Abstract
Non-coding uridine-rich small nuclear RNAs (UsnRNAs) have emerged in recent years as effective tools for exon skipping for the treatment of Duchenne muscular dystrophy (DMD), a degenerative muscular genetic disorder. We recently showed the high capacity of a recombinant adeno-associated virus (rAAV)-U7snRNA vector to restore the reading frame of the DMD mRNA in the muscles of DMD dogs. We are now moving toward a phase I/II clinical trial with an rAAV-U7snRNA-E53, carrying an antisense sequence designed to hybridize exon 53 of the human DMD messenger. As observed for genome-editing tools, antisense sequences present a risk of off-target effects, reflecting partial hybridization onto unintended transcripts. To characterize the clinical antisense sequence, we studied its expression and explored the occurrence of its off-target effects in human in vitro models of skeletal muscle and liver. We presented a comprehensive methodology combining RNA sequencing and in silico filtering to analyze off-targets. We showed that U7snRNA-E53 induced the effective exon skipping of the DMD transcript without inducing the notable deregulation of transcripts in human cells, neither at gene expression nor at the mRNA splicing level. Altogether, these results suggest that the use of the rAAV-U7snRNA-E53 vector for exon skipping could be safe in eligible DMD patients.
Keywords: exon skipping, off-target, RNA sequencing, Duchenne, U7snRNA
Introduction
Exon skipping is a therapeutic strategy relying, within an mRNA harboring a deleterious mutation, on the specific non-recognition of exon(s) by the splicing machinery and the subsequent restoration of a functional open reading frame (ORF). The mechanism behind exon skipping is the use of antisense sequences that typically target and hide recognition splice sites (donor or acceptor splice sites, branch points, or splicing regulatory sequences) from the spliceosome. The resulting skipped mRNA exhibits a modified reading frame which leads to the removal of a frameshift and restoration of a protein-of-interest expression, or on the contrary, which leads to the appearance of a premature stop codon causing the degradation of a targeted pre-mRNA through nonsense-mediated RNA decay (NMD).1, 2, 3
The most currently developed antisense sequences are synthetic, chemically modified antisense oligonucleotides (AONs), which are short single-stranded DNA or RNA-like molecules and which required regular (usually weekly) injections, even if their half-life is currently improved.4 The development and the optimization of their synthesis have led to a wide array of AON chemistries for the treatment of various disorders, including Duchenne muscular dystrophy (DMD).5, 6, 7 Two synthetic AONs, Drisapersen and Eteplirsen, indeed showed a great potential for DMD via skipping of the exon 51 of the DMD messenger. They have reached the final clinical phases in DMD patients.6, 7 However, in early 2016, the US Food and Drug Administration (FDA) rejected Drisapersen because of insufficient evidence of clinical efficacy associated with serious safety issues.8 Finally, in September 2016, after intensive hesitation, the FDA decided to give an accelerated approval for Eteplirsen, but future trials will be required to confirm its clinical benefit.8
Exon skipping can also be achieved through the expression of in vivo antisense sequences cloned in genes of uridine-rich small nuclear RNA (UsnRNA). U1 and U7snRNAs have previously been used as carriers of antisense sequences to correct splicing in models of several diseases, including β-thalassemia, DMD, HIV infections, spinal muscular atrophy (SMA), and cardiomyopathies.9, 10, 11, 12, 13, 14 When inserted in UsnRNA, antisense sequences can be continuously produced in the targeted tissue and can accumulate in the nuclear compartment, being protected from damages by their inclusion in snRNP (small nuclear ribonucleoprotein particle).15 U7snRNA, the most frequently used snRNA in exon skipping approaches, is a nonspliceosomal snRNA involved in the 3′ maturation of histone pre-mRNAs.16 The optimization of the specific Sm-binding site17 facilitated conversion from a nonspliceosomal to a spliceosomal snRNA and the use of this molecule as an antisense tool to modify splicing.9, 10 The small size of U7snRNA facilitates the incorporation of this molecule into recombinant vectors derived from adeno-associated virus (rAAV). Even if some immune responses against these vectors can sometimes impede their long-term action,18 they remain today very attractive tools to obtain an efficient gene transfer and stable expression of the transgene over time, after one single injection in the targeted tissue.19, 20 We, among others, have evaluated the use of an rAAV-U7snRNA-based strategy for exon skipping as a potential treatment for DMD.10, 21, 22, 23
DMD is a severe X-linked neuromuscular disorder that affects approximately 1 in 5,000 males at birth.24 This disease is characterized by the progressive degeneration of all the skeletal muscle tissues, eventually reaching the diaphragm and the cardiac muscle, and leading to early death prior to the fourth decade.25 DMD results from mutations in the DMD gene encoding dystrophin, a cytoskeleton protein essential for the integrity of muscular fibers.26
Recently, we demonstrated that an rAAV8-U7snRNA vector restores the expression and function of the dystrophin protein in the muscles of golden retriever muscular dystrophy (GRMD) dogs.23 These results prompted a phase I/II clinical trial in DMD patients treatable through exon skipping. In dogs, exons 6 and 8 were targeted to restore the reading frame by exon skipping. Because exon skipping reaction is specific to the targeted sequences, and thus of each species, a specific antisense sequence has been designed to target the human DMD mRNA. Because 8%–13.5% of DMD patients carrying a genomic deletion within their DMD gene are eligible for the skipping of exon 53 (deletion spanning exons 52, 45–52, 47–52, 48–52, 59–52, and 50–52),27 we developed a recombinant AAV vector (rAAV8-U7snRNA-E53) carrying an antisense sequence targeting exon 53 of the human DMD messenger as a therapeutic product for DMD patients.
The permanent and abundant presence of the therapeutic U7snRNA could result in the partial hybridization to undesired targets, leading to unexpected side effects. Therefore, during the pre-clinical translation of this therapeutic approach, we considered it mandatory to characterize: (1) the expression profile of the therapeutic transgene in the targeted tissue (skeletal muscles) and in non-targeted tissues (especially the liver), and (2) the potential off-target events in these tissues.
Off-target effects have been extensively studied and were observed after the use of reverse genetics tools, such as small interfering RNAs (siRNAs),28 or genome-editing tools, such as ZFNs (zing finger nucleases),29 the recently developed CRISPR/Cas9 system,30 and to a lesser extent, TALENs (transcription activator-like effector nucleases).31 However, the risk of off-target effects of AON-based tools, historically considered as low, has not been extensively investigated. Among the very few published studies, the results differ showing alternatively a high off-target rate32 or no off-targets effects.33, 34, 35 The potential off-target effects have been even less investigated for the antisense sequences used for exon skipping.34 Moreover, the methodologies used in off-target studies largely comprised microarrays or molecular validations of potential off-target sites predicted in silico. To assess whether the antisense sequences designed for exon skipping could induce sequence-specific off-targets, comprehensive studies and high-throughput methodologies are needed. Here, we demonstrated the expression and efficacy to induce exon skipping of the clinical transgene U7snRNA-E53 in muscular and hepatic human materials. We next set up an exhaustive methodology combining RNA sequencing (RNA-seq) and in silico filtering to identify the sequence-specific off-target effects of the transgene in human cells. Using this approach, we did not detect any major modification of splicing, except for DMD, nor the modification of gene expression associated with U7-E53 expression in both muscular and hepatic human contexts. These results suggest that rAAV-U7snRNA-E53 is a safe therapeutic potential for the treatment of DMD patients.
Results
The U7snRNA-E53 RNA Is Expressed Both in Muscular and in Hepatic Human Cells
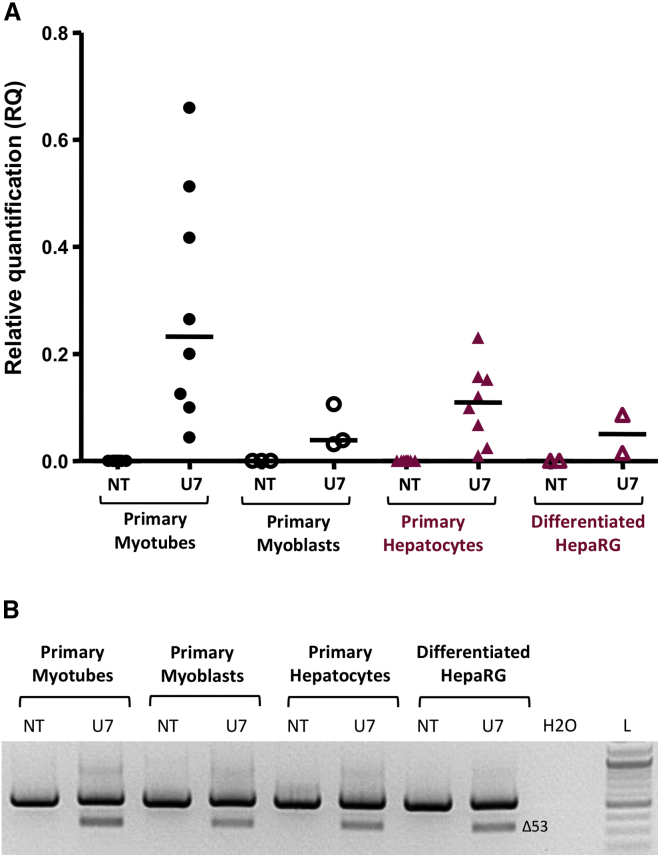
To analyze the expression pattern, the efficacy to induce exon skipping, and the off-target effects of our therapeutic product (rAAV-U7-E53) in muscular and hepatic human cells, we developed relevant cellular models that were subsequently transduced with rAAV-U7-E53. We used human primary hepatocytes from healthy subjects and HepaRG cells, a hepatoma human cell line that differentiates in hepatocyte-like cells and represents a relevant surrogate to primary cells.36 We also used human primary myoblasts and differentiated myotubes obtained from healthy subjects. The muscular or hepatic transcriptomic pattern of each cell type was confirmed by RT-PCR analyses of some muscle-specific genes (myosin [MYHC3] and myogenin [MYOG] for myotubes and myoD [MYOD1] for myoblasts; Figure S1A) or liver-specific genes (albumin [ALB], aldolase B [ALDOB], CYP2C9, and CYP3A4; Figure S1B). As expected, the expression of MYHC3 and MYOG mRNAs were observed only in primary myotubes, while MYOD mRNA was solely expressed in primary myoblasts (Figure S1A). At the protein level, the expression of the myosin heavy chain was confirmed in human primary myotubes using immunofluorescence analysis (Figure S2). All primary hepatocyte samples and differentiated HepaRG cells exhibited the expression of ALB, ALDOB, and CYP2C9 mRNA. CYP3A4 gene expression was weaker and detected in only two of the three batches of primary hepatocytes (Figure S1B), as previously described.36 Knowing that rAAV serotypes 2 and 8 led to poor transduction levels of human primary hepatocytes, we used the rAAV serotype 3b,37 which efficiently transduced both human primary myoblasts and hepatocytes, with respectively ≈90% and ≈75% of GFP-positive cells 3 days after transduction at MOI 2 × 105 (Figure S3). Several different batches of human primary myoblasts and hepatocytes were then transduced with the rAAV3b-U7-E53 vector. Three days after transduction, we confirmed that the muscular and hepatic patterns of the cells were not modified through the transduction event, confirming the relevance of the cellular models used for this study (Figure S1). To determine whether the transduction of rAAV3b-U7-E53 drove the expression of U7-E53 RNA in different cell types, we assessed the U7-E53 RNA levels using qRT-PCR (Figure 1A). The U7-E53 RNA was detected in transduced human primary myoblasts and myotubes, human primary hepatocytes, and differentiated HepaRG cells, with the highest levels detected in primary myotubes and primary hepatocytes. To examine the activity of U7-E53 RNA on the human DMD messenger, we analyzed the region of DMD surrounding exon 53 using specific nested RT-PCR. An RT-PCR product of 556 base pairs (bp) corresponding to the inclusion of exon 53 in the DMD messenger was observed in non-transduced (NT) samples. Following the transduction of rAAV3b-U7-E53, a RT-PCR product of 344 bp was detected in myotubes and hepatic samples corresponding to an mRNA devoid of exon 53 (confirmed by sequencing of the PCR products) (data not shown) (Figure 1B). These results demonstrated that U7-E53 RNA induces the skipping of exon 53 in the DMD mRNA, and that rAAV3b-U7-E53 is able to drive the expression of active U7-E53 RNA in tissues other than human skeletal muscle and particularly in the liver, a non-targeted tissue.
Figure 1.
Analysis of U7-E53 RNA Expression and Activity on DMD mRNA Splicing in Human Cellular Models
(A) Relative quantification of U7-E53 RNA in human cells through qRT-PCR. The results are represented as scattered dots and median value (central bar) of U7-E53 RNA relative quantification (RQ) in human primary myotubes (n = 8), human primary myoblasts (n = 3), human primary hepatocytes (n = 8), and differentiated HepaRG cells (n = 2) non-transduced (NT) or transduced with rAAV3b-U7-E53 vector (U7). (B) Analysis of exon skipping of the DMD mRNA in human cells using nested RT-PCR. One representative result is presented for each cell type. The DMD messenger was amplified from exons 51–54. The RT-PCR product (556 bp) detected in non-transduced samples (NT) corresponds to DMD mRNA with the exon 53. After transduction with rAAV3b-U7-E53 vector (U7), the new RT-PCR product (344 bp) detected corresponds to out-of-frame mRNA lacking exon 53. H2O, water; L, DNA ladder.
Identification of U7-E53 Off-Target Events through RNA-Seq and In Silico Target Predictions
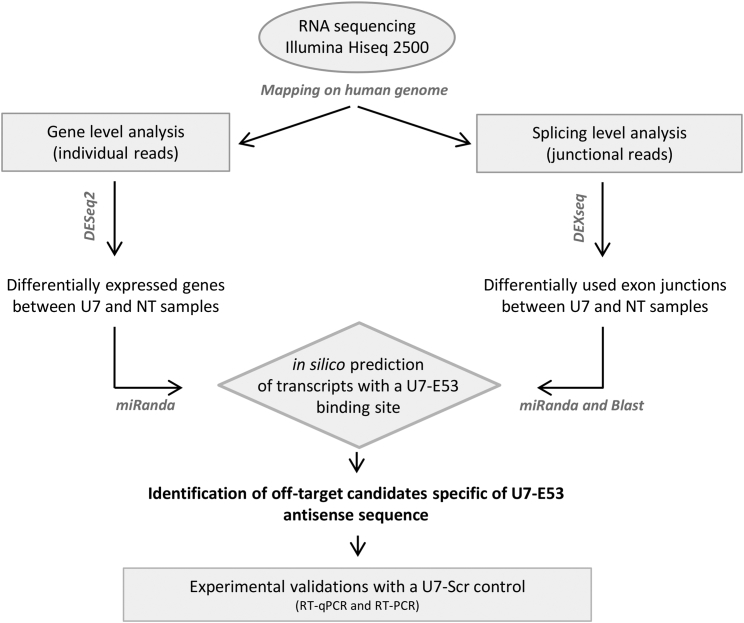
We previously showed through biodistribution analysis of the vector in GRMD dogs that skeletal muscle and liver were the most transduced tissues after locoregional injection in one forelimb of a therapeutic rAAV8-U7snRNA vector.23 Because we showed in the present study that U7-E53 is expressed and active in human muscle and hepatic cells, off-target events could therefore occur in both tissues in humans. The antisense sequence harbored within U7-E53 is 40 bases long and designed to specifically target a sequence of exon 53 in the human DMD pre-mRNA. However, cross-hybridization to other human transcripts, even incomplete, could lead to splicing alterations favoring exon skipping or inclusion, or to binding to mature mRNAs inducing the modification of expression levels through, for example, an siRNA-like effect or competition with the binding of regulatory factors. To determine the overall off-target effects induced through U7-E53 expression on the human transcriptome, we performed an exhaustive RNA-seq analysis of transduced and control cells, combined with the in silico identification of potential off-target sites (Figure 2). We focused on human primary myotubes and human primary hepatocytes, the most pertinent cellular models previously used for U7-E53 RNA analysis. Three independent biological replicates were analyzed for each cell type: three batches of human primary myotubes from three different healthy subjects (MT_1, MT_2, and MT_3) and three batches of human primary hepatocytes from three other different subjects (HEP_1, HEP_2, and HEP_3), each batch being NT or transduced with the rAAV3b-U7-E53 vector. The cells were harvested at 3 days after transduction for RNA-seq analysis. The cDNA libraries were generated and sequenced as stranded, paired-end 2 × 100 base reads, a protocol particularly adapted to identify splicing variants. Reads mapping to the human genome (GRCh38 primary assembly) were obtained with STAR 2.4.2a.38 A high proportion of the reads (86.82%–93.81% including spliced reads) were uniquely aligned to the human genome (Table S1).
Figure 2.
Global Strategy for Off-Target Analysis through RNA Sequencing
Reads were uniquely mapped on the human genome (hg38). Differentially expressed genes were identified using DESeq2. Junctional read dataset was analyzed with DEX-seq to quantify the differentially used junctions. Alignment tools miRanda and Blast enable in silico predictions of U7-E53 binding sites. Off-target gene candidates with a splicing modification or a differential expression were defined after comparing the in silico results and DEX-seq or DEseq2 results, respectively. Experimental validations with samples transduced using a rAAV3b-U7-E53 or a rAAV3b-U7-Scramble vector enabled the final identification of U7-E53-specific off-target events.
To address U7-E53 off-target effects, we analyzed two different aspects of gene expression: (1) global gene expression variations between transduced and NT samples were compared in a gene-level analysis with DESeq2;39 and (2) splicing modifications based on differential junction usage were determined with DEX-seq.40 This last analysis was performed after considering only junctional reads, i.e., reads mapping onto exon-exon junctions41 (Figure 2).
Because we compared rAAV3b-U7-E53 transduced samples with NT samples, part of the observed differential expression either at the gene or junction levels could reflect a non-specific effect of rAAV transduction alone. To specifically focus on U7-E53 off-targets, we performed in silico filtrations of all transcripts containing a predictive degenerated binding site for the antisense sequence harbored in the U7-E53, using the alignment tools miRanda and Blast for splicing analysis and miRanda alone for gene analysis. RNA-seq results and in silico predictions were subsequently compared with the final set of off-targets candidates, most likely specific for the U7-E53 (Figure 2). Moreover, experimental validations were performed through qRT-PCR or nested RT-PCR on the off-target candidates identified through genome-wide analysis. For these final validations, some supplemental controls were included to confirm the U7-E53-related specificity of the off-target events. These controls were generated through the transduction of human primary myotubes and hepatocytes with the rAAV3b-U7-Scramble (Scr) vector, which carries a scrambled sequence instead of the U7-E53 antisense sequence.
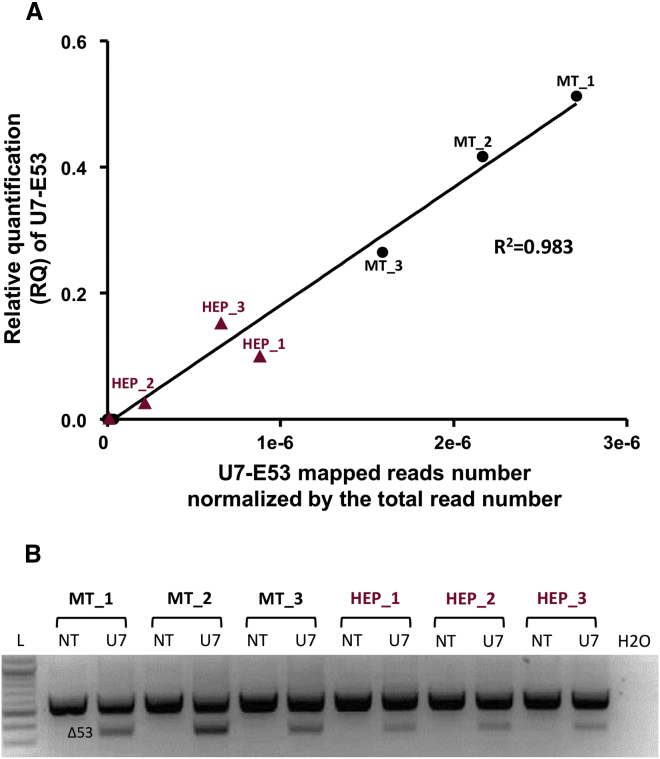
We first assessed whether the RNA-seq analysis enabled the quantification of U7-E53 RNA expressed in transduced cells from reads that were not mapped on the human genome. Therefore, among the unmapped reads, we counted raw reads specifically mapped on the U7-E53 sequence (Figure 3A) and compared it with qRT-PCR results. As observed by qRT-PCR, the expression of U7-E53 was lower in transduced hepatocytes than in transduced myotubes. An adequate correlation (R2 = 0.983) was observed between U7-E53 expression measured through qRT-PCR and the number of RNA-seq reads mapped on the U7-E53 sequence (Figure 3A). We further confirmed that the expression of U7-E53 in these samples induced the skipping of exon 53 in the DMD messenger in all transduced samples (Figure 3B). Albeit not strictly quantitative, the skipping of exon 53 was weaker in hepatocytes than in myotubes samples, consistent with the weaker U7-E53 RNA levels in these cells. Of note, we also assessed the ratios of DMD transcripts versus U7-E53 RNA expressed in both cell types by evaluating their relative RPKM (reads per kilobase per million mapped reads) values in each sample (Table S2). In adequacy with the lower exon skipping events, a slightly weaker DMD transcript/U7-E53 RNA ratio was observed in hepatocytes. Altogether, these results validated the use of these primary myotube and primary hepatocyte samples to assess the off-target effects after rAAV3b-U7-E53 transduction.
Figure 3.
Validation of the Samples Used for U7-E53 Off-Target Analysis through RNA-Seq
(A) Correlation between U7-E53 RNA expression analyzed through qRT-PCR and the number of RNA-seq reads mapped on the U7-E53 RNA sequence normalized by the total reads number. Relative quantification (RQ) of U7-E53 RNA was obtained through qRT-PCR analysis on human primary myotubes (MT_1, 2, and 3, circle points) and human primary hepatocytes (HEP_1, 2, and 3, triangle points) transduced with rAAV3b-U7-E53 (identified points) or non-transduced samples (non-identified points). (B) Analysis of exon skipping of the DMD messenger in the different samples used for RNA-seq analysis. The DMD messenger was amplified using nested RT-PCR from exons 51–54 in three replicates of human primary myotubes (MT_1, 2, and 3) and of human primary hepatocytes (HEP_1, 2, and 3). The PCR product (556 bp) detected in non-transduced samples (NT) corresponds to DMD mRNA with the exon 53. After transduction with rAAV3b-U7-E53, a new PCR product (344 bp) was detected, corresponding to out-of-frame mRNA without exon 53. H2O, water; L, DNA ladder.
U7-E53 RNA Caused Limited Gene Expression Variations in Human Primary Myotubes and Hepatocytes
First, to determine whether the RNA-seq data could discriminate between the different samples, we performed a principal component analysis (PCA) on the raw expression data obtained from human primary myotubes and hepatocytes transduced or not with rAAV3b-U7-E53 (Figure S4). Interestingly, the principal source of variance reflected the origin of the samples, liver versus muscle cells. The second source of variance reflected the origin of the healthy subjects. These observations support the idea that transduction with rAAV3b-U7-E53 did not significantly impact the overall gene expression.
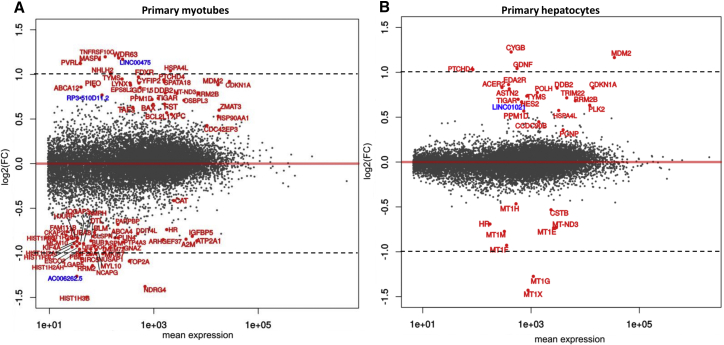
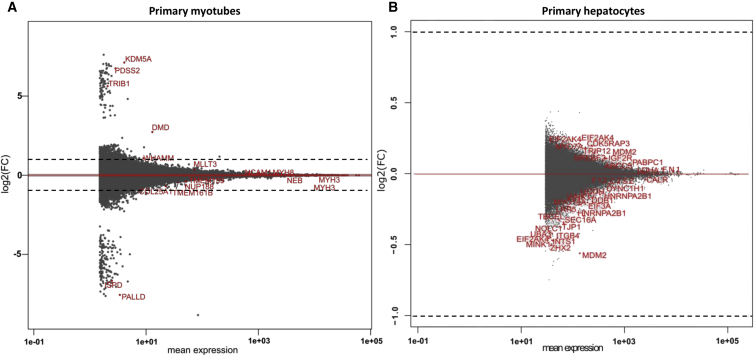
Using DESeq2, we analyzed the differential gene expression in transduced versus NT primary myotube and hepatocyte samples. MAplots showing, by gene (protein coding genes and linear non-coding RNAs [lincRNAs]), the fold-change (FC) values relative to the mean expression level are represented in Figure 4. The statistically differentially expressed genes are listed in Table S3 (myotubes and hepatocytes data). Overall, in both cell types, the amplitude of gene expression variations was low, with only a few genes exhibiting an over- or under-expression of more than 2-fold between transduced and NT samples. Indeed, the log2(FC) values varied between −1.52 to +1.19 for primary myotubes (Figure 4A) and −1.42 to +1.22 for primary hepatocytes (Figure 4B). Based on the sole p value (p < 0.05) criteria, while 80 and 30 genes are considered differentially expressed in primary myotubes and hepatocytes, respectively, 63/80 genes in myotubes and 24/30 in hepatocytes presented a log2(FC) below 1 (or −1), representing a variation of less than a factor of 2 between transduced and NT samples (Table S3). Therefore, we concluded that neither the expression of the U7-E53 RNA nor the transduction with the rAAV3b vector significantly disturbed the expression of genes in the transduced samples.
Figure 4.
Gene-Level Analysis of Differential Gene Expression by DESeq2
Analysis was done in myotubes (A) and hepatocytes (B) transduced with rAAV3b-U7-E53 and compared with non-transduced samples. MAplots represented log2(FC) of each gene (between transduced and non-transduced samples) function of gene mean expression (in reads number). Each point represents a gene, and statistically (padj ≤ 0.05) differentially expressed genes are identified and colored in red (coding genes) or blue (lincRNA). Dotted lines represent an FC value of a factor 2 [log2(FC) = 1 or −1].
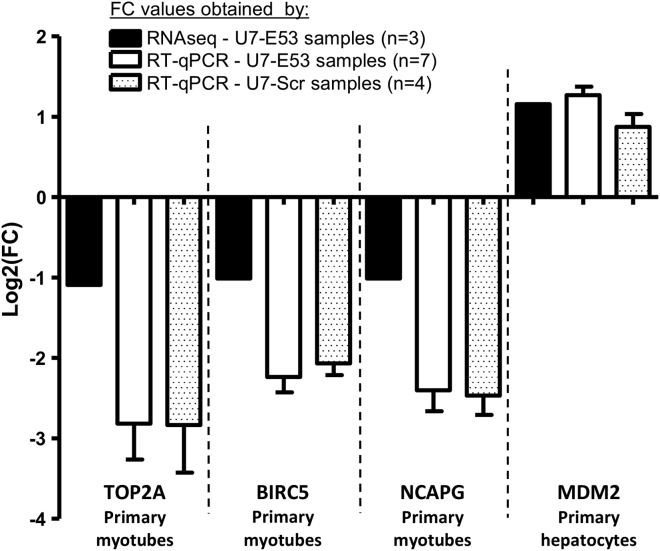
Among these differentially expressed genes, we identified the transcripts harboring a predicted binding site (i.e., a sequence at least partially complementary) for the antisense sequence expressed through the U7-E53. We used the alignment tool miRanda, specifically developed for miRNA target design,42 and subsequently crossed the results with those of the differential analysis described above (Table S3). Among the 38 differentially expressed genes containing a putative degenerated U7-E53 binding site, only 8 genes (2 lincRNAs and 5 coding genes in myotubes and 1 coding gene in hepatocytes) exhibited a more than 2-fold variation in gene expression between transduced and NT samples. Among these genes, we performed a qRT-PCR assay on TOP2A, BIRC5, and NCAPG genes from primary myotubes and the MDM2 gene from primary hepatocytes to validate the quantification using an alternative method. As presented in Figure 5, we compared the gene expression between NT samples, samples transduced with the rAAV3-U7-E53 vector, and samples transduced with the rAAV3-U7-Scr vector carrying a scrambled antisense sequence. As explained above, this latter supplemental control was included to confirm the U7-E53-related specificity of the potential off-target events. After assessing whether U7-E53 and U7-Scr were expressed in all samples (data not shown), we confirmed the differential expression for the four tested genes, with the under-expression of TOP2A, BIRC5, and NCAPG and the overexpression of MDM2 (Figure 5). Notably, all four genes were similarly deregulated in the U7-Scr samples, leading us to conclude that none of these off-target effects specifically reflected the antisense sequence harbored in the U7-E53, but rather reflected rAAV3b transduction per se. Altogether, with this experimental design, these results suggested a minimal off-target impact of our clinical rAAV-U7-E53 product on gene expression levels.
Figure 5.
Validation of Some Differentially Expressed Genes through qRT-PCR in Human Primary Myotubes and Hepatocytes Transduced or Not with rAAV3b-U7-E53 or rAAV3b-U7-Scr
The results are represented as the mean and SD of log2(FC) values obtained using the 2−ΔΔCt methodology (normalization of qRT-PCR by POLIIR human transcript) between non-transduced samples and samples transduced with rAAV3b-U7-E53 (n = 7, white bars) or rAAV3b-U7-Scr (n = 4, punctuated bars). In comparison, the results obtained through RNA-seq in three samples of primary myotubes and three samples of primary hepatocytes transduced with rAAV3b-U7-E53 are represented as black bars.
U7-E53 RNA Triggers the Specific Exon Skipping of the DMD Target with Low Impact on Global mRNA Splicing
Because U7-E53 was designed as an exon skipping tool presumably localized in the nucleus, an exhaustive analysis of splicing modifications was also needed to identify the potential impact of U7-E53 expression on non-targeted mRNAs. This analysis at the exon junction level was performed using DEX-seq, facilitating the comparison of the abundance of reads overlapping the exon-exon junctions between transduced and NT samples. Instead of an exon-centric analysis of splicing, we selected a junctional reads analysis, which is more prone to identify subtle changes in the location of splicing events and more adapted to identify new splicing events arising from experimental or pathological conditions, rather than relying solely on the available gene annotation.
MAplots showing, for each exon junction, the FC values relative to the mean expression level (in read number) are represented in Figure 6. The statistically differentially used exon junctions are listed in Table S4 (myotubes and hepatocytes data). A positive FC represents an increase in exon junction usage, while a negative FC shows a decrease in exon junction usage. Overall, in both primary myotube and hepatocyte samples, few changes in junction usage were detected.
Figure 6.
Exon Junction Usage Analyzed Using DEXseq
Analysis was done in myotubes (A) and hepatocytes (B) transduced with rAAV3b-U7-E53 and compared with non-transduced samples. MAplots represented log2(FC) of each junction (between transduced and non-transduced samples) function of junction mean expression (in reads number). Each point represents a junction, and statistically differentially used junctions (padj ≤ 0.05) are identified in red (coding genes). Dotted lines represent an FC value of a factor 2 [log2(FC) = 1 or −1].
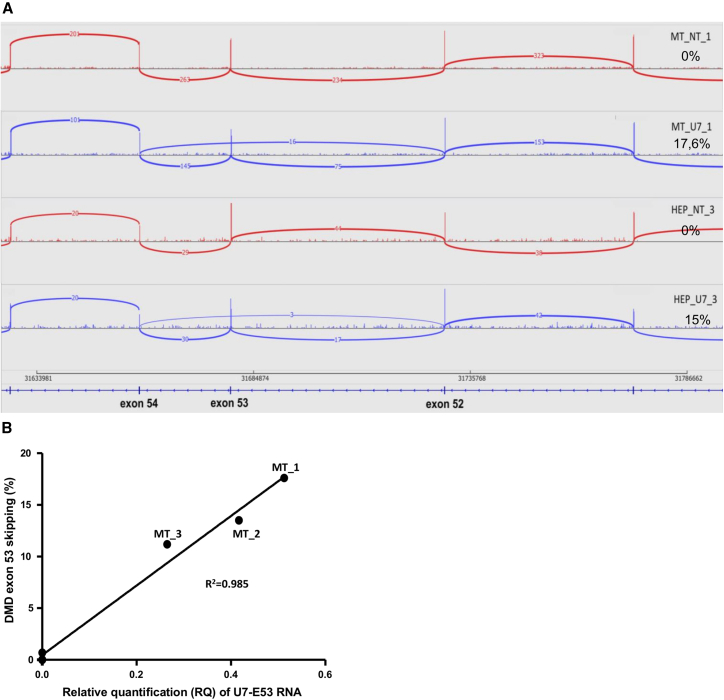
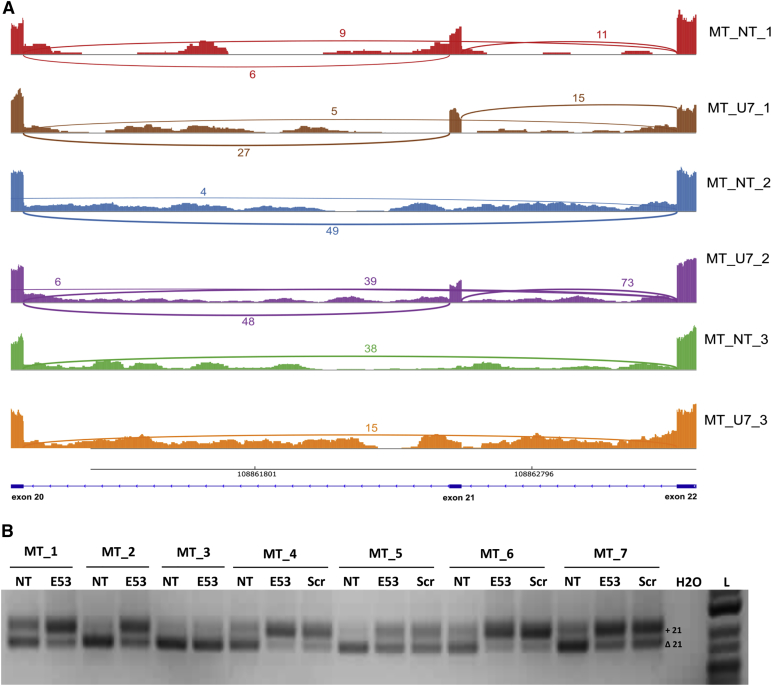
In myotube samples, only 24 differentially used exons junctions (corresponding to 17 transcripts) were detected (Figure 6A) and harbored different profiles. The first profile, representing a group of five junctions, exhibited a relatively important FC but a low expression level without any biological significance (junction mean expression between 2.1 and 4.1 reads). On the contrary, the second profile was represented by junctions with a higher expression level but a weaker FC, close to zero, indicating a moderate, even negligible effect. Only one exon junction presented a third different profile, characterized by a higher expression level and a relatively important FC. This junction was identified as the exon 52 to exon 54 junction of the DMD gene, and its use was significantly increased in the transduced versus the NT samples [p = 0.000; log2(FC) = 2.73], consistent with the exclusion of DMD exon 53 observed through nested RT-PCR. A sashimi plot43 representing the mapped RNA-seq data within the locus of the DMD gene surrounding exon 53 is shown in Figure 7A. The percentage of skipped exon 53 [J52-54/(J52-54+J52-53)] was calculated for all six myotubes samples and compared with the expression level of the U7-E53 measured using qRT-PCR (Figure 7B). A high correlation was observed between the expression of U7-E53 and the level of exon 53 skipping, illustrating a dose-effect response of the therapeutic product.
Figure 7.
Skipping Quantification of Exon 53 of the DMD Messenger through RNA-Seq
(A) Sashimi plot (software Integrative Genomics Viewer) showing the reads data mapped on the DMD locus (exons 51–55), in representative examples of human primary myotube (MT) and hepatocyte (HEP) samples non-transduced (NT) or transduced with rAAV3b-U7-E53 (U7). Alignments on exons are represented as read densities (spikes) and reads spanning exon-exon junctions are represented as arcs between exons. The number of reads mapped on each junction is indicated. Percentages of exon 53 skipping were estimated from these data and indicated for each sample. (B) Correlation between exon 53 skipping percentages obtained through RNA-seq and U7-E53 expression analyzed through qRT-PCR on human primary myotubes (MT_1, 2, and 3) transduced with rAAV3b-U7-E53 (identified points) or non-transduced samples (non-identified points).
In hepatocyte samples, the dynamics of splicing modifications were lower than in myotubes [log2(FC) between −0.75 and +0.5 in hepatocytes compared with −8 to +8 for myotubes] (Figure 6B). Of note, these results could be correlated with the lower U7-E53 expression in hepatocytes, when compared with primary myotubes (Figure 3). Globally, the extent of differential exon usage was low in hepatocytes because the majority of the significantly differentially spliced genes were observed within the bulk of the result of the MAplot. A total of 36 differentially used junctions (corresponding to 31 transcripts) were detected, but only 1 junction in the MDM2 gene was particularly modified in transduced hepatocytes. However, because MDM2 global gene expression was also statistically decreased in hepatocyte samples (Figures 4 and 5), the modification of exon junction usage in this gene was not interpretable. Notably, even when some reads mapped on exon 52 to the exon 54 junction of the DMD gene in the transduced samples (Figure 7A), this junction was not statistically differentially used in the hepatocyte samples (Figure 6B; Table S4). In hepatocytes, the DMD gene was primarily expressed from a promoter located downstream of exon 53 (i.e., the Dp71 promoter, just before exon 6244); therefore, very few reads mapped upstream of exon 62. This promoter usage is likely sufficient to explain why no statistically significant value was obtained for the exon 52-exon 54 junction during the differential analysis, despite the identification of transcripts with exon 53 skipping using nested RT-PCR.
Among all transcripts deemed differentially spliced (in human primary myotubes and hepatocytes), we used in silico predictions to highlight those that had a predicted binding site for the antisense sequence harbored in U7-E53. Alterations in splicing could reflect the interaction of the antisense sequence with regulatory elements located in exons or introns up to 100 bp from the exon.45 We therefore searched for sequences at least partially complementary to U7-E53 antisense sequence in “extended exons” comprising the exon plus 100 bp of flanking intronic sequences on both sides. We used miRanda42 and Blast (NCBI), which provided complementary results with the notion of contiguous identical bases between the antisense sequence and the pre-mRNA target. After comparing these in silico data with the differentially used junctions, we identified a small number of off-target events potentially altered through U7-E53 binding (Table S4). In myotubes, we showed that only three differentially used junctions overlapped a partial U7-E53 fixation motif, including two junctions in the TTN gene and one junction in NUP188 (Table S4, myotubes data). In hepatocytes, four other differentially used junctions overlapped the U7-E53 fixation motif (Table S4, hepatocytes data) in genes ZHX2, LRP5, DDB1, and CTSZ. However, using nested RT-PCR and qRT-PCR analyses, we were not able to validate the differential splicing of these few off-target candidates. While analyzing the sashimi plots representing the mapped RNA-seq data within the locus of these different junctions, we showed that differentially used junctions often included two adjacent exons (data not shown). This splicing modification pattern, and low FC values obtained between transduced and NT samples, explained the inability to confirm the RNA-seq results using alternative molecular approaches.
However, to confirm that RNA-seq is adapted to detect splicing modifications, we used RT-PCR to analyze the differential junction usage of another unrelated gene, COL25A1. This gene did not exhibit any potential fixation site for the antisense sequence harbored by U7-E53, but rather showed a junction differentially used with a log2(FC) = −0.69 in human primary myotubes (Table S4). Using raw reads data, we identified that the junction between exons 20 and 22 was statistically less used in transduced myotubes, suggesting an inclusion of exon 21 within the COL25A1 mature mRNA (Figure 8A). We subsequently developed a specific nested RT-PCR to amplify COL25A1 messengers from exons 19–23 and analyzed the splicing pattern between NT samples, samples transduced with rAAV3b-U7-E53 vector, and samples transduced with rAAV3b-U7-Scr vector (Figure 8B). Both isoforms with or without exon 21 could be detected, but the one with exon 21 was less intense or even absent in NT myotubes (Figure 8B). In transduced myotubes samples (rAAV3b-U7-E53 or rAAV3b-U7-Scr), the relative proportion of COL25A1 isoform without or with exon 21 was inverted, indicating the inclusion of exon 21 in the COL25A1 messenger. This result was confirmed after sequencing the PCR products (data not shown).
Figure 8.
Validation of a Differentially Used Junction in COL25A1
(A) Sashimi plot (software Integrative Genomics Viewer) showing the raw reads data mapped on COL25A1 (exons 20–22) in human primary myotubes non-transduced (NT) or transduced with rAAV3b-U7-E53 (U7). Alignments on exons were represented as read densities (spikes), and alignments on junctions were represented as arcs between exons. The exact number of reads mapped on each junction was specified. (B) Splicing modifications of COL25A1 messenger analyses using nested RT-PCR. The COL25A1 messenger was detected from exons 19–23 in three replicates of primary myotubes sequenced through RNA-seq and four supplemental primary myotube samples transduced with rAAV3b-U7-E53 or rAAV3b-U7-Scr. Two PCR products of 210 and 165 bp were detected, corresponding, respectively, to COL25A1 isoforms with and without exon 21. H2O, water; L, DNA ladder.
In conclusion, while differential splicing events could be specifically detected, the U7-E53 therapeutic antisense sequence showed a weak off-target impact on non-targeted mRNAs.
Discussion
The present study was aimed at the comprehensive analysis of the potential specific off-target effects of a gene therapy clinical product, U7snRNA-E53, designed to skip exon 53 of the human DMD messenger for the treatment of DMD patients. The objectives were also to propose a methodology as proof of principle to evaluate the specific toxicity (i.e., off-target effects) of an antisense sequence designed for exon skipping, using a transcriptomic analysis through RNA-seq, in human primary cells transduced (or not) with the clinical candidate rAAV-U7-E53. The major findings were that the clinical antisense sequence harbored in U7-E53 did not significantly disturb gene expression or mRNA splicing in human primary myotubes and primary hepatocytes transduced with a rAAV3b-U7-E53, and that no specific off-target was identified using this methodology.
Off-target effects have been extensively investigated for siRNAs and genome-editing nucleases. Most studies typically used in silico analyses to predict potential unintended DNA or RNA targets, using a predefined cutoff value of identity with the antisense sequence of interest (i.e., predefined number of mismatches tolerated between the antisense sequence and its target).46 These putative off-targets were subsequently typically validated through molecular techniques, such as PCR, qPCR, or targeted sequencing.29, 30, 31, 32, 47 Concerning AONs, despite the FDA approval of a product for DMD treatment by exon skipping (Eteplirsen), few studies have investigated their off-targets effects,33, 34 and some authors considered the off-target risk as negligible.46 This off-target assessment is indeed not yet required by standard FDA clinical trials protocols, but it could be the case in the near future, favored by the development of more sensitive high-throughput sequencing tools. Moreover, it has been described that siRNA and nuclease off-targets were dependent on the site of hybridization, the nature per se of the antisense sequence, and the number of mismatches tolerated between the antisense sequence and its target.29, 48, 49 It has also been shown that AONs of only 15 nucleotides could effectively induce exon skipping.50 We think that in silico analyses alone could not be pertinent enough because the results are heavily dependent on: (1) the database or reference genome used, (2) the software or algorithm used, and (3) the cutoff values applied to identify complementary sequences. Particularly, off-target analyses should be preferentially conducted in a highly sensitive and comprehensive manner with high-throughput strategies.35, 49, 51 Therefore, in the present study, we designed a methodology based on RNA-seq to identify, at the transcriptomic level, all the potential off-target effects induced through the antisense sequence harbored in our clinical product, U7-E53.
The choice of the experimental model is also essential. As the exon skipping reaction itself, off-targets are specific for the antisense sequence, and the tissue and species in which the analyses are performed. Therefore, the use of tissue samples from treated GRMD dogs injected with high doses of rAAV8-U7snRNA23 would not have been relevant for this off-target analysis because the antisense sequences used in dogs (E6 and E8) were not the same as our clinical antisense sequence E53, specifically designed for exon skipping of the human dystrophin messenger. Moreover, in comparing in parallel siRNA off-targets in mice and human cells, Burchard and colleagues52 suggested that in vitro analyses in human cells are the most relevant (i.e., better than analyses performed in vivo in another species) to identify off-target effects, when evaluating a clinical product.52 Therefore, we used human in vitro models to analyze U7-E53-specific off-targets. Moreover, we assumed that the use of primary cells rather than immortalized strains of myoblasts or hepatocytes would enable a better prediction of potential off-target genes in patients. Because DMD myoblasts are impossible to maintain in culture reflecting early senescence53 and because the collection of DMD hepatocytes was not feasible, we used primary cells obtained from healthy subjects. The healthy context raised some concerns about the efficiency of exon skipping, reflecting the fact that exon 53 skipping leads to a frameshift and such aberrant transcripts are subjected to nonsense-mediated decay (NMD).54 Indeed, in this experimental model, exon 53 skipping levels did not exceed 17.6% in transduced myotubes and were even lower in transduced hepatocytes. Yet, like for exon skipping efficiency, off-target effects could be expected to be dose dependent. These levels of exon 53 skipping could be increased using a higher MOI, requiring a higher titer rAAV3b batch. However, despite these limitations, we detected U7-E53 expression and exon 53 skipping within the targeted DMD messenger in all transduced cells, the prerequisite criterion for a relevant off-target analysis. Moreover, such levels of exon 53 skipping (≈10%–20%) were recently shown to be sufficient to obtain a significant increase in dystrophin protein expression over baseline (F. Muntoni et al., 2017, World Muscle Society, conference).
The differential gene expression analysis through DESeq2 showed that neither rAAV3b transduction nor U7-E53 expression adversely impacts the transcriptome of the targeted human cells. These results are consistent with previous studies showing that rAAV transduction did not lead to a massive deregulation of gene expression, contrary to adenovirus.55, 56
Similarly, the analysis of splicing junction’s usage alterations through DEX-seq identified few differentially used junctions, but the vast majority exhibited low FC values, with the exception of the DMD gene and of some poorly expressed junctions. Overall, weakly expressed junctions are more differentially used than junctions that are highly expressed. This phenomenon, giving a typical triangular shape to MAplots, is commonly observed in RNA-seq datasets as a direct consequence of the use of reads count data in which the results are biased when the number of reads is low.39 Moreover, MAplots showed a symmetric representation of junctions, meaning an equivalent number of junctions statistically lower and more used in transduced samples, likely reflecting background splicing noise.
Filtrations through in silico software (miRanda and/or Blast) following RNA-seq analysis facilitated the selection, among identified potential off-target candidates, of transcripts harboring a potential fixation site for the antisense sequence harbored in the U7-E53. To minimize biases and false-negative results potentially induced through miRanda and/or Blast programs,46, 57 we did not apply cutoff values regarding the percentages of identity or the number of mismatches tolerated between the antisense sequence harbored in the U7-E53 and candidates transcripts. Notably, such a strategy does not enable the identification of potential indirect off-target effects (i.e., on transcripts that do not share any complementarity with U7-E53). The supplemental control samples transduced with a scrambled antisense sequence (U7-Scr) could address this last issue, but these samples were not analyzed through RNA-seq.
To validate the RNA-seq results using alternative molecular approaches, we opted for qRT-PCR and nested RT-PCR analyses, and we included some U7-Scr control samples to distinguish events related to rAAV transduction per se or to the antisense sequence harbored in U7-E53. These experimental validations confirmed the results obtained through RNA-seq, supporting the adequate correlation between RNA-seq and molecular techniques, in particular qRT-PCR.58, 59 Yet, RT-PCR was not adapted for our experimental validations, because the differentially used junctions often included two adjacent exons. For these particular cases, we developed qRT-PCR assays designed between the two exons of interest. Unfortunately, the lack of sensitivity of qRT-PCR prevented the analysis of the differential expression of genes and junctions with a FC value <2 or those too weakly expressed in cells. The power and sensitivity of RNA-seq enable the identification of all modifications of gene expression or splicing, including the weakest that could not be validated by conventional molecular techniques. We consider that these weakest modifications may reflect background noise and consequently probably may not be relevant from a biological point of view. It highlights the need to set up cutoff values in RNA-seq analyses that could enable a focus only on significant events. Moreover, the safety of this therapeutic approach would depend on: (1) the off-target effects’ amplitude, which is obviously low in our samples; (2) the function of the off-target genes, and (3) the translation of the transcriptomic effects at the protein level. Indeed, transcriptomic modifications should be interpreted with caution because multi-omic studies described a weak-to-moderate correlation between mRNA and protein expression.60, 61
In conclusion, the present study described a multi-strategy approach for the analysis of potential off-target effects induced by an antisense sequence designed for exon skipping. When this methodology requires optimization to reduce biases and analysis time, we propose a relevant transgene-specific in vitro toxicology study for exon-skipping-based gene therapies. Indeed, the active substance of exon skipping products remains the antisense sequence per se, whose efficacy and toxicity are sequence and species dependent. In rAAV-based exon skipping approaches, conventional regulatory toxicology studies in animal models are essential to anticipate potential adverse effects, including immune responses developed against the rAAV capsid, one of the remaining challenges for rAAV-based gene therapy,62 but these toxicology studies are not adapted to evaluate the specific toxic effects of the antisense sequence per se. Off-target analyses mediated through RNA-seq in human cells are more relevant to evaluate the toxicity of antisense sequences and could be rapidly suitable for each new antisense sequence designed to target other mutations in the same gene.
Using this approach, it appears that the U7-E53 therapeutic antisense sequence has a negligible off-target impact, making it suitable for controlling DMD splicing in a clinical context.
Materials and Methods
Cell Culture and rAAV3b Transduction
rAAV3b-U7-E53 was produced by the Vector Core of the Nantes University Hospital by transfection of HEK293 cells and purified by cesium chloride density gradients. All of the cells were grown at 37°C and 5% CO2. Human primary myoblasts were obtained from three muscle biopsies of healthy subjects (from quadriceps or gluteal muscle) and provided by the immortalization platform of the Institute of Myology (Paris, France). These cells were grown in muscle cell growth medium (Promocell, Heidelberg, Germany) supplemented with complements (supplement pack; Promocell), 20% fetal calf serum (Dominique Dutscher, Brumath, France), and 1% penicillin/streptomycin (Life Technologies, Saint Aubin, France). For rAAV vector transduction, 140,000 myoblasts were seeded in each well of a 12-well plate. After 15 hr, the cells were transduced with rAAV3b-U7-E53 or rAAV3b-U7-Scr at MOI 1 × 105 in a total volume of 750 μL of medium. After 6 hr of incubation, 750 μL of medium was added per well. Cells were incubated during 3 days before harvesting for analysis.
Differentiation into myotubes was induced at 3 days post-transduction of myoblasts preliminary plated on gelatin-coated 12-well plates in DMEM high GlutaMAX (Life Technologies) supplemented with 100 μg/mL human apolipotransferrin (Sigma-Aldrich, Saint Quentin Fallavier, France), 10 μg/mL human insulin (Sigma-Aldrich), and 1% penicillin/streptomycin (Life Technologies). Differentiating cells were incubated during 4 days (i.e., 7 days post-transduction) before harvesting for analysis.
Human primary hepatocytes were obtained from liver of different human donors provided by Biopredic International (Rennes, France) and grown in a specific medium from Biopredic. For rAAV transduction, 300,000 hepatocytes already seeded in a 24-well plate coated with rat tail type I collagen were transduced the day of reception with rAAV3b-U7-E53 or rAAV3b-U7-Scr at MOI 1 × 105 in a total volume of 500 μL of medium. After 6 hr of incubation, 500 μL of medium was added per well. Cells were incubated during 3 days before harvesting for analysis. HepaRG cells were grown in a 24-well plate in William’s E medium (Sigma-Aldrich) supplemented with several complements as described before.36 Differentiation of HepaRG into hepatocyte-like cells was induced by adding DMSO (Sigma-Aldrich).36 For rAAV vector transduction, confluent cells were transduced with rAAV3b-U7-E53 at MOI 1 × 105 in a total volume of 500 μL of medium. After 6 hr of incubation, 500 μL of medium was added per well. Cells were incubated during 3 days before harvesting for analysis. Harvest of cells was performed in adding a small volume of trypsin (trypsin-EDTA; Sigma-Aldrich) in each well. Cells were then washed out and centrifuged to obtain dry pellets. Finally cells were frozen in liquid nitrogen and stocked at −80°C.
U7-E53 RNA and Off-Target Candidate Expression Analyses through qRT-PCR
Total RNA was extracted from human cell pellets with a Nucleospin RNA kit (Macherey Nagel, Hoerdt, France) and treated with RNase free DNase I from the TURBO DNA-free kit (Life Technologies) according to the manufacturer’s instructions. Five hundred nanograms of RNA was reverse-transcribed using a RT2 first strand kit (QIAGEN, Les Ulis, France) according to the manufacturer’s instructions. qPCR analyses were conducted on a StepOne Plus (Life Technologies) using a 15-fold diluted cDNA.
For U7-E53 expression analysis, a reaction of TaqMan-based qPCR was performed in a final volume of 20 μL containing template cDNA, gene expression master mix (Applied, Life Technologies, Saint Aubin, France), 0.4 μL of ROX reference dye (Ozyme, Montigny-le-Bretonneux, France), 0.25 μmol/L of each primer (forward: 5′-TCAACTGTTGCCTCCGGTTC-3′ and reverse: 5′-GGTTTTCCGACCGAAGTCAG-3′; Life Technologies), and 0.2 μmol/L TaqMan probe (5′-TCTTGTACAATTTTT GGAGCAGGTT-3′; Life Technologies). For each sample, Ct values were compared with those obtained by qRT-PCR for the reference gene (Rg) POLR-IIL [Polymerase (RNA) II polypeptide L] (forward: 5′-CCTGGAGAAGTGACCACGCT-3′, reverse: 5′-AATTCGGGGCAGGACGCTCAG-3′ and probe: 5′-CACCCACCCGCTGTGCTGACCAT-3′; Life Technologies). To validate qPCR efficacies, a range of dilutions of a plasmid containing the sequence of interest was dropped in parallel of samples. Relative quantification (RQ) was calculated with the 2−ΔCt method (RQ = 2−[Ct target − Ct Rg]).
For off-target candidates (differential expression analysis) expression assessment, reactions of Sybr-green-based qPCR were performed in a final volume of 20 μL containing template cDNA, Sybr-green reagent (Applied, Life Technologies) and 0.2 μmol/L of each specific primer. To validate qPCR efficacies, a range of dilutions of a cDNA obtained from NT myotubes or hepatocytes was dropped in parallel of samples. Normalization was also performed by POLR-IIL qRT-PCR as described above. The FC between transduced (U7) and NT samples was calculated with the 2−ΔΔCt method, FC = 2−([Ct target U7 − Ct Rg U7] − [Ct target NT − Ct Rg NT]).
DMD and COL25A1 mRNAs Splicing Analyses through Nested RT-PCR
Detection of the DMD messenger was done using a nested PCR from exons 51–54, using GoTaq DNA polymerase (Promega, Charbonnieres, France) and following amplification conditions: the first reaction was performed with specific primers (forward: 5′-GTTACTCTGGTGACACAACC-3′ and reverse: 5′-ATGTGGACTTTTCTGGTATC-3′) for 25 cycles (95°C/1 min; 50°C/1 min; 72°C/1 min). Then, 1 μL of the first reaction was amplified for 30 cycles (forward: 5′-ACTAGAAATGCCATCTTCCT-3′ and reverse: 5′-CAAGTCATTTGCCACATCTA-3′).
Detection of the COL25A1 messenger was done using a nested PCR from exons 19–23, using GoTaq DNA polymerase (Promega) and following amplification conditions: the first reaction was performed with specific primers (forward: 5′-TAAGGCTTCGTGGAGGTTGC-3′ and reverse: 5′-TCCTCAAGGAGAACCAGGCT-3′) for 25 cycles (95°C/1 min; 60°C/1 min; 72°C/1 min). Then, 1 μL of the first reaction was amplified for 30 cycles (forward: 5′-TGATCTCAGTGGCTCCTTGA-3′ and reverse: 5′-GAACAAAAGGTGAACGGGGG-3′). Final PCR products were migrated on a 2% agarose gel and revealed with ethidium bromide staining.
RNA-Seq Libraries Preparation
Total RNA was isolated as described above, and RNA integrity was measured using a model 2100 Bioanalyzer (Agilent, Massy, France): all samples have an RNA integrity number (RIN) score of at least 7.5. “Ribozero” RNA-seq libraries were prepared using “TruSeq Stranded Total RNA with Ribo-Zero Gold Prep Kit” (RS-122-2301; Illumina, Paris, France). In brief, starting with 300 ng of total RNA, the first step involved the removal of cytoplasmic and mitochondrial rRNA using biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads. Following purification, the RNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers followed by second strand cDNA synthesis using DNA polymerase I and RNase H. The double-stranded cDNA fragments were blunted using T4 DNA polymerase, Klenow DNA polymerase, and T4 PNK. A single ‘A’ nucleotide was added to the 3′ ends of the blunt DNA fragments using a Klenow fragment (3′ to 5′ exo minus) enzyme. The cDNA fragments were ligated to double-stranded adapters using T4 DNA Ligase. The ligated products were enriched by PCR amplification (30 s at 98°C; [10 s at 98°C, 30 s at 60°C, 30 s at 72°C] × 12 cycles; 5 min at 72°C). Then surplus PCR primers were removed by purification using AMPure XP beads (Beckman Coulter). Final cDNA libraries were checked for quality and quantified using 2100 Bioanalyzer (Agilent). Two libraries were loaded per lane of the Illumina flow cell at a concentration of 7 pM, and clusters were generated using the Cbot and sequenced on the HiSeq 2500 (Illumina) as stranded, paired-end 100 base reads following Illumina’s instructions. Image analysis and base calling were performed using RTA 1.18.61 and CASAVA 1.8.2. Sequencing was performed by the IGBMC Microarray and Sequencing platform, a member of the “France Génomique” consortium (ANR-10-INBS-0009).
Data Preprocessing
The human genome sequence primary assembly, transcript sequences, and comprehensive gene annotation files were all retrieved from the Gencode consortium website (Release23-GRCh38.p3 accessible at https://www.gencodegenes.org/releases/23.html). Raw BCL data for the samples were de-multiplexed with CASAVA (Illumina) according to their barcodes and were stored in independent fastq files (data available at the European Nucleotide Archive website PRJEB13980). A subset of reads was aligned to select genomes/DNA sequences to assess the presence of contamination within the sample using FastQScreen 0.5.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/). Following a general sequence quality control using FastQC v0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), fastq files were trimmed with Cutadapt 1.8.1 (https://github.com/marcelm/cutadapt) in paired-end mode to clean up sequences with low-quality extremities (-q 30,30) and those containing sequencing adapters, polyA/T (-a file:adapters.fa -A file:adapters.fa) or N bases (–trim-n). The fasta sequences contained in the adapters.fa file are as follows.
> polyA
AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
> polyT
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT
> Truseq_indexed_adapter
AGATCGGAAGAGCACACGTCTGAACTCCAGTCACNNNNNNATCTCGTATGCCGTCTTCTGCTT
After trimming, sequences shorter than 25 bp were removed from the datasets (-m 25).
Alignment against Human Genome
The short read alignment was performed with STAR 2.4.2a.38 First an index was built using recommended options for the human genome with a gff annotation file (STAR–runMode genomeGenerate–genomeDir ./–genomeFastaFiles GRCh38.primary_assembly.genome.fa–sjdbGTFfile gencode.v23.primary_assembly.annotation.gff3–sjdbOverhang 100–sjdbGTFtagExonParentTranscript Parent). The alignment step, the post-processing of reads with samstat 1.5 and bedtools 2.17.0, and the read count per gene with HTSeq 0.6.1 were automatized with a dedicated bash script available at https://github.com/a-slide/BASH_NGS_Tools/blob/master/RNAseq_U7_pipeline.sh. Except for the minimal mapping quality (MAPQ) that was set up to 30, all of the other options are hard-coded directly in the script.
Differential Gene Expression Analysis
Read counts per gene were generated using HTSEQ63 based on the annotation gencode.v23.primary_assembly.annotation.gff3. Differential gene expression was performed using the DESeq2 package.39 Protein coding genes and lincRNAs were included in the analysis. Differentially expressed genes were selected based on a Wald test adjusted for multiple tests (false discovery rate [FDR] < 0.05).
Differential Splicing Analysis
Differential splicing analysis was based on the use of counting the reads overlapping a junction, as described in Noiret et al.64 This analysis was called junction-centric41 in opposition to the exon-centric analysis more commonly used, where the relative abundance of exons is compared. The DEX-seq package initially developed for the exon-centric approach was used without modification (https://bioconductor.org/packages/release/bioc/html/DEXSeq.html). In brief, the junctions identified by RNA-seq were quantified during the mapping step with STAR. Then junctions were annotated according to their chromosomal positions (chromosome name, start/end of genomic coordinates, and strand information) and assigned to a gene. Only unambiguous junctions (i.e., not assigned to two different genes) were used. Differentially used junctions were selected based on Wald test adjusted for multiple tests (FDR < 0.05).
In Silico Prediction of U7-E53 Binding Sites
All the bioinformatic tools used for prediction of U7-E53 binding sites are available as part of the TargetPredict package, which is available at http://a-slide.github.io/TargetPredict/. As stated previously, all reference files were obtained from the Gencode consortium website (Release23-GRCh38.p3).
For the differential expression analyses (DESeq2), the reference human transcript fasta file was enriched with additional information gathered from the GFF reference annotation file with the FastaEnhanceFromGff script. This fasta dataset is referred as “gencode_v23_transcripts_enhanced”. Regarding the differential splicing analyses (DEX-seq), extended exon sequences were extracted from the primary human genome assembly sequence (GRCh38 primary assembly), based on the gff files (Gencode v23) using GffFastaExtractor. Exons sequences were extended upstream and downstream by 100 bp to include intronic splicing regulatory elements. Overlapping extended exonic regions were fused so as to avoid multiple reports of the same genomic targets. This fasta dataset is referred to as “GRCh38_gencode_Offset-100_Features-exon_Chr-all_fused.”
From these two datasets, imperfect targets of the recognition sequence (U7-E53) were found in the subject sequences using TargetPredict based on hits found by Miranda v3.3a and NCBI Blast 2.2.28. TargetPredict reports containing the parameters used to run the program and raw results are available in Table S5 for the gencode_v23_transcripts_enhanced dataset and in Table S6 for the GRCh38_gencode_Offset-100_Features-exon_Chr-all_fused dataset.
Author Contributions
C.D., P.M., M.M., L.S., T.V., and C.L.G. designed the study. C.D., M.A., and V.F. conducted the experiments to evaluate the expression analyses in human cells. C.D., A.L., E.L., and Y.A. conducted the bioinformatics analyses of the RNA-seq data. C.D., Y.A., and C.L.G. wrote the manuscript. All authors reviewed the manuscript.
Conflicts of Interest
V.F. is involved in a patent application for the U7-E53 antisense sequence (FR1562036, dated 12/09/2015).
Acknowledgments
We would like to acknowledge Vincent Mouly and Anne Bigot (Institute of Myology, Paris, France) for providing us with human primary myoblasts. We also thank all the personnel of the Atlantic Gene Therapies institute vector core for rAAV3b productions. We thank France Pietri-Rouxel (Institute of Myology) for providing us the U7-Scr plasmid. This project was supported by FRM (Fondation pour la Recherche Médicale; grant FDT20140931046), by AFM-Téléthon (Association Française contre les Myopathies), and by ADNA (Advanced Diagnostics for New Therapeutic Approaches), a program dedicated to personalized medicine, coordinated by Institut Mérieux, and supported by research and innovation aid from the French public agency, Bpifrance.
Footnotes
Supplemental Information includes four figures and six tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.008.
Contributor Information
Claire Domenger, Email: claire.domenger@gmail.com.
Caroline Le Guiner, Email: caroline.le-guiner@univ-nantes.fr.
Supplemental Information
References
- 1.Havens M.A., Duelli D.M., Hastings M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA. 2013;4:247–266. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward A.J., Cooper T.A. The pathobiology of splicing. J. Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.H.P., Lim S., Wong W.S.F. Antisense oligonucleotides: from design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006;33:533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- 5.Saleh A.F., Arzumanov A.A., Gait M.J. Overview of alternative oligonucleotide chemistries for exon skipping. Methods Mol. Biol. 2012;867:365–378. doi: 10.1007/978-1-61779-767-5_23. [DOI] [PubMed] [Google Scholar]
- 6.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., Eteplirsen Study Group Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 7.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., De Kimpe S.J., Eagle M., Guglieri M., Hood S. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 8.Niks E.H., Aartsma-Rus A. Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2017;17:225–236. doi: 10.1080/14712598.2017.1271872. [DOI] [PubMed] [Google Scholar]
- 9.Gorman L., Suter D., Emerick V., Schümperli D., Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc. Natl. Acad. Sci. USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyenvalle A., Vulin A., Fougerousse F., Leturcq F., Kaplan J.C., Garcia L., Danos O. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 11.Denti M.A., Rosa A., D’Antona G., Sthandier O., De Angelis F.G., Nicoletti C., Allocca M., Pansarasa O., Parente V., Musarò A. Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model. Proc. Natl. Acad. Sci. USA. 2006;103:3758–3763. doi: 10.1073/pnas.0508917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asparuhova M.B., Marti G., Liu S., Serhan F., Trono D., Schümperli D. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J. Gene Med. 2007;9:323–334. doi: 10.1002/jgm.1027. [DOI] [PubMed] [Google Scholar]
- 13.Geib T., Hertel K.J. Restoration of full-length SMN promoted by adenoviral vectors expressing RNA antisense oligonucleotides embedded in U7 snRNAs. PLoS ONE. 2009;4:e8204. doi: 10.1371/journal.pone.0008204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gedicke-Hornung C., Behrens-Gawlik V., Reischmann S., Geertz B., Stimpel D., Weinberger F., Schlossarek S., Précigout G., Braren I., Eschenhagen T. Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO Mol. Med. 2013;5:1128–1145. doi: 10.1002/emmm.201202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 16.Mowry K.L., Steitz J.A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 17.Grimm C., Stefanovic B., Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basner-Tschakarjan E., Bijjiga E., Martino A.T. Pre-clinical assessment of immune responses to adeno-associated virus (AAV) vectors. Front. Immunol. 2014;5:28. doi: 10.3389/fimmu.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera V.M., Gao G.P., Grant R.L., Schnell M.A., Zoltick P.W., Rozamus L.W., Clackson T., Wilson J.M. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105:1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- 20.Nathwani A.C., Reiss U.M., Tuddenham E.G.D., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bish L.T., Sleeper M.M., Forbes S.C., Wang B., Reynolds C., Singletary G.E., Trafny D., Morine K.J., Sanmiguel J., Cecchini S. Long-term restoration of cardiac dystrophin expression in golden retriever muscular dystrophy following rAAV6-mediated exon skipping. Mol. Ther. 2012;20:580–589. doi: 10.1038/mt.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vulin A., Barthélémy I., Goyenvalle A., Thibaud J.-L., Beley C., Griffith G., Benchaouir R., le Hir M., Unterfinger Y., Lorain S. Muscle function recovery in golden retriever muscular dystrophy after AAV1-U7 exon skipping. Mol. Ther. 2012;20:2120–2133. doi: 10.1038/mt.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Guiner C., Montus M., Servais L., Cherel Y., Francois V., Thibaud J.-L., Wary C., Matot B., Larcher T., Guigand L. Forelimb treatment in a large cohort of dystrophic dogs supports delivery of a recombinant AAV for exon skipping in Duchenne patients. Mol. Ther. 2014;22:1923–1935. doi: 10.1038/mt.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendell J.R., Shilling C., Leslie N.D., Flanigan K.M., al-Dahhak R., Gastier-Foster J., Kneile K., Dunn D.M., Duval B., Aoyagi A. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 25.Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., DMD Care Considerations Working Group Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 27.Popplewell L.J., Adkin C., Arechavala-Gomeza V., Aartsma-Rus A., de Winter C.L., Wilton S.D., Morgan J.E., Muntoni F., Graham I.R., Dickson G. Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: implications for future clinical trials. Neuromuscul. Disord. 2010;20:102–110. doi: 10.1016/j.nmd.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 29.Gupta A., Meng X., Zhu L.J., Lawson N.D., Wolfe S.A. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamola P.J., Kitson J.D.A., Turner G., Maratou K., Eriksson S., Panjwani A., Warnock L.C., Douillard Guilloux G.A., Moores K., Koppe E.L. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43:8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obad S., dos Santos C.O., Petri A., Heidenblad M., Broom O., Ruse C., Fu C., Lindow M., Stenvang J., Straarup E.M. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid F., Hiller T., Korner G., Glaus E., Berger W., Neidhardt J. A gene therapeutic approach to correct splice defects with modified U1 and U6 snRNPs. Hum. Gene Ther. 2013;24:97–104. doi: 10.1089/hum.2012.110. [DOI] [PubMed] [Google Scholar]
- 35.Chen J.C., King O.D., Zhang Y., Clayton N.P., Spencer C., Wentworth B.M., Emerson C.P., Jr., Wagner K.R. Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol. Ther. 2016;24:1405–1411. doi: 10.1038/mt.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guillouzo A., Corlu A., Aninat C., Glaise D., Morel F., Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Glushakova L.G., Lisankie M.J., Eruslanov E.B., Ojano-Dirain C., Zolotukhin I., Liu C., Srivastava A., Stacpoole P.W. AAV3-mediated transfer and expression of the pyruvate dehydrogenase E1 alpha subunit gene causes metabolic remodeling and apoptosis of human liver cancer cells. Mol. Genet. Metab. 2009;98:289–299. doi: 10.1016/j.ymgme.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Rao X., Mattox W.W., Amos C.I., Liu B. RNA-seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS ONE. 2015;10:e0136653. doi: 10.1371/journal.pone.0136653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz, Y., Wang, E.T., Silterra, J., Schwartz, S., Wong, B., Mesirov, J.P., Airoldi, E.M., and Burge, C.B. (2013). Sashimi plots: quantitative visualization of RNA sequencing read alignments. arXiv:1306.3466.
- 44.Bar S., Barnea E., Levy Z., Neuman S., Yaffe D., Nudel U. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem. J. 1990;272:557–560. doi: 10.1042/bj2720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X.H.-F., Leslie C.S., Chasin L.A. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindow M., Vornlocher H.-P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
- 47.Meng X., Noyes M.B., Zhu L.J., Lawson N.D., Wolfe S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Summerton J.E. Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr. Top. Med. Chem. 2007;7:651–660. doi: 10.2174/156802607780487740. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aartsma-Rus A., De Winter C.L., Janson A.A.M., Kaman W.E., Van Ommen G.-J.B., Den Dunnen J.T., Van Deutekom J.C. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15:284–297. doi: 10.1089/oli.2005.15.284. [DOI] [PubMed] [Google Scholar]
- 51.Gabriel R., Lombardo A., Arens A., Miller J.C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C.C., Wang J., Friedman G. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 52.Burchard J., Jackson A.L., Malkov V., Needham R.H.V., Tan Y., Bartz S.R., Dai H., Sachs A.B., Linsley P.S. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Webster C., Blau H.M. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat. Cell Mol. Genet. 1990;16:557–565. doi: 10.1007/BF01233096. [DOI] [PubMed] [Google Scholar]
- 54.Aartsma-Rus A., Janson A.A., Kaman W.E., Bremmer-Bout M., den Dunnen J.T., Baas F., van Ommen G.J., van Deutekom J.C. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- 55.Stilwell J.L., Samulski R.J. Role of viral vectors and virion shells in cellular gene expression. Mol. Ther. 2004;9:337–346. doi: 10.1016/j.ymthe.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 56.McCaffrey A.P., Fawcett P., Nakai H., McCaffrey R.L., Ehrhardt A., Pham T.-T.T., Pandey K., Xu H., Feuss S., Storm T.A., Kay M.A. The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver. Mol. Ther. 2008;16:931–941. doi: 10.1038/mt.2008.37. [DOI] [PubMed] [Google Scholar]
- 57.Snøve O., Jr., Holen T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 58.Chandramohan R., Wu P.-Y., Phan J.H., Wang M.D. Benchmarking RNA-seq quantification tools. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013;2013:647–650. doi: 10.1109/EMBC.2013.6609583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dapas M., Kandpal M., Bi Y., Davuluri R.V. Comparative evaluation of isoform-level gene expression estimation algorithms for RNA-seq and exon-array platforms. Brief. Bioinform. 2017;18:260–269. doi: 10.1093/bib/bbw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier T., Güell M., Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 61.Roberts T.C., Johansson H.J., McClorey G., Godfrey C., Blomberg K.E.M., Coursindel T., Gait M.J., Smith C.I., Lehtiö J., El Andaloussi S., Wood M.J. Multi-level omics analysis in a murine model of dystrophin loss and therapeutic restoration. Hum. Mol. Genet. 2015;24:6756–6768. doi: 10.1093/hmg/ddv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandamme C., Adjali O., Mingozzi F. Unraveling the complex story of immune responses to AAV vectors trial after trial. Hum. Gene Ther. 2017;28:1061–1074. doi: 10.1089/hum.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noiret M., Méreau A., Angrand G., Bervas M., Gautier-Courteille C., Legagneux V., Deschamps S., Lerivray H., Viet J., Hardy S. Robust identification of Ptbp1-dependent splicing events by a junction-centric approach in Xenopus laevis. Dev. Biol. 2017;426:449–459. doi: 10.1016/j.ydbio.2016.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.