Abstract
Paper-based lateral flow immunoassays (LFIAs) are one of the most widely used point-of-care (PoC) devices; however, their application in early disease diagnostics is often limited due to insufficient sensitivity for the requisite sample sizes and the short time frames of PoC testing. To address this, we developed a serum-stable, nanoparticle catalyst-labeled LFIA with a sensitivity surpassing that of both current commercial and published sensitivities for paper-based detection of p24, one of the earliest and most conserved biomarkers of HIV. We report the synthesis and characterization of porous platinum core–shell nanocatalysts (PtNCs), which show high catalytic activity when exposed to complex human blood serum samples. We explored the application of antibody-functionalized PtNCs with strategically and orthogonally modified nanobodies with high affinity and specificity toward p24 and established the key larger nanoparticle size regimes needed for efficient amplification and performance in LFIA. Harnessing the catalytic amplification of PtNCs enabled naked-eye detection of p24 spiked into sera in the low femtomolar range (ca. 0.8 pg·mL–1) and the detection of acute-phase HIV in clinical human plasma samples in under 20 min. This provides a versatile absorbance-based and rapid LFIA with sensitivity capable of significantly reducing the HIV acute phase detection window. This diagnostic may be readily adapted for detection of other biomolecules as an ultrasensitive screening tool for infectious and noncommunicable diseases and can be capitalized upon in PoC settings for early disease detection.
Keywords: lateral flow immunoassay, porous platinum core−shell nanoparticles, broad dynamic range, enzyme mimic, nanobodies, biorthogonal chemistry, point-of-care, HIV detection
The emergence of infectious diseases is one of the greatest threats to human health. The recent outbreaks of Zika and Ebola viruses and the ongoing HIV pandemic continue to highlight the need for diagnostic tools that are affordable, sensitive, simple, and able to rapidly detect pathogens under challenging circumstances. These demands have led to the uptake of paper-based lateral flow immunoassays (LFIAs) as the most prominent rapid point-of-care (PoC) diagnostic tests. Although meeting many of these criteria, their sensitivity generally falls short of other benchtop protein and nucleic acid detection methods.1 The continuing hurdle of developing PoC diagnostics is that compromises must be made between sensitivity, simplicity, speed, and cost. A great challenge is the development of diagnostics for use in resource-limited settings that require exceptional performance in all these categories but also resilience toward a lack of cold-chain support.
Sensitive diagnostic platforms often include signal or target amplification mechanisms, such as polymerase-chain reaction (PCR) and isothermal nucleic acid amplification for detection of nucleic acids or enzyme-catalyzed signal amplification and immuno-PCR for detection of protein targets.2,3 However, these techniques are relatively resource demanding, requiring technical expertise to run and interpret (e.g., purification steps) and utilize equipment that is often incompatible with use in resource-limited settings where unreliable power supplies and maintenance challenges restrict their use.4,5 Due to their high catalytic efficiency and extraordinary stability in harsh environments, such as high temperature and extreme pH, mixed noble-metal catalytic nanoparticles have emerged as promising materials for signal amplification in colorimetric immunoassays.6−11 We show here that incorporating inorganic nanoparticle catalysts in lateral-flow testing formats, through an investigation of their size and porosity, provides a pathway to significantly improve the sensitivity of simple to read, absorbance-based LFIAs. This has resulted in a simple and cost-effective paper based diagnostic test with a level of performance far in advance of current standards for LFIA.
Here, we integrate peroxidase-mimicking porous platinum core–shell nanocatalysts (PtNCs) into a LFIA for sensitive detection of p24, the viral capsid protein of HIV. Acute HIV infection poses a unique challenge in diagnosis as it encompasses a period from initial infection to complete seroconversion (the period over which patient antibodies against proteins of HIV develop). During this window period, there is a lack of detectable circulating biomarkers, with antibodies appearing ca. 28 days after infection.12,13 HIV p24 is the earliest protein biomarker post infection, and a test capable of detecting p24 earlier would be transformative in key target settings, such as the diagnosis of infants whose mothers are known to be HIV positive, where antibody tests are unreliable, and also to ensure early intervention with antiretroviral therapies where they have the potential to be most beneficial.14−17 However, clinically relevant ranges of p24 during acute infection are far below the detectable limit of the leading field-ready colorimetric LFIA tests (ca. 10–15 pg·mL–1),18−20 which are limited by the absorbance of the colored label and the affinities of the antibody recognition components.21
Through the design and optimization of a nanocatalyst amplification platform, high-affinity and orthogonally designed binding components, and lateral flow architecture, we demonstrate a high level of sensitivity and broad dynamic range previously unattainable in LFIA formats. The catalytic amplification process developed herein allows the test, in spiked serum, to surpass the published sensitivity of the leading commercial rapid tests for p24 (Alere Determine HIV-1/2 Ag/Ab Combo and Alere HIV Combo, for example) by a factor of up to 20. Here, we show detection of low femtomolar concentrations of p24 in spiked serum (ca. 32.5 fM), and naked-eye detection of acute stage HIV in clinical plasma samples in under 20 min.
Results and Discussion
The LFIA is used to quickly establish whether a target analyte is present in a complex sample (blood, plasma, sera, etc.) using a simple colorimetric readout. The core process in a LFIA initiates when a detection label (typically an intensely colored nanoparticle) is hydrated by the sample, drawn along the paper membrane by capillary action and bound, via a target analyte, to a region of the strip called the test line. To achieve this, both the nanoparticles and the test line must be decorated with affinity ligands (typically antibodies) capable of binding discrete regions of the target analyte. In order to achieve a high level of sensitivity in a LFIA format, the components must exhibit fast binding reaction kinetics toward the solid-phase to ensure that the particle labels can bind to the test line within the short time window during which the sample is flowing past. The use of a high affinity binding system like biotin–streptavidin (KDca. 10–15 M) for the test line binding capture step is essential for moving toward the goal where all target antigens are labeled with a nanoparticle at the test line.22 The advantage of this architecture means that the antibody-binding complex forms in solution such that the biotin–streptavidin interaction is the rapid binding event which immobilizes the complex at the test line. This circumvents the often poor performance of conventional antibody capture, as the biotin–streptavidin binding affinity is over 4 orders of magnitude higher than a conventional antibody–antigen interaction.23
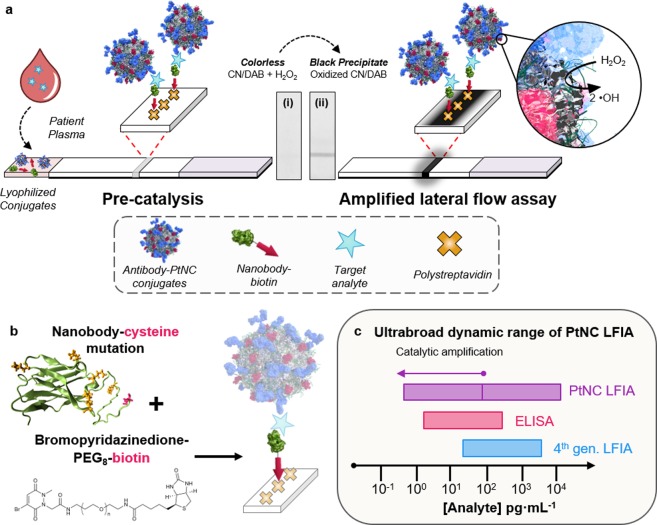
ur PtNC amplified LFIA takes advantage of the simple, low cost LFIA platform but incorporates a strong amplification mechanism through the catalytic activity of the nanoparticle itself to increase the signal intensity through local deposition of dyes at the test line. The schematic in Figure 1a highlights the essential components and configuration of the PtNC amplified LFIA. During the assay, a plasma (or serum) sample is introduced into a vessel containing both lyophilized antibody modified PtNCs and agile, orthogonally biotinylated camelid antibody fragments (nanobody-biotin) capable of binding to discrete epitope regions of the p24 capsid protein. Next, a lateral flow strip, composed of a nitrocellulose reaction membrane and an absorbent pad, is used to draw the solution up the strip toward a streptavidin bearing test line by capillary action. Where present, p24 in the sample can then be sandwiched between the antibody–PtNC conjugate and a biotinylated antibody fragment, forming a biotinylated complex that is captured at the streptavidin coated test line. At high target concentrations (100−10000 pg·mL–1 p24), a clear black line is visible with the naked eye due to the intrinsic absorbance of the particles bound at the test line, indicating a positive test result. Lower concentrations of the target are visualized by taking advantage of the catalytic amplification of the PtNCs that act to disproportionate H2O2, enabling the oxidation of a chromogenic substrate.24−27 After running the plasma sample through, followed by a chase buffer, the test line is exposed to a mixture of hydrogen peroxide and a chromogenic substrate. Any PtNC captured on the test line will oxidize the substrate, generating an insoluble colored product. This amplification step increases the window of sensitivity and lowers the detection limit by 2 orders of magnitude. Because of the unambiguous colored product deposited at the test line, the assay result can be easily read by eye or captured by a mobile phone camera for analysis, with the potential for semiquantitative result readout with a pre- and postamplification signal regime.
Figure 1.
(a) Scheme showing amplified LFIA, where functionalized Pt nanocatalysts (PtNCs) and biotinylated nanobody fragments are mixed with a plasma or serum sample. In the presence of a target, PtNCs become biotinylated through complexation with the target, and rapid high affinity biotin–streptavidin binding enables a [target] dependent deposition of PtNC at the test line. PtNCs bound at the test line catalyze the oxidation of CN/DAB (4-chloro-1-naphthol/3,3′-diaminobenzidine, tetrahydrochloride) substrate in the presence of hydrogen peroxide producing an insoluble black product which is clearly visible with the naked eye. (b) Scheme for site-selective chemical modification of a nanobody with an exposed cysteine mutation (red), where lysine residues are highlighted in orange on the structural model (left), and cartoon of oriented elements at the streptavidin test line. (c) Schematic comparing the dynamic ranges of fourth-generation LFIA, ELISA, and PtNC LFIA.
Figure 1b outlines the site-selective modification strategy used to prepare robust antibody fragments with oriented biotin presentation in the assay. Specifically, we have integrated a thermally robust and high-affinity nanobody component into the nanocatalyst-labeled LFIA. Nanobodies are single domain antibody fragments that contain a complete antigen-binding site but are one tenth the size of a conventional antibody. In addition to their small size and stable conformation, they can be efficiently mass produced in bacteria with site-selective mutations that enable biorthogonal chemistry.28,29 Because of the ease and control over nanobody modification and purification, we chose to biotinylate a p24-binding nanobody validated against a broad spectrum of HIV subtypes to facilitate PtNC–analyte complex binding to the streptavidin test line.30
An anti-p24 nanobody containing a solvent-accessible terminal cysteine residue was expressed to enable site-selective biotinylation. The position of the cysteine residue, at the end of a flexible 6-mer HIS-tag linker, was chosen to maximize the distance between the subsequent modification and the paratope of the nanobody (structural model in Figure 1b), thus minimizing disruption.30 Site-selective biotinylation of the cysteine residue was achieved through reaction with a biotin-functionalized pyridazinedione (PD), the first example of nanobody modification using PD chemistry (Supplementary Methods and Characterization). Analysis of the nanobody postmodification using LCMS confirmed addition of only a single molecule of biotin, evidencing no undesirable off-target conjugation. Pyridazinediones have a proven track record in antibody modification, and hold many advantages over other cysteine-selective reagents such as maleimides and haloacetamides, which have been shown to react with lysine residues andw often display poor long-term stability.31−100 Through this approach, we have specifically engineered the nanobody to present biotin away from the p24 binding region, enabling oriented presentation in our assay.
We examined the binding kinetics of the modified and unmodified nanobodies with p24 and found that the site-selective modification had no significant impact on the dissociation constant, confirming that the modification does not interfere with the antigen binding region (Supplementary Table 1). The site-selectively biotinylated nanobody circumvents the nonspecific modification of the nanobody paratope, which is rich with lysine residues (highlighted in orange in Figure 1b), where standard carbodiimide cross-linker modification strategies will risk interfering with the binding site and thus knock out activity. The site-selective biotinylation of the cysteine presenting nanobody preserves epitope recognition and ensures efficient use of the p24 available in the sample. We selected a monoclonal antibody (anti-HIV-1/2, Capricorn products), which is a p24 binding pair with the nanobody–biotin, to physically adsorb onto the nanocatalysts.30 This coupling of orthogonally modified nanobody and physisorbed whole IgG antibody enabled the rapid engineering of a PtNC amplified assay. Nanobody mutation, expression, and orthogonal modification can be readily achieved within several days where the preparation of the appropriate PtNC catalysts can be performed simultaneously for use on a standard, commercially available streptavidin lateral flow membrane (Supplementary Figure 1).
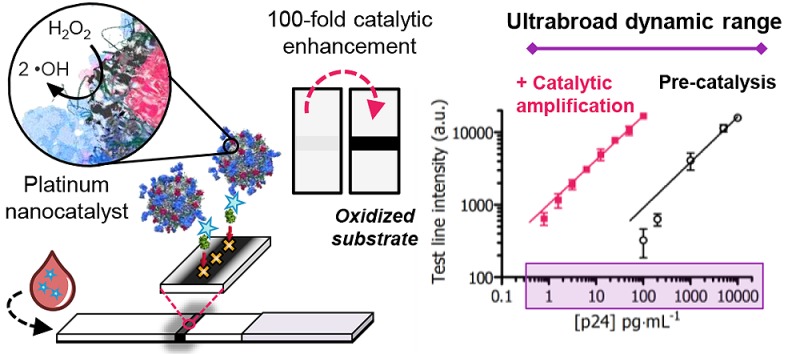
Figure 1c highlights the broad dynamic range of PtNC amplified LFIA, which spans over 4 orders of magnitude. This dual sensitivity regime is enabled by the inherent coloration of the PtNC (for detection of 100–10000 pg·mL–1 analyte) and their ability to catalytically generate a colored product to decorate the test line, accessing a second regime from 100 to less than 1 pg·mL–1 analyte. This is ca. 2 orders of magnitude broader dynamic range than standard colorimetric ELISAs or fourth generation LFIAs, which are limited to either postcatalysis or to the coloration of a particle label, respectively.35
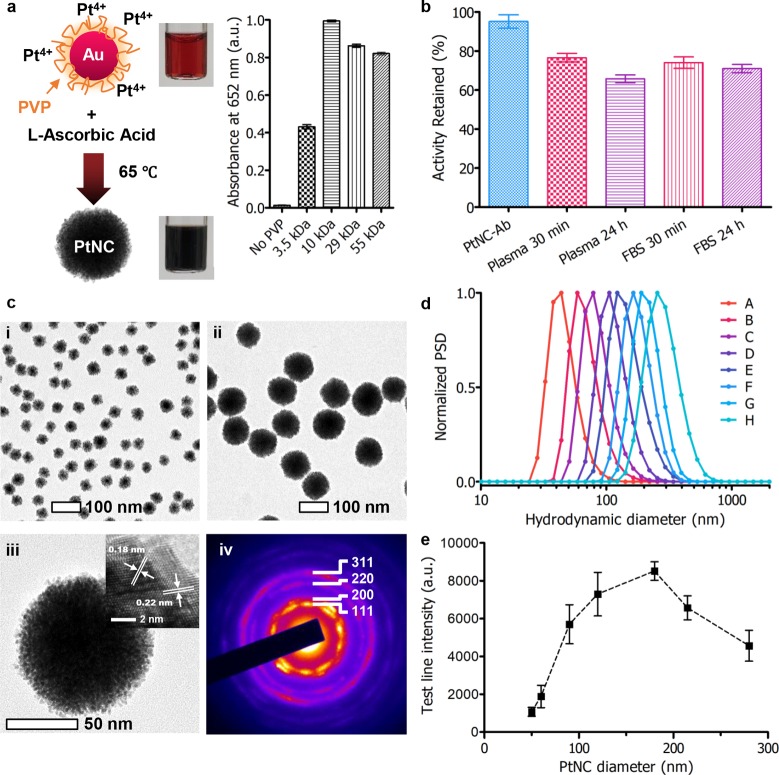
In order to produce PtNCs with high catalytic activity and effective antibody modification, their synthesis required careful optimization. At the early stages of production, the particles are capped with polyvinylpyrrolidone (PVP) to ensure colloidal stability (Supplementary Figure 2) and to reduce the extent of fouling that occurs upon exposure to complex media during LFIA. The molecular weight of PVP used during the particle synthesis played an important role in dictating the catalytic activity of the PtNCs (before antibody modification) as highlighted by their ability to oxidize tetramethylbenzidine (TMB) in Figure 2a, with 10 kDa PVP-capped particles performing optimally. A key performance requirement of the antibody-capped PtNCs is that they can retain their catalytic activity following exposure to complex environments such as patient serum, which contains ca. 7 wt % protein. Indeed, PtNCs when prepared in this manner with 10 kDa PVP and antibody modification were found to retain a significant amount of the initial particle activity (ca. 75%) over a period of 24 h (Figure 2b), far in excess of the exposure time expected during a serological measurement. Although protein adsorption to the PtNCs can reduce the catalytic activity of the particles, it has the benefit of making their modification with antibodies simple, where mixing the PtNCs with antibodies in the correct stoichiometry and at an appropriate pH yields PtNCs with efficiently conjugated antibodies (Supplementary Figure 3).
Figure 2.
(a) Scheme showing synthesis of Au–Pt core–shell structure (PtNC), where 15 nm gold nanoparticles are used as seeds for subsequent platinum overgrowth in the presence of polyvinylpyrrolidone (PVP) as a stabilizer and l-ascorbic acid as a reducing agent. Influence of PVP molecular weight on PtNC catalytic activity, measured by the absorbance at 652 nm corresponding to the oxidation of TMB by H2O2 (n = 3). In the absence of PVP, significant aggregation occurred. (b) Catalytic activity of PtNCs incubated in serological and protein-rich environments up to 24 h (n = 6). (c) Transmission electron micrographs of PtNCs synthesized with varying AuNP seed concentrations to control size: (i) 5 nM seeds and (ii) 0.3 nM seeds. (iii) High-resolution TEM image of an individual PtNC formed from 0.3 nM seed concentration. Inset shows the lattice fringes corresponding to platinum (111) and (200). (iv) Selected area electron diffraction (SAED) pattern taken from a single PtNC (ca. 120 nm) with diffraction spots consistent with polycrystalline platinum with an FCC lattice. (d) Number distribution of the hydrodynamic diameter of PtNCs formed by varying [Au]:[Pt] measured by dynamic light scattering. Batches were synthesized in the presence of different gold nanoparticle seed concentrations: A, 5 nM; B, 1.2 nM; C, 0.6 nM; D, 0.3 nM; E, 0.15 nM; F, 0.08 nM; G, 75 pM ca. 120 nm PtNCs as seeds; H, 40 pM ca. 120 nm PtNCs as seeds. (e) Intensity of test line for antibody modified PtNCs (150 pM) varying in size from ca. 50 to 280 nm (mean number distribution by DLS) for detection of 100 pg·mL–1 p24 in FBS with 5 min development in CN/DAB and H2O2. All data are averaged from ≥3 independent measurements where error bars represent the standard deviation from the mean.
TEM images in Figure 2c and Supplementary Figure 4 show the precise control over PtNC size and monodispersity possible using the seeded synthesis. PtNCs could be produced with porous, polycrystalline platinum shells.37,38 We have observed that there is a significant contribution to the PtNC catalytic activity that arises from the internal catalytic surface area of the particles as shown by assessing the role of gold core size for 15 and 40 nm cores (Supplementary Figure 5). This additional surface area is accessible through nanometer-scale pores (ca. 1–2 nm by TEM image analysis) within the platinum shell. This likely renders the pores inaccessible to larger molecules (e.g., antibodies and serum proteins), but accessible to smaller molecules such as H2O2, making them well suited for use in catalytic amplification processes following exposure to protein-rich serum environments.
Selecting the smaller 15 nm gold nanoparticles as seeds, PtNCs were produced with diameters up to approximately 280 nm by modulating the concentration of gold seeds used for subsequent overgrowth (Figure 2d). Particles larger than ca. 150 nm were produced via a layer-by-layer deposition of platinum on as-synthesized ca. 120 nm PtNCs. High-angle annular dark-field STEM (HAADF-STEM) images of PtNCs of varying size with corresponding energy dispersive X-ray point spectra (EDS) confirm core–shell structure, with Au core and Pt confined to the external porous shell (Supplementary Figure 6 and 7). Thanks to the well-defined concentration of nucleation points for overgrowth with platinum, the polydispersity as measured by dynamic light scattering was maintained at a low level (0.01 < PDI < 0.1) ensuring reproducibility in their synthesis (Supplementary Figure 8). The fine size control of this seeded synthesis allows for control over subsequent antibody modification.
PtNCs provide an ideal platform for assessing the role of catalytic particle size on sensitivity in LFIA by virtue of their core–shell configuration and their high porosity and the resulting large surface area for catalysis that is retained independent of the particle size. The nanometer-scale roughness and morphology of the particles produced during this seeded synthesis appeared unchanged with increasing size (Supplementary Figures 4 and 6). The size of the nanocatalyst has a distinct effect on the test line signal intensity obtained for PtNC sizes up to ca. 280 nm when employed in the LFIA. Larger particles with increased surface area for catalytic reactions would be expected to generate a higher signal in LFIA format, and indeed, up to a point, this was observed experimentally. PtNC sizes ranging 50 to 280 nm were synthesized and modified with an optimal surface density of antibody (Supplementary Figure 3). Keeping the PtNC concentrations constant (150 pM), the different sized PtNCs were used as catalytic labels in the LFIA format for serum samples spiked with 100 pg·mL–1 p24 and the biotinylated nanobody. The resulting variation in signal intensity as a function of particle size is shown in Figure 2e. Larger particles were found to result in increased signal intensity reflecting the increased surface area available for catalytic reactions. However, we observed that this effect was most prominent approaching a threshold in the region of 150 nm, above which a reduced performance was observed. This reduced signal intensity for larger particles may be due to a reduction in diffusivity and a concomitant decrease in internal porosity (and therefore catalytic activity), resulting from the layer-by-layer synthesis (Supplementary Figure 7). An optimal performance regime was exhibited by PtNCs in the 120–200 nm diameter range, so due to their facile one-step synthesis, low nonspecific binding, and minimizing the need to use excessive platinum, 120 nm PtNCs were used for further experiments.
Looking toward the challenges of translation, we investigated both the reproducibility and stability of the test reagents. Figure 3a shows the test line intensity produced by five independently synthesized batches of ca. 120 nm PtNCs for detection of 50 pg·mL–1 p24 spiked sera (Supplementary Figure 8). The red line indicates the average test line intensity across batches. The coefficient of variation between five independently synthesized and antibody modified batches was 2.4%, indicating that PtNC amplified LFIA is reproducible through the synthesis, antibody conjugation, and assay running steps. This suggests that our nanocatalyst platform holds great promise for reliable clinical sample detection.
Figure 3.
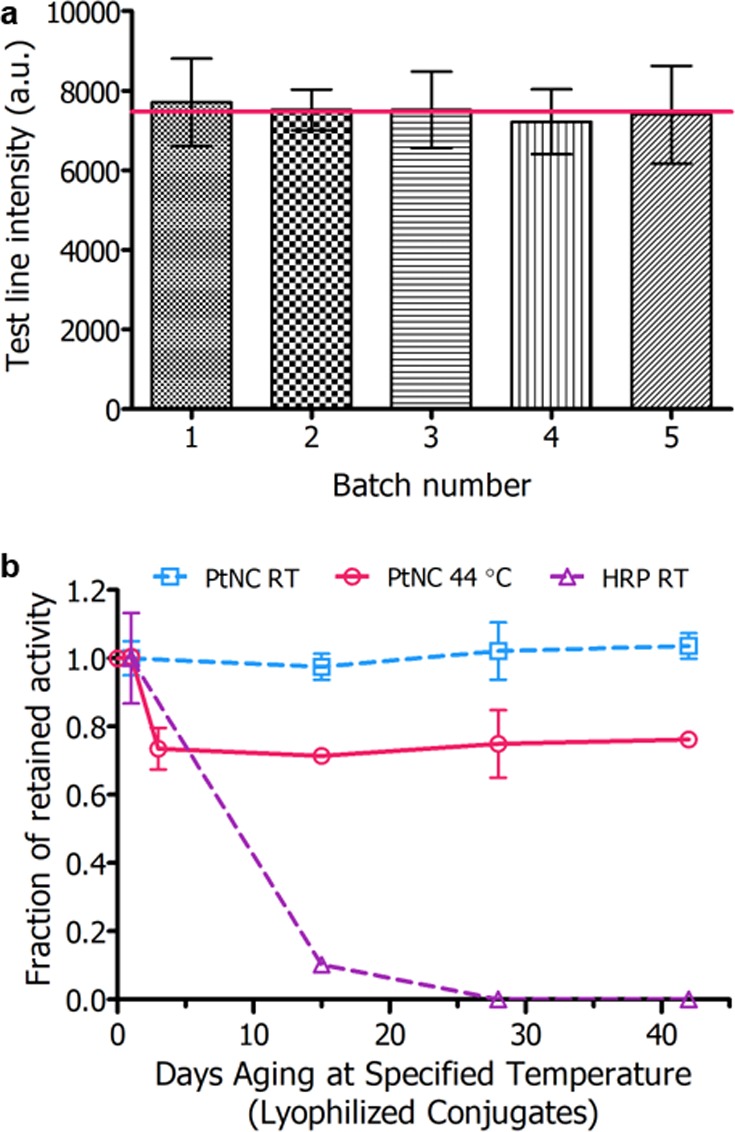
(a) Intensity of test line for five independently synthesized batches of PtNCs (ca. 120 nm) for detection of 50 pg·mL–1 p24 in FBS with 5 min development in CN/DAB and H2O2. Red line indicates average test line intensity for triplicate measurements across all batches. Coefficient of variation (n = 5) was 2.4%. (b) Intensity of test line (for detection of 500 pg·mL–1 p24-spiked plasma) against days of test component aging at 44 °C or room temperature (RT). For the aging experiment, nanobody–biotin and either PtNC or HRP–antibody conjugates were lyophilized in appropriate buffers and incubated at specified temperature. At aging time points, spiked plasma was added to the vessel of freeze-dried components and flowed up the LFIA test strip followed by catalytic amplification.
Stability is a key aspect of PoC devices where the performance of both the detection labels and the antibody components must be independent of the storage conditions the test might be exposed to. Many assays require controlled temperature storage conditions, which imposes cost on the supply chain and may prevent use in high temperature regions, thus limiting the test’s use in the developing world. Enzymes and antibody reagents are typically the most susceptible components to long-term storage, although with appropriate drying conditions and matrices, long-term dried storage is possible.36 Accelerated aging studies, shown in Figure 3b, were carried out on lyophilized reagents–comparing the stability of peroxidase-mimicking PtNC labels to their biological counterpart horseradish peroxidase (HRP), the most common enzyme used for catalytic amplification in immunoassays. For aging studies, nanobody–biotin and either antibody-conjugated HRP or PtNC were lyophilized in buffer mixtures containing sucrose, BSA, PVP, and Tween-20. After freeze-drying and aging at 44 °C or room temperature over a period of 6 weeks, p24 spiked plasma was added to the dried samples and flowed up the strip, and the test line intensity was quantified after catalytic amplification. PtNC-amplified LFIA showed no notable change in test line intensity over 6 weeks aging at room temperature, and only minimal change in test line intensity when incubated at 44 °C for 6 weeks (6-month room temperature aging equivalent). An initial ca. 25% decrease in signal intensity was observed after 3 days of incubation at 44 °C, after which no additional signal was lost over a 42-day period. In contrast, anti-HIV-1/2 antibody-functionalized HRP labels were found to lose 90% of signal intensity after 2 weeks of room temperature dry storage when run in LFIA (these samples did not withstand elevated temperature aging conditions). The resilience of these components is critical in determining the systems suitability for PoC use, especially in resource-limited settings where cold-chain transport is not guaranteed. In particular, this highlights the benefit of using robust inorganic catalytic nanoparticles in the place of conventional enzymes where the catalytic amplification obtained using the porous PtNCs is highly resilient to rapid aging. Conventional antibodies can be readily physisorbed onto metallic nanoparticles and show significant thermal stability when lyophilized. We also observe excellent long-term stability of the biorecognition components in our amplified LFIA, where we employ a combination of a conventional antibody and a biotinylated nanobody. However, we note that nanobodies exhibit significantly greater thermal stability than conventional immunoglobulins (Supplementary Figure 9) and we see great potential in pairing two nanobody components in this system to further improve the thermal stability for use in the most adverse conditions.37−41
The sensitivity of the nanocatalyst-labeled LFIA (employing ca. 120 nm PtNC labels) before and after catalysis is highlighted in Figure 4a with a plot of the signal intensity and the concentration of p24 in spiked sera before and after catalytic amplification. Test line signal intensity was captured by a mobile phone camera, a readily available tool for image acquisition. Unlike enzyme-labeled immunoassays which require catalysis for visualization, the PtNC-labeled LFIA achieves two windows of sensitivity, pre- and postcatalysis, taking advantage of both the absorbance of the particles and their peroxidase-mimicking activity (Supplementary Figure 10). This empowers the assay with a dual range diagnostic regime, quantifiable by use of mobile phone-based detection and image analysis. Our assay exhibits a linear dynamic range across over 4 orders of magnitude from 10000 to <1 pg·mL–1 protein target (Figure 4b) through measurement pre- and post-amplification. This can enable a broader potential quantitative range of the test if an image is taken before and after chromogen amplification. PtNC-amplified LFIA can enable the detection of p24 in spiked sera down to a level of 0.8 pg·mL–1 (ca. 32.5 fM) in under 20 min, which is within the clinically relevant range for [p24] during acute stage infection when the individual is most infectious but also when early intervention with antiretroviral therapies is most beneficial.17
Figure 4.
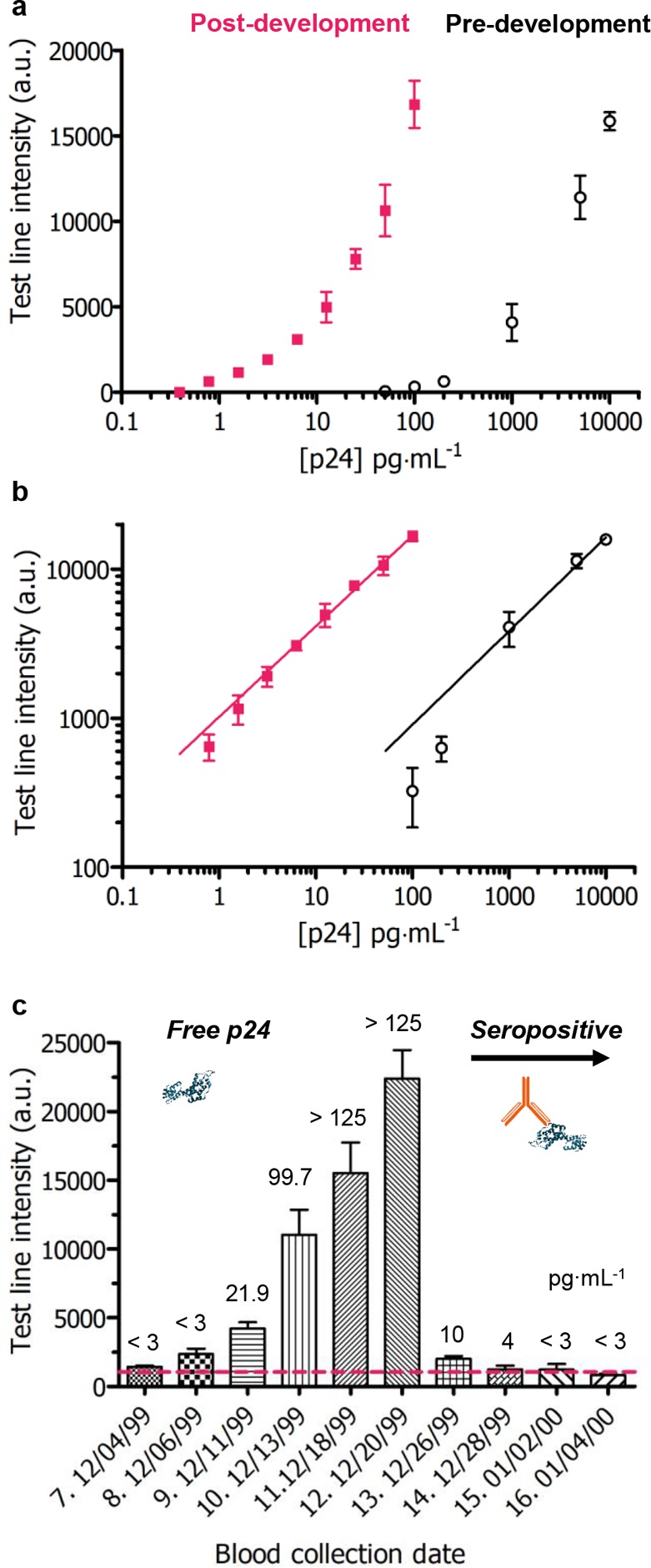
(a) Test line intensity obtained by mobile phone camera images for a dilution series of p24 spiked into sera before (black) and after amplification (red) in a mixture of CN/DAB and H2O2 for 5 min using ca. 120 nm PtNC labels. Error bars represent standard deviation from three independent experiments. (b) Illustration of the broad linear dynamic range across 4 orders of magnitude from 10000 to 1 pg·mL–1 achieved with the present method with nonlinear (log scale) regression least-squares fit (predevelopment: 0.62x + 1.7, R2 = 0.9813; postdevelopment: 0.61x + 3.0, R2 = 0.9853). (c) Clinical sample HIV seroconversion panel results for a single donor showing the change in test line intensity as a function of sample blood collection date covering the seroconversion period. 50 μL of each clinical sample was added to a vessel containing lyophilized test reagents and run up the LFIA strip followed by catalytic amplification. Red dotted line indicates cutoff (average test line intensity of 3 nonspiked blank plasma samples plus three times standard deviation). Error bars represent standard deviation from three independent experiments.
We evaluated the performance of the PtNC LFIA in clinical human plasma samples using an HIV seroconversion panel collected by ZeptoMetrix (Panel Donor No. 75062). Longitudinal clinical samples from the same donor spanning a one-month blood collection period encompassing seroconversion were assessed. In a blinded experiment, PtNC amplified LFIA could successfully detect positive antigen levels in infected plasma from ZeptoMetrix panel samples 7 through 14 corresponding to increasing levels of antigen in the blood during acute phase infection, followed by immune complex formation (Figure 4c). Panel samples 13 and 14 correspond to low levels of p24: 10.8 pg·mL–1 ∼ 211000 copies/mL HIV-1 RNA and 4.6 pg·mL–1 ∼ 46300 copies/mL, respectively (verified by ZeptoMetrix HIV p24 antigen ELISA and Roche Cobas Amplicor HIV-1 Monitor). A fourth-generation HIV LFIA test incorporates an antibody detection line to ensure the test can detect HIV infection across the seroconversion regime. We envisage a similar approach with the PtNC-amplified LFIA which would still benefit from the increased dynamic range for immune response monitoring but also exceptional levels of sensitivity toward p24 at the early stages of infection.
Conclusions
Here we report one of the simplest and most sensitive absorbance-based LFIA for protein detection to date, demonstrate its utility in a clinically significant range for p24 detection in spiked sera and plasma, and detection of acute phase HIV infection in serial clinical samples. We achieve sensitivity in a simple paper-based platform by pairing porous catalytic inorganic nanoparticles and thermally robust antibody fragments with easily modifiable functional groups for oriented presentation of the solid phase recognition elements. Capitalizing on the porous nature of the catalyst, we establish the optimal size regime for catalytic nanoparticles as amplifying labels in LFIA. We balance the high catalytic activity of the noble metal PtNCs toward peroxide disproportionation with the simultaneous tendency for biomolecules to adsorb and block the reactive surface by careful tailoring of PVP coatings and antibody conjugation. We demonstrate the utility of both nanobody and nanocatalyst components in LFIA to move toward a simple, robust, and highly amplifying system. The result is a PtNC amplified LFIA that is easy to use with a colorimetric output that can be read by the naked-eye or by mobile phone camera. The applications for this rapid test with broad dual dynamic range extend far beyond HIV diagnostics. This sensitive platform can be readily adapted for detection of other biomarkers across multiple stages of disease progression and monitoring of therapeutic efficacy. Our versatile and modular platform incorporates highly efficient inorganic catalytic labels into existing LFIA technology, enhancing its potential uptake into PoC diagnostics for the detection of any protein target.
Materials and Methods
Synthesis of Gold Nanoparticle Seeds
Gold nanoparticle seeds with a diameter of ca. 15 nm were synthesized by sodium citrate reduction of HAuCl4. In a typical synthesis, 10 mL of gold(III) chloride trihydrate aqueous solution (20 mM, Sigma) was added to 180 mL of purified water under reflux at 100 °C. The temperature was then reduced to 70 °C, and the reaction initiated by fast injection of 10 mL of trisodium citrate dihydrate (68 mM, BDH Chemicals) with vigorous stirring and refluxed for 5 min. The resulting ca. 15 nm gold nanoparticle (AuNP) seeds were cooled to room temperature and subsequently stored at 4 °C (Supplementary Figure 2). Seed concentration was determined by TEM size analysis and initial gold concentration.
Synthesis of Porous Platinum Core–Shell Nanoparticles (PtNCs)
120 nm PtNCs were synthesized via reduction of chloroplatinic acid hydrate on gold seeds adapted from Gao et al.(9) In a typical synthesis, 31 μL (10 nM) of 15 nm Au seeds were mixed with 969 μL of purified water (Milli-Q), followed by addition of 20 μL of 20 wt % poly(vinylpyrrolidone) (PVP MW 10 kDa, Sigma). The solution was vortexed briefly and incubated for 5 min for the polymer to coat and stabilize particles. l-Ascorbic acid (40 μL, 100 mg·mL–1, Sigma) was then added to the mixture followed by 40 μL of chloroplatinic acid hydrate (100 mM, Sigma), mixed, and immediately incubated at 65 °C for 30 min until the color of the solution changed from red to brown/black indicating successful deposition of platinum (Supplementary Figure 2). PtNCs were then cooled to room temperature in a water bath, and excess reagents were removed through four sequential washing cycles at 1250 rcf for 12 min with resuspension into purified water. PtNC size in the range of 50–120 nm was controlled by changing the volume of Au seed solution while keeping the amount of PVP, ascorbic acid, and H2PtCl6 constant.
PtNCs larger than 120 nm (mean number distribution by DLS) were synthesized via a stepwise layer-by-layer platinum reduction and deposition approach. 120 nm PtNCs were used as seeds for subsequent deposition of platinum layers iterating on the above synthesis to achieve colloidally stable PtNCs up to ca. 280 nm.
Characterization of Nanoparticles
Dynamic light scattering (DLS, Zeta Sizer Nanoseries, Malvern Instruments, Ltd.) was used to characterize the hydrodynamic radius of nanoparticles. Absorption measurements were recorded on a SpectraMax M5 multimode microplate reader (Molecular Devices, Ltd.). For electron microscopy characterization, samples were drop-casted onto carbon-coated copper grids (Electron Microscopy Sciences), and TEM imaging was performed using a JEOL 2100Plus or JEOL 2100F operating at 200 kV. High angle annular dark field (HAADF) images were acquired in scanning transmission electron microscopy (STEM) mode using a camera length of 10–12 cm (HAADF5) on JEOL 2100F. The microscope was equipped with Gatan Orius SC 1000 (2 × 4k) high-angle annular dark field detector, Gatan annular dark field detector/bright field detector. Small camera length ensured that diffraction contrast was excluded from the imaging and contrast was related to the atomic number (Z) or density of the particles. Corresponding elemental compositional mapping of PtNCs was determined by energy-dispersive X-ray spectroscopy (EDS) in STEM mode.
Preparation of PtNC–antibody Conjugates
In a typical conjugation, 200 μL of 120 nm PtNC (300 pM) was mixed with 20 μL of HEPES buffer (pH 6, 0.1 M) followed by addition of 2 μL of a 23 μM solution of monoclonal anti-HIV-1/2, purified (Capricorn Products, HIV-018-48303); optimization of [Ab]:[PtNC] is outlined in Supplementary Figure 3. The mixture was incubated for 3.5 h under shaking (800 rpm) at 20 °C for antibody physisorption. Modified particles were subsequently blocked by addition of 200 μL of blocking solution: 2 wt % β-casein from bovine milk (Sigma) in PBS for 1.5 h under shaking (800 rpm) at 20 °C. Excess reagents were removed through three wash steps in assay running buffer (0.2 wt % β-casein, 0.2 wt % Tween20 [Sigma] in 50 mM phosphate buffer pH 7.2). Particles were ultimately resuspended in particle storage buffer (10 wt % sucrose [Sigma], 1 wt % bovine serum albumin [BSA, Sigma], 1 wt % PVP 10 kDa, 0.1 wt % beta-casein, 0.1 wt % Tween20 in 50 mM phosphate buffer pH 7.2).
Horseradish peroxidase was also modified with anti-HIV-1/2 antibody following Lightning-Link Horseradish Peroxidase kit (Innova Biosciences) with no deviation from the manufacturer’s protocol.
Evaluation of Peroxidase-like Activity
All assays were carried out at room temperature in a 96-well plate (Corning UK). For stability and activity of PtNCs in complex environments, 10 μL of PtNCs or antibody modified PtNCs (300 pM) were mixed with 90 μL of either deionized water, 2 wt % β-casein in PBS, fetal bovine serum (FBS, Sigma), or human plasma pool (Lee Biosciences) for up to 24 h at room temperature. PtNCs were further diluted in purified water to 5 pM for catalytic activity test. For activity tests, 10 μL of PtNCs (5 pM) diluted in purified water were added to a microwell plate followed by addition of 200 μL of 1-Step Ultra TMB ELSA Substrate Solution (Thermo Scientific). The absorbance of the reaction solution at 652 nm was monitored up to 25 min after addition of substrate.
Production of Cysteine-Presenting Nanobody via Site-Directed Mutagenesis of an Existing Nanobody
The C-terminus of a validated nanobody against p24, 59H10, characterized in Gray et al., comprises an alanine spacer, myc and His6 tags.30 To insert a Cys residue in this region to generate ···HHHHHHCZ the following primers were used with the QuikChange Site-Directed Mutagenesis kit (Agilent) following the manufacturer’s instructions, verified by sequencing, and the resultant cysteine presenting nanobody purified as described previously.30
F: GCCGCACATCATCATCATCATCATTGCTAATGAGAATTCACTGG
R: CCAGTGAATTCTCATTAGCAATGATGATGATGATGATGTGCGGC
Mutations were verified by sequencing.
Site-Selective Modification of Nanobody
Site-selective modification of the cysteine presenting nanobody was achieved through reaction of the reduced nanobody with bromopyridazinedione-PEG8-biotin. Synthesis and characterization (NMR, IR, LRMS, HRMS) of pyridazinedione–biotin, and subsequent site-selective modification of the cysteine-mutated nanobody (shown by LCMS, SDS-PAGE) is described in the Supporting Information.
Characterization of Antibody Components
Affinity constants for antibody components were measured on Octet RED96 instrument (Pall ForteBio LLC, as previously described in Gray et al.).30
Preparation of PtNC-Amplified Lateral Flow Immunoassay
The lateral flow strips with polystreptavidin test line were manufactured by Mologic Ltd. (UK) (Batch Number ML-ICL-130317). The strips were 33 mm long and 5 mm wide. The distance between the test line and the bottom of the strip was 7 mm. Clinical utility of assay was tested on human plasma from HIV Seroconversion Panel Donor No. 75062 (ZeptoMetrix Corp., product code: HIV9079). Donor source plasma was collected from a U.S. licensed blood collection facility. Donor was untreated and asymptomatic throughout the collection period. A 50 μL portion of each panel sample was run on PtNC LFIA (outlined below).
All PtNC lateral flow assays for p24 detection were performed by submerging a polystreptavidin strip into a 96-well plate containing the following solution in each well: 10 μL of nanobody–biotin (200 nM), 50 μL p24 spiked into: FBS; plasma; or infected patient clinical plasma sample; and 15 μL PtNC antibody conjugate (300 pM). When the solution had fully wicked up the strip (ca. 10 min), the strip was moved into another well filled with 100 μL of assay running buffer for 3 min to wash through any residual serum (which may contain peroxidases that can also perform catalysis). Next, the strip was immersed in another well for 5 min filled with 330 μL (enough solution to cover test line on strip in well) freshly prepared PtNC development solution containing a modified Pierce CN/DAB (4-chloro-1-naphthol/3,3′-diaminobenzidine, tetrahydrochloride) Substrate Kit (Thermo Scientific) adjusted with hydrogen peroxide solution 30% (w/w) (Sigma) to a final added peroxide concentration of 4 M. Finally, the strip was moved into a well containing 330 μL purified water for 1 min to stop the reaction. Strips were imaged with an iPhone 6 mobile phone camera for all experiments, except the clinical samples, which were imaged using a camera (Canon PowerShot G15) approved for use in contained environment for handling infectious samples. Images of strips were acquired 5 min after removal from the water.
Preparation of Aged Samples for Accelerated Stability Testing
An accelerated aging test was carried out to simulate real-time shelf life aging. All aging experiments were performed in a nonbinding 96-well plate (Corning, UK). In each well, 15 μL of either PtNC–antibody conjugate (300 pM) or HRP–antibody conjugate (50 nM) in particle storage buffer was added, followed by 10 μL nanobody–biotin (200 nM) and 50 μL of particle storage buffer. Plates were immediately frozen at −80 °C for 30 min and freeze-dried overnight. When removed from the freeze-dryer, plates were stored in a sealed plastic bag filled with silica bead desiccant at either room temperature or 44 °C for accelerated aging study. Aging at 44 °C for ca. 42 days is functionally equivalent to room temperature (23 °C) aging for 6 months. Aging conditions were selected following guidelines of ASTM F1980 Standard Guide for Accelerated Aging of Sterile Medical Device Packages and Westpak Accelerated Aging Time Calculator.42,43 Accelerated aging techniques are based on the assumption that the chemical reaction involved in the deterioration of materials follow the Arrhenius reaction rate function, which states that a 10 °C increase in temperature will cause the reaction rate to double.
To examine long-term stability of reagents, 50 μL spiked serum or plasma was added to freeze-dried reagents. After dissolution of reagents in sample, strip was inserted into the well following above protocol for wash and development.
Structural Model of Nanobody
The structural model was predicted using the i-tasser Web server, http://zhanglab.ccmb.med.umich.edu/I-TASSER/, for the sequence provided.44
Image Analysis
Images of lateral flow strips were acquired using an Apple iPhone 6 and analyzed with ImageJ software.45 ImageJ Gel Analyzer plug-in was used to plot intensity profiles of LFIA strips and analyze test line intensities (Supplementary Figure 11).
Acknowledgments
C.N.L., M.R.T., E.R.G, D.A.R, B.S.M., J.C.B, V.C., R.A.M., and M.M.S acknowledge support from the i-sense Engineering and Physical Sciences Research Council (EPSRC) IRC in Early Warning Sensing Systems for Infectious Diseases (EP/K031953/1; www.i-sense.org.uk). M.R.T., R.A.M., and M.M.S. acknowledge support from the Medical Research Council (MRC) grant “m-Africa” (MR/P024378/1). C.N.L. acknowledges support from the Marshall Aid Commemoration Commission. M.M.S. acknowledges support from the ERC Seventh Framework Programme Consolidator grant “Naturale CG” under grant agreement no. 616417 and the EPSRC grant “Bio-functionalised nanomaterials for ultrasensitive biosensing” [EP/K020641/1]. R.A.M. acknowledges the Royal Society Wolfson Research Merit Award and the National Institute for Health Research University College London Hospitals Biomedical Research Centre. B.S.M acknowledges the UCL studentship. D.A.R. and V.C. gratefully acknowledges EPSRC (EP/M01792X/1). We acknowledge use of characterization facilities within the Harvey Flower Electron Microscopy Suite, Department of Materials, Imperial College London. We thank Y. Lin, C. Parolo, and C. Spicer for insightful discussions. We also thank S. Hemmington, D. Hall, and J. Denton (Mologic, Ltd.) for the preparation of dipstick test strips. We acknowledge T. Verrips (QVQ) and R. Weiss for access to 59H10 nanobody. Raw data is available online at DOI: doi: 10.5281/zenodo.1098208..
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b06229.
Characterization of nanocatalysts (reaction conditions, TEM, UV–vis, HAADF-STEM, EDS, DLS), optimization of antibody modification and lateral flow parameters, details of image analysis, characterization of nanobody and site-selective modification (synthesis procedure, NMR, LCMS), and affinity constants for biorecognition components (PDF)
Author Contributions
⊥ C.N.L. and M.R.T. contributed equally.
The authors declare the following competing financial interest(s): M.R.T., C.N.L., and M.M.S. are inventors on the priority patent application number 1703818.3.
Supplementary Material
References
- O'Farrell B. O. Lateral Flow Immunoassays Systems: Evolution from the Current State of the Art to the next Generation of Highly Sensitive, Quantitative Rapid Assays. Immunoassay Handbook 2013, 89–107. 10.1016/B978-0-08-097037-0.00007-5. [DOI] [Google Scholar]
- Niemeyer C. M.; Adler M.; Wacker R. Detecting Antigens by Quantitative Immuno-PCR. Nat. Protoc. 2007, 2, 1918–1930. 10.1038/nprot.2007.267. [DOI] [PubMed] [Google Scholar]
- Barletta J. M.; Edelman D. C.; Constantine N. T. Lowering the Detection Limits of HIV-1 Viral Load Using Real-Time Immuno-PCR for HIV-1 p24 Antigen. Am. J. Clin. Pathol. 2004, 122, 20–27. 10.1309/529T2WDNEB6X8VUN. [DOI] [PubMed] [Google Scholar]
- Mauk M.; Song J.; Bau H. H.; Gross R.; Bushman F. D.; Collman R. G.; Liu C. Miniaturized Devices for Point of Care Molecular Detection of HIV. Lab Chip 2017, 17, 382–394. 10.1039/C6LC01239F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.; Blumenfeld N. R.; Laksanasopin T.; Sia S. K. Point-of-Care Diagnostics: Recent Developments in a Connected Age. Anal. Chem. 2017, 89, 102–123. 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.; Zhuang J.; Nie L.; Zhang J.; Zhang Y.; Gu N.; Wang T.; Feng J.; Yang D.; Perrett S.; et al. Intrinsic Peroxidase-like Activity of Ferromagnetic Nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- Duan D.; Fan K.; Zhang D.; Tan S.; Liang M.; Liu Y.; Zhang J.; Zhang P.; Liu W.; Qiu X.; et al. Nanozyme-Strip for Rapid Local Diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- Kim M.; Kim M. S.; Kweon S. H.; Jeong S.; Kang M. H.; Kim M. Il; Lee J.; Doh J. Simple and Sensitive Point-of-Care Bioassay System Based on Hierarchically Structured Enzyme-Mimetic Nanoparticles. Adv. Healthcare Mater. 2015, 4, 1311–1316. 10.1002/adhm.201500173. [DOI] [PubMed] [Google Scholar]
- Gao Z.; Xu M.; Lu M.; Chen G.; Tang D. Urchin-like (Gold Core)@(platinum Shell) Nanohybrids: A Highly Efficient Peroxidase-Mimetic System for in Situ Amplified Colorimetric Immunoassay. Biosens. Bioelectron. 2015, 70, 194–201. 10.1016/j.bios.2015.03.039. [DOI] [PubMed] [Google Scholar]
- Gao Z.; Ye H.; Tang D.; Tao J.; Habibi S.; Minerick A.; Tang D.; Xia X. Platinum Decorated Gold Nanoparticles with Dual Functionalities for Ultrasensitive Colorimetric In Vitro Diagnostics. Nano Lett. 2017, 17, 5572–5579. 10.1021/acs.nanolett.7b02385. [DOI] [PubMed] [Google Scholar]
- Xia X.; Zhang J.; Lu N.; Kim M. J.; Ghale K.; Xu Y.; McKenzie E.; Liu J.; Ye H. Pd-Ir Core-Shell Nanocubes: A Type of Highly Efficient and Versatile Peroxidase Mimic. ACS Nano 2015, 9, 9994–10004. 10.1021/acsnano.5b03525. [DOI] [PubMed] [Google Scholar]
- Branson B. M.; Stekler J. D. Detection of Acute HIV Infection: We Can’t Close the Window. J. Infect. Dis. 2012, 205, 521–524. 10.1093/infdis/jir793. [DOI] [PubMed] [Google Scholar]
- Rosenberg N. E.; Pilcher C. D.; Busch M. P.; Cohen M. S. How Can We Better Identify Early HIV Infections?. Curr. Opin. HIV AIDS 2015, 10, 61–68. 10.1097/COH.0000000000000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher C. D.; Louie B.; Facente S.; Keating S.; Hackett J.; Vallari A.; Hall C.; Dowling T.; Busch M. P.; Klausner J. D.; et al. Performance of Rapid Point-of-Care and Laboratory Tests for Acute and Established HIV Infection in San Francisco. PLoS One 2013, 8, e80629. 10.1371/journal.pone.0080629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandori M. W.; Hackett J.; Louie B.; Vallari A.; Dowling T.; Liska S.; Klausner J. D. Assessment of the Ability of a Fourth-Generation Immunoassay for Human Immunodeficiency Virus (HIV) Antibody and p24 Antigen to Detect Both Acute and Recent HIV Infections in a High-Risk Setting. J. Clin. Microbiol. 2009, 47, 2639–2642. 10.1128/JCM.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone P. D.; Bartlett A. H. Diagnosing Acute HIV Infection. Pediatr. Ann. 2017, 46, e47–e50. 10.3928/19382359-20170118-01. [DOI] [PubMed] [Google Scholar]
- Rutstein S. E.; Ananworanich J.; Fidler S.; Johnson C.; Sanders E. J.; Sued O.; Saez-cirion A.; Pilcher C. D.; Fraser C.; Cohen M. S. Clinical and Public Health Implications of Acute and Early HIV Detection and Treatment: A Scoping Review. J. Int. AIDS Soc. 2017, 20, 21579. 10.7448/IAS.20.1.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tienen C.; Rugebregt S.; Scherbeijn S.; Götz H.; GeurtsvanKessel C. The Performance of the Alere HIV Combo Point-of-Care Test on Stored Serum Samples; Useful for Detection of Early HIV-1 Infections?. Sex. Transm. Infect. 2017, sextrans-2016-052818. 10.1136/sextrans-2016-052818. [DOI] [PubMed] [Google Scholar]
- Miedouge M.; Grèze M.; Bailly A.; Izopet J. Analytical Sensitivity of Four HIV Combined Antigen/antibody Assays Using the p24 WHO Standard. J. Clin. Virol. 2011, 50, 57–60. 10.1016/j.jcv.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Vallefuoco L.; Mazzarella C.; Portella G. Fourth Generation Assays for HIV Testing. Expert Rev. Mol. Diagn. 2016, 16, 723–732. 10.1080/14737159.2016.1179115. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Garcia-D’Angeli A.; Brennan J. P.; Huo Q. Predicting Detection Limits of Enzyme-Linked Immunosorbent Assay (ELISA) and Bioanalytical Techniques in General. Analyst 2014, 139, 439–445. 10.1039/C3AN01835K. [DOI] [PubMed] [Google Scholar]
- Gonzalez M.; Bagatolli L. A.; Echabe I.; Arrondo J. L. R.; Argarana C. E.; Cantor C. R.; Fidelio G. D. Interaction of Biotin with Streptavidin. J. Biol. Chem. 1997, 272, 11288–11294. 10.1074/jbc.272.17.11288. [DOI] [PubMed] [Google Scholar]
- Holstein C. A.; Chevalier A.; Bennett S.; Anderson C. E.; Keniston K.; Olsen C.; Li B.; Bales B.; Moore D. R.; Fu E.; et al. Immobilizing Affinity Proteins to Nitrocellulose: A Toolbox for Paper-Based Assay Developers. Anal. Bioanal. Chem. 2016, 408, 1335–1346. 10.1007/s00216-015-9052-0. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wu H.; Li M.; Yin J.-J.; Nie Z. pH Dependent Catalytic Activities of Platinum Nanoparticles with Respect to the Decomposition of Hydrogen Peroxide and Scavenging of Superoxide and Singlet Oxygen. Nanoscale 2014, 6, 11904–11910. 10.1039/C4NR03848G. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Xia X.; Peng H. C. Shape-Controlled Synthesis of Colloidal Metal Nanocrystals: Thermodynamic versus Kinetic Products. J. Am. Chem. Soc. 2015, 137, 7947–7966. 10.1021/jacs.5b04641. [DOI] [PubMed] [Google Scholar]
- Ye H.; Liu Y.; Chhabra A.; Lilla E.; Xia X. Polyvinylpyrrolidone (PVP)-Capped Pt Nanocubes with Superior Peroxidase-Like Activity. ChemNanoMat 2017, 3, 33–38. 10.1002/cnma.201600268. [DOI] [Google Scholar]
- Panchenko A.; Koper M. T. M.; Shubina T. E.; Mitchell S. J.; Roduner E. Ab Initio Calculations of Intermediates of Oxygen Reduction on Low-Index Platinum Surfaces. J. Electrochem. Soc. 2004, 151, A2016–A2027. 10.1149/1.1809586. [DOI] [Google Scholar]
- Richards D. A.; Maruani A.; Chudasama V. Antibody Fragments as Nanoparticle Targeting Ligands: A Step in the Right Direction. Chem. Sci. 2017, 8, 63–77. 10.1039/C6SC02403C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer T.; Muyldermans S.; Depicker A. Nanobody-Based Products as Research and Diagnostic Tools. Trends Biotechnol. 2014, 32, 263–270. 10.1016/j.tibtech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Gray E. R.; Brookes J. C.; Caillat C.; Turbé V.; Webb B. L. J.; Granger L. A.; Miller B. S.; McCoy L. E.; El Khattabi M.; Verrips C. T.; et al. Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights. ACS Infect. Dis. 2017, 3, 479–491. 10.1021/acsinfecdis.6b00189. [DOI] [PubMed] [Google Scholar]
- Chudasama V.; Smith M. E. B.; Schumacher F. F.; Papaioannou D.; Waksman G.; Baker J. R.; Caddick S. Bromopyridazinedione-Mediated Protein and Peptide Bioconjugation. Chem. Commun. 2011, 47, 8781. 10.1039/c1cc12807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson E.; Nunes J. P. M.; Vassileva V.; Maruani A.; Nogueira J. C. F.; Smith M. E. B.; Pedley R. B.; Caddick S.; Baker J. R.; Chudasama V. Pyridazinediones Deliver Potent, Stable, Targeted and Efficacious Antibody–drug Conjugates (ADCs) with a Controlled Loading of 4 Drugs per Antibody. RSC Adv. 2017, 7, 9073–9077. 10.1039/C7RA00788D. [DOI] [Google Scholar]
- Lee M. T. W.; Maruani A.; Richards D. A.; Baker J. R.; Caddick S.; Chudasama V. Enabling the Controlled Assembly of Antibody Conjugates with a Loading of Two Modules without Antibody Engineering. Chem. Sci. 2017, 8, 2056–2060. 10.1039/C6SC03655D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. T. W.; Maruani A.; Baker J. R.; Caddick S.; Chudasama V. Next-Generation Disulfide Stapling: Reduction and Functional Re-Bridging All in One. Chem. Sci. 2016, 7, 799–802. 10.1039/C5SC02666K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene M. K.; Richards D. A.; Nogueira J. C. F.; Campbell K.; Smyth P.; Fernandez M.; Scott C. J.; Chudasama V. Forming next-generation antibody–nanoparticle conjugates through the oriented installation of non-engineered antibody fragments. Chem. Sci 2018, 9, 79–87. 10.1039/C7SC02747H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild D. G.Immunoassay Handbook - Theory and Applications of Ligand Binding, ELISA and Related Techniques; 4th ed.; Elsevier, 2013. [Google Scholar]
- Ramachandran S.; Fu E.; Lutz B.; Yager P. Long-Term Dry Storage of an Enzyme-Based Reagent System for ELISA in Point-of-Care Devices. Analyst 2014, 139, 1456–1462. 10.1039/C3AN02296J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Linden R. H.; Frenken L. G.; de Geus B.; Harmsen M. M.; Ruuls R. C.; Stok W.; de Ron L.; Wilson S.; Davis P.; Verrips C. T. Comparison of Physical Chemical Properties of Llama VHH Antibody Fragments and Mouse Monoclonal Antibodies. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1999, 1431, 37–46. 10.1016/S0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Gorlani A.; Brouwers J.; McConville C.; van der Bijl P.; Malcolm K.; Augustijns P.; Quigley A. F.; Weiss R.; De Haard H.; Verrips T. Llama Antibody Fragments Have Good Potential for Application as HIV Type 1 Topical Microbicides. AIDS Res. Hum. Retroviruses 2012, 28, 198–205. 10.1089/aid.2011.0133. [DOI] [PubMed] [Google Scholar]
- Saerens D.; Conrath K.; Govaert J.; Muyldermans S. Disulfide Bond Introduction for General Stabilization of Immunoglobulin Heavy-Chain Variable Domains. J. Mol. Biol. 2008, 377, 478–488. 10.1016/j.jmb.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Dolk E.; Van Vliet C.; Perez J. M. J.; Vriend G.; Darbon H.; Ferrat G.; Cambillau C.; Frenken L. G. J.; Verrips T. Induced Refolding of a Temperature Denatured Llama Heavy-Chain Antibody Fragment by Its Antigen. Proteins: Struct., Funct., Genet. 2005, 59, 555–564. 10.1002/prot.20378. [DOI] [PubMed] [Google Scholar]
- Yamada K.; Shibata H.; Suzuki K.; Citterio D. Toward Practical Application of Paper-Based Microfluidics for Medical Diagnostics: State-of-the-Art and Challenges. Lab Chip 2017, 17, 1206–1249. 10.1039/C6LC01577H. [DOI] [PubMed] [Google Scholar]
- ASTM. Standard Guide for Accelerated Aging of Sterile Medical Device Packages. ASTM F1980-02, 2002.
- Yang J.; Yan R.; Roy A.; Xu D.; Poisson J.; Zhang Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.