsystems) with controlled temperature and CO2.
Flow Cytometry
D7 EBs were dissociated with 1× TrypLe Select and D14 EBs with 10× TrypLe Select and percentages of G+/M−, G−/M+, double-positive, or double-negative cells were immediately quantified with a MACSQuant VYB flow cytometer (Miltenyi Biotech). See the Supplemental Experimental Procedures for extended description on antibodies.
FACS
G+/M+ were purified into four populations using a BD ARIA III flow cytometer after exclusion of dead cells and debris according to side and forward scatter. After sorting, cells were immediately lysed for RNA isolation or re-plated for functional analysis onto Matrigel-coated glass coverslips into TID medium (Birket et al., 2015).
Transcriptional Analysis
Microarrays were performed on Human HT-12 v4 Expression Bead Chips at ServiceXS, and analysis was performed with GeneSpring GX (Agilent Technologies). qPCR was carried out using SYBR Green (Applied Biosystems) and the CFX384 real-time PCR detection system. Statistical analysis was carried out with GraphPad Prism 7. Ordinary one-way ANOVA with Tukey's multiple comparisons test was applied for differences in means between groups. Data are expressed as means ± SEM. Statistical significance was defined as p < 0.05. GO was performed using DAVID (https://david.ncifcrf.gov) (Huang et al., 2008, Huang et al., 2009). Additional information can be found in the Supplemental Experimental Procedures.
Cellular Electrophysiology
APs were recorded at 36°C ± 0.2°C using the amphotericin-B perforated patch-clamp in silico technique (Giles and Noble, 2016, Hoekstra et al., 2012, Meijer Van Putten et al., 2015). An in silico inward rectifier K+ current with kinetics of Kir2.1 channels was injected through a dynamic clamp to overcome the depolarized state of the cells. See the Supplemental Experimental Procedures for an extended description. Data are presented as means ± SEM. Statistical analysis was carried out with SigmaStat 3.5 software. Normality and equal variance assumptions were tested with the Kolmogorov-Smirnov and the Levene median tests, respectively. Two groups were compared with unpaired t test or, in case of a failed normality and/or equal variance test, Mann-Whitney rank-sum test. More than two groups were compared using one-way ANOVA followed by a Student-Newman-Keuls method post hoc test. In case of non-normally distributed parameters, Kruskal-Wallis test followed by pairwise comparisons with Dunn's method was performed p < 0.05 was considered statistically significant.
Immunostaining and Confocal Imaging
Cells were stained for troponin I, mCherry, SMA, PECAM, or AFP, using primary antibodies followed by secondary conjugated antibodies to Alexa Fluor 647, 555, or Cy-3. Confocal images were analyzed using Leica Microsystems. See the Supplemental Experimental Procedures for extended description.
Western Blot
For western blotting, samples were lysed in standard RIPA buffer, and 100 μg protein was analyzed on an 8% polyacrylamide gel. For detection, mouse IgG anti-COUP-TF I (1:500; Perseus Proteomics), mouse IgG anti-COUP-TFII (isoform a; 1:500; Perseus Proteomics), rabbit IgG anti-COUP-TFII (isoform a + b + possible c, 1:500; clone D16C4, Cell Signaling Technology), rabbit IgG anti-GAPDH (1:500; Millipore), and mouse IgG anti-α-actinin (A7811, Sigma-Aldrich) were used in combination with the corresponding secondary antibodies goat anti-mouse- or anti-rabbit-IgG HRP (1:2,000; Cell Signaling Technology).
Author Contributions
V.S. designed and performed experiments, analyzed data, and wrote the manuscript. A.O.V. performed and analyzed perforated patch-clamp experiments and approved the manuscript. M.M. performed experiments concerning the reporter knockin. J.J.M.K. performed immunostainings and imaging. H.D.D. provided input and approved the manuscript. C.L.M. provided input and approved the manuscript. V.V.O. and K.A. prepared the targeting construct and approved the manuscript. R.P.D. contributed by designing and performing reporter knockin, designing correction experiments, and approving the manuscript. R.P. designed the study, analyzed data, and wrote and approved the manuscript.
Acknowledgments
Funding for this study is gratefully acknowledged from the following sources: LUMC (BW-plus) doctoral grant to V.S., ERC-AdG STEMCARDIOVASC and EU FP7 to C.L.M., ERC-StG STEMCARDIORISK and VIDI-917.15.303 (the Netherlands Organisation for Scientific Research [NWO]) to R.P.D., the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 602423: Plurimes to V.V.O., C.L.M., and K.A., Eurostars Cardiopredict E!9527 to V.S., H.D.D., and R.P., and ZonMw-MKMD-40-42600-98-036 to R.P. The authors would like to thank Dorien Ward-Van Oostwaard for technical advice on the culture of human stem cells and cardiac differentiation, Joop C. Wiegant and Berend J. van Meer for technical assistance during microscopy, the Department of Molecular Cell Biology of the LUMC for karyotyping, Sabrina Veld and Guido M. de Roo for assistance with cell sorting, and Christian B. Schwach for illustrations.
Published: November 22, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, three tables, and four movies and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.10.024.
Accession Numbers
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar, 2002) and are accessible through accession number GEO: GSE100199.
Supplemental Information
References
- Achatz G., Hölzl B., Speckmayer R., Hauser C., Sandhofer F., Paulweber B. Functional domains of the human orphan receptor ARP-1/COUP-TFII involved in active repression and transrepression. Mol. Cell. Biol. 1997;17:4914–4932. doi: 10.1128/mcb.17.9.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Turki S., Manickaraj A.K., Mercer C.L., Gerety S.S., Hitz M.-P., Lindsay S., D’Alessandro L.C.A., Jawahar Swaminathan G., Bentham J., Arndt A.-K. Rare variants in NR2F2 cause congenital heart defects in humans. Am. J. Hum. Genet. 2014;94:574–585. doi: 10.1016/j.ajhg.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beqqali A., Van Eldik W., Mummery C., Passier R. Human stem cells as a model for cardiac differentiation and disease. Cell. Mol. Life Sci. 2009;66:800–813. doi: 10.1007/s00018-009-8476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:1–13. doi: 10.1016/j.celrep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam S.R., Tertoolen L., van de Stolpe A., Meyer T., Passier R., Mummery C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5eGFP/w hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Giles W.R., Noble D. Rigorous phenotyping of cardiac iPSC preparations requires knowledge of their resting potential(s) Biophys. J. 2016;110:278–280. doi: 10.1016/j.bpj.2015.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M., Mummery C.L., Wilde A.A.M., Bezzina C.R., Verkerk A.O. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front. Physiol. 2012;3:346. doi: 10.3389/fphys.2012.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer D., Gambardella L., Bernard W.G., Serrano F., Mascetti V.L., Pedersen R.A., Talasila A., Sinha S. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development. 2015;142:1528–1541. doi: 10.1242/dev.119271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josowitz R., Lu J., Falce C., D’Souza S.L., Wu M., Cohen N., Dubois N.C., Zhao Y., Sobie E.A., Fishman G.I. Identification and purification of human induced pluripotent stem cell-derived atrial-like cardiomyocytes based on sarcolipin expression. PLoS One. 2014;9:3–10. doi: 10.1371/journal.pone.0101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kim B.J., Takamoto N., Yan J., Tsai S.Y., Tsai M.-J. Chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) regulates growth and patterning of the postnatal mouse cerebellum. Dev. Biol. 2009;326:378–391. doi: 10.1016/j.ydbio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse S.W., Suino-Powell K., Zhou X.E., Kretschman J.E., Reynolds R., Vonrhein C., Xu Y., Wang L., Tsai S.Y., Tsai M.J. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:2002–2015. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.T., Li L., Takamoto N., Martin J.F., DeMayo F.J., Tsai M., Tsai S.Y. The nuclear orphan receptor COUP-TFII is required for limb and skeletal muscle development. Mol. Cell. Biol. 2004;24:10835–10843. doi: 10.1128/MCB.24.24.10835-10843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.J., You L.R., Yu C.T., Hsu W.H., Tsai M.J., Tsai S.Y. Endocardial cushion morphogenesis and coronary vessel development require chicken ovalbumin upstream promoter-transcription factor II. Arterioscler. Thromb. Vasc. Biol. 2012;32:135–146. doi: 10.1161/ATVBAHA.112.300255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddah M., Heidmann J.D., Mandegar M.A., Walker C.D., Bolouki S., Conklin B.R., Loewke K.E. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Reports. 2015;4:621–631. doi: 10.1016/j.stemcr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer Van Putten R.M.E., Mengarelli I., Guan K., Zegers J.G., Van Ginneken A.C.G., Verkerk A.O., Wilders R. Ion channelopathies in human induced pluripotent stem cell derived cardiomyocytes: a dynamic clamp study with virtual IK1. Front. Physiol. 2015;6:1–16. doi: 10.3389/fphys.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa S., Wang Y., Chen J., Ishikawa Y., Chien K.R., Matsuoka R. Atrial chamber-specific expression of sarcolipin is regulated during development and hypertrophic remodeling. J. Biol. Chem. 2003;278:9570–9575. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- Mummery C., Zhang J., Ng E., Elliott D., Elefanty A.G., Kamp T.J. Differentiation of human ES and iPS cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F.A., Yuhong Q., Zhou G., Tsai M.-J., Tsai S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., Stainier D.Y.R. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 2016;174:3749–3765. doi: 10.1111/bph.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S., Stainier D.Y.R. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development. 2014;141:3103–3104. doi: 10.1242/dev.112003. [DOI] [PubMed] [Google Scholar]
- Tang L.S., Alger H.M., Lin F., Pereira F.A. Dynamic expression of COUP-TFI and COUP-TFII during development and functional maturation of the mouse inner ear. Gene Expr. Patterns. 2005;5:587–592. doi: 10.1016/j.modgep.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Tang K., Xie X., Park J.-I., Jamrich M., Tsai S., Tsai M.-J. COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development. 2010;137:725–734. doi: 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Y., Tsai M.J. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): Coming of age. Endocr. Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- van Meer B.J., Tertoolen L.G.J., Mummery C.L. Measuring physiological responses of human pluripotent stem cell derived cardiomyocytes to drugs and disease. Stem Cells. 2016;34:2008–2015. doi: 10.1002/stem.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Cheng C.-M., Lanz R.B., Wang T., Respress J.L., Ather S., Chen W., Tsai S.-J., Wehrens X.H.T., Tsai M.-J. Atrial identity is determined by a COUP-TFII regulatory network. Dev. Cell. 2013;25:417–426. doi: 10.1016/j.devcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Suehiro J., Miyazaki H., Minami T., Kodama T., Miyazono K., Watabe T. The COUP-TFII variant lacking a DNA-binding domain inhibits the activation of the Cyp7a1 promoter through physical interaction with COUP-TFII. Biochem. J. 2013;452:345–357. doi: 10.1042/BJ20121200. [DOI] [PubMed] [Google Scholar]
- You L.-R., Lin F.-J., Lee C.T., DeMayo F.J., Tsai M.-J., Tsai S.Y. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Yu C.-T., Tang K., Suh J.M., Jiang R., Tsai S.Y., Tsai M.-J. COUP-TFII is essential for metanephric mesenchyme formation and kidney precursor cell survival. Development. 2012;139:2330–2339. doi: 10.1242/dev.076299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Bennoun M., Gogard C., Bossard P., Leclerc I., Kahn A., Vasseur-Cognet M. Expression of COUP-TFII in metabolic tissues during development. Mech. Dev. 2002;119:109–114. doi: 10.1016/s0925-4773(02)00286-1. [DOI] [PubMed] [Google Scholar]
- Zheng J., Yancey D.M., Ahmed M.I., Wei C.-C., Powell P.C., Shanmugam M., Gupta H., Lloyd S.G., McGiffin D.C., Schiros C.G. Increased sarcolipin expression and adrenergic drive in humans with preserved left ventricular ejection fraction and chronic isolated mitral regurgitation. Circ. Heart Fail. 2014;7:194–202. doi: 10.1161/CIRCHEARTFAILURE.113.000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.