Abstract
We report a novel multi-omic resource generated by applying quantitative trait locus (xQTL) analyses to RNA sequence, DNA methylation, and histone acetylation data from the dorsolateral prefrontal cortex of 411 older adult individuals that have all three data types. We identify SNPs significantly associated with gene expression, DNA methylation, and histone modification levels. Many of these SNPs influence multiple molecular features, and we demonstrate that SNP effects on RNA expression are fully mediated by epigenetic features in 9% of these loci. Further, we illustrate the utility of our new resource, xQTL Serve, by using it to prioritize the cell type(s) most affected by an xQTL. We also re-analyze published genome wide association studies (GWAS) using a xQTL-weighted analysis approach and identify 18 new schizophrenia and 2 new bipolar susceptibility variants, which is more than double the number of loci that can be discovered with a larger blood-based eQTL resource.
Introduction
Genome wide association studies (GWAS) have identified thousands of SNPs that are associated with various human diseases1. However, the majority of identified SNPs fall in the non-coding regions of the genome2. Connecting these regulatory changes to specific genes or to molecular pathways that may be implicated in human diseases is not straightforward. Suggestive evidence indicate that many more such SNPs exist, but they are difficult to detect due to their typically small effect sizes and the challenge of multiple testing burden in genome-wide assessment of common genetic variation3.
Expression quantitative trait locus (eQTL) analyses4–6 have been very useful in understanding the functional consequences of trait- and disease-associated variants and in identifying genes that are likely to be affected by a risk allele. Recently, QTL analyses have been extended to other molecular phenotypes, such as DNA methylation (mQTL)7,8 and histone modification (haQTL)9. Overall, SNPs associated with molecular phenotypes (xQTLs) are over-represented among SNPs that are linked to various traits and diseases6,10, and previous studies have used eQTL hits to prioritize associations in GWAS, leading to improved detection sensitivity11–13. While a few datasets exist for brain tissue, large datasets measuring all three of these epigenomic and transcriptomic features have only recently been generated from the same brain region of the same individuals.
Here, we present a new Resource for the neuroscience community by performing xQTL analyses on a multi-omic dataset that consists of RNA sequence (RNA-seq), DNA methylation, and histone acetylation (H3K9Ac ChIP-seq) data derived from the dorsolateral prefrontal cortex (DLPFC) of up to 494 subjects (411 subjects having all three data types available). Samples are collected from participants of the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP), which are two longitudinal studies of aging designed by the same group of investigators. These studies share the same sample and data collection procedures, which naturally permits joint analyses14,15. At its heart, the Resource presents a list of SNPs associated with cortical gene expression, DNA methylation, and/or histone modification levels that reflects the impact of genetic variation on the transcriptome and epigenome of aging brains. While our xQTLs replicated well in both brain and blood, a notable portion is specific to genes that are only expressed in brain. Also, many SNPs influence multiple molecular features, with a small number of them having their impacts on gene expression mediated through epigenetics. Further, we apply a computational approach to prioritize the cell types that may be driving the tissue-level effect, a critical piece of information for designing follow-up molecular experiments in which an in vitro or in vivo target cell type needs to be selected. Finally, we illustrate the efficacy of an “xQTL-weighted GWAS” approach for applying our xQTLs. We show that this approach increases the statistical power of GWAS, resulting in the detection of a number of new susceptibility variants for several diseases. All data used in this study are available from www.radc.rush.edu, and the xQTL results and analysis scripts can be accessed through our online portal, xQTL Serve, at http://mostafavilab.stat.ubc.ca/xQTLServe.
Results
xQTL Discovery
Genotype data16 were generated from 2,093 individuals of European-descent. Of these individuals, gene expression (RNA-seq)(n=494), DNA methylation17 (450K Illumina array)(n=468), and histone modification data (H3K9Ac ChIP-seq)(n=433) were derived from post-mortem frozen samples of a single cortical region, the dorsolateral prefrontal cortex (DLPFC) (Figure 1A). 411 individuals have all four data types. Demographics of the analyzed individuals are summarized in Table S1. Although some of these data have been previously published with respect to analysis of aging brain phenotypes (see Table S2), here we report genome-wide xQTL analyses for these datasets for the first time. Genotype imputation was performed using BEAGLE 3.3.218 with the 1000 genome reference panel19, yielding 7,321,515 SNPs for analysis. For the molecular phenotype data, 13,484 expressed genes, 420,103 methylation sites, and 26,384 acetylation peaks remained after quality control (QC) analyses (Figure S1–S3). The effects of known and hidden confounding factors were removed from the molecular phenotype data using linear regression (Supplementary Information). Consistent with previous studies, we observed that accounting for hidden confounding factors greatly enhances the statistical power of cis eQTL detection20, and we confirm that this observation holds true for cis mQTL and cis haQTL detection (Figure S4).
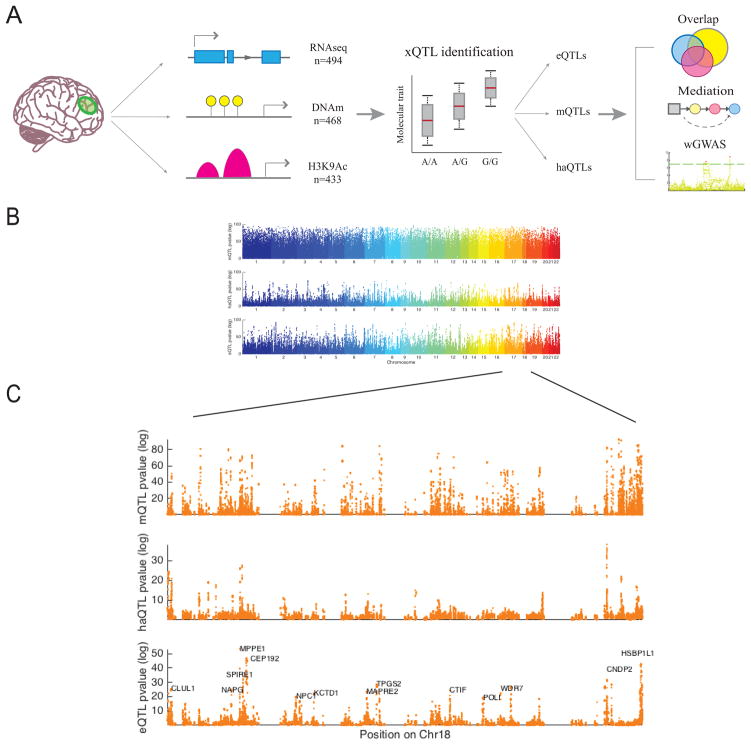
Figure 1. Overview of xQTL analysis.
(A) Graphical summary of our data and analyses. We first associate genetic variation with each data type separately to establish our xQTL reference. We then use these xQTLs to assess whether a given SNP influences more than one data type, whether epigenomic features mediate the effects of SNPs on gene expression, and whether our xQTLs can be leveraged to discover new susceptibility loci. (B) −log10 p-value of Spearman’s correlation between SNPs and DNA methylation (mQTL), histone acetylation (haQTL), and gene expression (eQTL) vs. the SNPs’ physical positions in the genome. Each dot represents the strongest association within a cis window for each SNP. (C) Zoomed in Manhattan plot of chromosome 18 to illustrate p-value distribution of xQTLs at a higher resolution.
We employed Spearman’s rank correlation to estimate the association strength between alleles of each SNP and gene expression (n=494), DNA methylation (n=468), and histone acetylation levels (n=433). We refer to the measurement unit of each molecular phenotype data as a feature and a significant association between a SNP and a feature as an xQTL (i.e. an xQTL is a SNP-feature pair). Based on the results of prior studies, we performed cis xQTL analysis between SNPs and each feature by defining a window size of 1Mb for eQTL analysis and haQTL analysis, and a 5Kb window for mQTL analysis21,22. The 1Mb window for haQTL analysis was motivated by the possibility that SNPs in enhancer regions, which are far away, can indeed impact gene regulation through interaction with promoter regions (e.g. chromatin looping). The much smaller window for the mQTL analysis was selected since the majority of cis mQTLs with the strongest correlation lie within a window of this size22. Also, the smaller window size helps reduce the multiple testing burden, given the much larger number of DNA methylation features.
Using a Bonferroni corrected p-value threshold (αFWER = 0.05, two-tailed), we found (1) 3,388 genes associated with eQTL SNPs (p<8x10−10), (2) 56,973 CG dinucleotides linked to mQTL SNPs (p<5x10−9), and 1,681 H3K9Ac peaks influenced by haQTL SNPs (p<4x10−10) (Figure 1B–C, Table 1). Among the eQTL genes, 133 of them correspond to lincRNAs out of a total of 391 lincRNAs tested. The complete lists of eQTLs, mQTLs, and haQTLs are provided through the xQTL Serve webpage: http://mostafavilab.stat.ubc.ca/xQTLServe.
Table 1.
Summary of xQTL associations.
| No. associations (SNP-gene pairs) | No. features | No. SNPs | ||||
|---|---|---|---|---|---|---|
| Tested | Significant | Tested | Significant | Tested | Significant | |
| eQTLs (1Mb) | 60,456,556 | 405,429 | 12,979 | 3,388 | 6,442,864 | 313,467 |
| mQTLs (5Kb) | 9,939,236 | 693,696 | 412,152 | 56,973 | 2,358,873 | 383,920 |
| haQTLs (1Mb) | 125,100,450 | 156,693 | 25,720 | 1,681 | 6,756,597 | 119,778 |
Replication and cross-tissue comparisons
We evaluated the extent to which our xQTLs replicate brain eQTLs and mQTLs found in prior studies. We focused on eQTL and mQTL replication since relevant large-sample datasets are only available for these two xQTL types. Specifically, we assessed the replication rate of brain eQTLs, discovered in the CommonMind23 and Braineac24 studies, and brain mQTLs in a fetal brain study8, in our dataset using the π1 statistics25, which estimates the proportion of these eQTLs (mQTLs) that are also significant in our dataset. π1 of the eQTLs are 0.91 and 0.56 for CommonMind and Braineac, respectively, and π1 of mQTLs is 0.87 for the fetal brain study. All of these results are greater than their respective empirical null mean of 0.11 and 0.33 for eQTLs and mQTLs (p < 0.0001, one-tailed, see Supplementary Information). The lower replication rate of Braineac eQTLs compared to CommonMind eQTLs could be due to its smaller sample size. Also, the Braineac eQTLs were based on false discovery rate (FDR) correction whereas CommonMind eQTLs were defined using Bonferroni correction, and stronger associations captured by more stringent correction are more likely to replicate26. We also assessed the replication rate of our eQTLs in the CommonMind data, and estimated similar replication rate (π1=0.90). For the mQTL replication analysis, we explored restricting our mQTL analysis to a 100Kb window, and observed similar replication rate (π1=0.87) on the fetal brain mQTLs8, which suggests a 5Kb window already captures majority of the stronger associations between SNP and DNA methylation.
For assessing cross-tissue replication, we used a large whole-blood eQTL dataset from the DGN study26 and two smaller eQTL datasets from the Immune Variation (ImmVar) study27 that consist of monocyte and T cell data. π1 of these eQTLs in our dataset are 0.63 (whole blood), 0.61 (monocytes), and 0.67 (T cells), which are greater than their empirical null mean of 0.10 (p < 0.0001 for all three datasets, one-tailed). Thus, a large proportion of blood eQTLs are present in our brain data. We also assessed the replication rate of our brain-derived eQTLs in the whole-blood DGN dataset (Figure 2A–B). When we consider SNP-gene pairs that can be tested in both studies, we observed a replication rate of 0.83 (Figure 2C), which is greater than its empirical null mean of 0.30 (p < 0.0001, one-tailed). This increase in replication rate may be due to the higher statistical power of the DGN study and the fact that cortical tissue consists of a large variety of cell types, which in aggregate, expresses a large proportion of the transcriptome. Since blood contains a mixture of cell types including immune cells that share characteristics with those in brain, we further assessed the replication rate on three additional tissues, namely subcutaneous adipose, visceral adipose, and liver from the GTEx study28. The replication rates are 0.51, 0.38, and 0.20, respectively, which are indeed lower than that of blood. Additional replication results for different tissues, window sizes, and xQTL types are provided in Table S3.
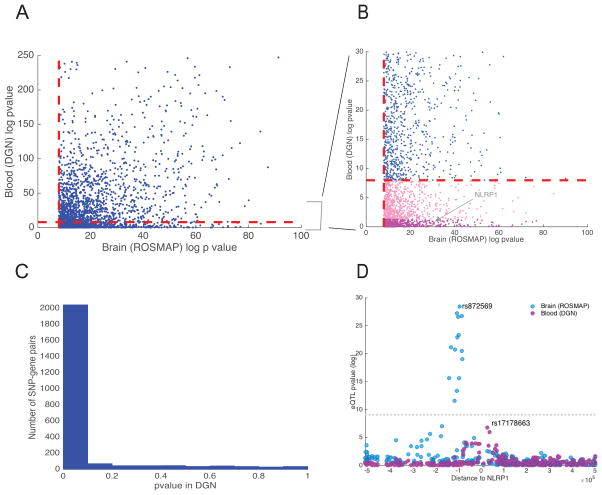
Figure 2. Cross-tissue replication analysis.
(A) Scatter plot of −log10 p-values of associations between the lead brain eQTL SNPs and their associated genes in brain and blood. The dashed red lines denote the significance threshold (αFWER=0.05 with Bonferroni correction). (B) −log10 p-value distribution of eQTLs that appear to be brain-specific (light and dark pink dots, the latter are specific to NLRP1). (C) Distribution of p-values from the DGN study restricted to brain eQTLs. Estimated replication rate (π1 statistics) between blood and brain eQTLs is 0.83. (D) eQTL p-values at NLRP1 locus. Each dot represents one SNP tested in either brain (ROSMAP) or blood (DGN). The x-axis corresponds to the distance between each assessed cis SNP and NLRP1’s TSS, and the y-axis corresponds to −log10 p-values for association between SNPs and NLRP1 expression. The LD between the lead SNP in blood and brain is r2 < 0.1.
An important question to answer with our data is whether and which of the detected xQTLs are brain-specific. However, without tissue samples from the same individuals, distinguishing between subject-specific and tissue-specific effects is not possible. Nonetheless, based on the sparsity of “population-specific” eQTLs27 and a lower replication rate of eQTLs in blood compared to brain, a notable fraction of our eQTLs are likely tissue-specific. For example, when we considered only eQTLs that consist of the top SNP for each gene, we found that, of the 2,416 eQTLs discovered in our cortical tissue study that are testable in the whole-blood dataset26, 433 eQTLs (18%) have an unadjusted p-value >0.05, indicating that this subset of brain eQTLs are unlikely to be present in blood (Figure 2B). As an example, NLRP1 RNA is expressed in both brain and blood (whole blood, monocytes, and T cells), but its expression is only associated with brain-specific eQTL SNPs (Figure 2D). NLRP1 is a member of the inflammasome complex that is implicated in inflammatory response in both immune cells (in particular myeloid cells) and in brain29. Interestingly, a few small-scale studies also linked polymorphisms in this gene with amyloid-beta secretion and Alzheimer’s disease (AD)30. In addition to the 2,416 eQTLs that are testable in both brain and blood, we identified 809 eQTL target genes from our brain eQTL analysis that were absent from the DGN blood eQTL analysis since these genes were not expressed in blood. As expected, this set of 809 brain-specific eQTL genes are enriched for brain-relevant functions (GSEA enrichment analysis, FDR<0.05, two-tailed) such as “Neuronal System”, “Potassium Channel Components”, and “Neurotransmitter Receptor Binding”.
Overall, the high cross-sample and cross-tissue replication rates suggest that a large number of SNPs that impact molecular phenotypes are likely shared across contexts. The degree of overlap between brain and blood eQTLs is quite high, with a π1 of ~0.8. Nevertheless, our results suggest some eQTLs are tissue-specific, and more tissue-specific effects would likely emerge from analyses of purified cell populations.
Genetic architecture of xQTL SNPs and sharing across molecular phenotypes
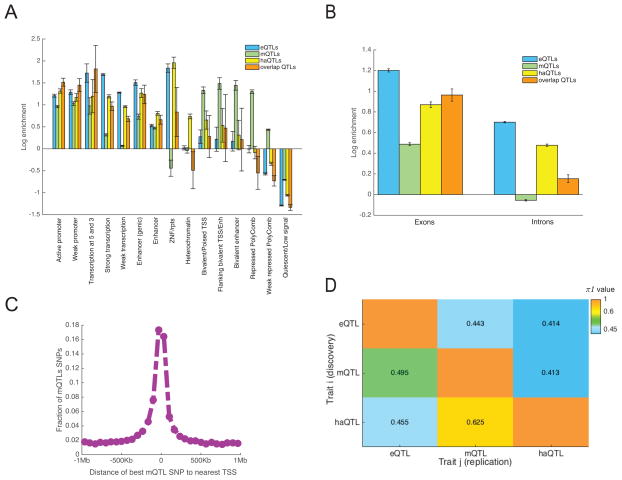
We used genomic annotations based on DLPFC tissue from ChromHMM31 and computed the log odds of an xQTL SNP belonging to 1 of 15 regulatory regions (annotated by chromatin states) as compared to all non-xQTL SNPs proximal to molecular features, i.e. within 1Mb, 5Kb, and 1Mb windows for eQTL, mQTL, and haQTL analyses with all SNPs tested in these analyses considered as proximal. As shown in Figure 3A, eQTL SNPs are mainly enriched in promoters and transcribed regions, conforming to our understanding of how SNPs at transcription factor (TF) binding sites can affect protein-DNA interactions32 and how SNPs in transcribed regions are known to affect mRNA processing and turnover33. haQTL SNPs are also largely enriched in promoter and transcribed regions, consistent with the role of H3K9Ac in transcriptional activation34. By contrast, mQTL SNPs are mainly enriched in bivalent regions (promoters and enhancers) and PolyComb repressed regions, which matches prior findings that a large portion of mQTL SNPs resides in chromatin regions that are developmentally regulated22. Also, suppressed gene expression in PolyComb repressed regions might partly explain why eQTL and haQTL SNPs derived from adult samples are scarce in these regions. Notably, xQTL SNPs that are shared across all three molecular phenotypes are mainly enriched close to the TSS as well as in the 5′ and 3′ transcribed regions. With respect to transcribed sequences, we saw enrichment for all types of xQTLs in exons relative to introns (Figure 3B), with this trend being most striking for mQTLs.
Figure 3. Genomic enrichment of xQTLs and their overlap.
(A) Log odds ratio of xQTL SNP enrichment in 15 different chromatin states31 as defined by the Roadmap Epigenomics project via applying ChromHMM to DLPFC samples from two cognitively non-impaired ROSMAP subjects. The error bars reflect standard deviation. (B) Log odds ratio of xQTL SNP enrichment in exons and introns. The error bars reflect standard deviation. (C) Distribution of distance between each lead mQTL SNP and its nearest TSS. (D) π1 statistics for assessing xQTL sharing across the three molecular features. Each cell (i,j) corresponds to the proportion of xQTLs of trait j that share the same xQTL SNPs identified in trait i.
To quantify the degree to which an xQTL SNP influences more than one molecular phenotype, we first identified the list of xQTL SNPs for a “discovery” phenotype and then estimated the π1 statistics of the SNP-feature associations for a “test” phenotype that share the same xQTL SNPs. Since an xQTL SNP might be tested for association with multiple cis features, e.g. an mQTL SNP was, on average, tested for association with 18 gene expression levels, a decision on which SNP-feature associations to include in the π1 estimation was necessary (see Supplementary Information). In particular, we examined the distance between each pair of “discovery” SNP and “test” feature, and found this distance to be a prime determinant of cross-phenotype sharing. For example, the strongest associated eQTL gene for each mQTL SNP is often the gene closest to the mQTL SNP (Figure 3C, Figure S5). Based on this observation, we estimated π1 to be 0.41–0.63 when we considered only the closest feature to each xQTL SNP (Figure 3D). Also, we examined the effect of window size by restricting the haQTL analyses to 2Kb, 40Kb, and 100Kb windows as well as changing the eQTL and mQTL analysis window to 100Kb, and found negligible differences in our estimates of xQTL sharing (Table S4).
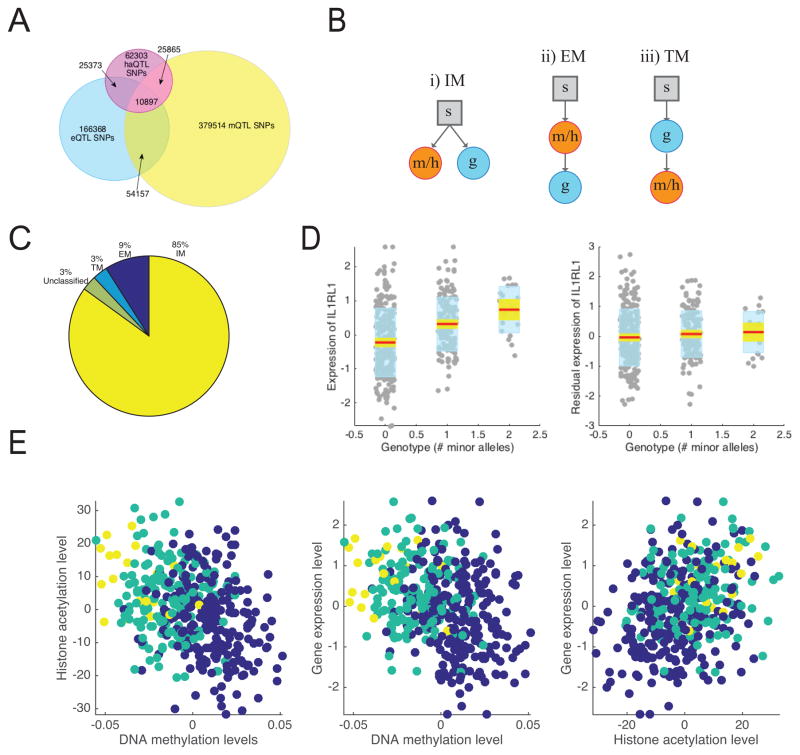
The availability of multi-omic data from the same individuals enabled us to go beyond “overlap analyses” (Figure 4A) and to investigate the cascading effect of genetic variation through the measured regulatory genomics layers. Specifically, we investigated whether the effect of a regulatory cis xQTL SNP is mechanistically mediated through its impact on epigenetic modification or gene expression using the casual inference test (CIT)35. This analysis was performed on 10,897 xQTL SNPs (impacting 629 genes based on the eQTL analysis) that are associated with all three molecular phenotypes, as only such SNPs satisfy the precondition for mediation analysis. With this analysis, we distinguished between three models for propagation of information from genetic variation: 1) independent effects of a SNP on cis gene expression and the cis epigenetic landscape (independent model or IM), 2) a propagation path from SNP to gene expression via epigenetic modifications (epigenetic mediation model or EM), or 3) a propagation path from SNP to the epigenome (namely DNA methylation) via gene expression (transcription mediation model or TM) (Figure 4B).
Figure 4. Epigenetic mediation of eQTLs.
(A) Sharing of SNPs between eQTLs, mQTLs, and haQTLs. 2,305,942 SNPs tested for all molecular phenotypes are considered. (B) Three models relating SNPs (s), epigenetic features (methylation/histone acetylation, m/h) and gene expression (g): i) independent model (IM) where effects of SNPs on epigenetic features and transcripts are unrelated, ii) epigenetic mediation model (EM) where epigenetic features mediate the effects of SNPs on gene expression, and iii) transcription mediation model (TM) where the effects of SNPs on epigenetics is mediated through its effect on gene expression. The causal inference test was used for assessing mediation35. (C) Proportion of shared xQTL SNPs that are consistent with each model. (D) Expression level of IL1RL1 vs. number of minor alleles present for rs13015714, which is a shared xQTL SNP that impacts IL1RL1 expression and nearby DNA methylation and histone acetylation levels. The red line corresponds to the mean. The yellow region corresponds to the 95% confidence interval of the mean. The edges of the blue region correspond to ±1 standard deviation. The SNP effect disappears after regressing out the effect of the mQTL probes and haQTL peaks associated with rs13015714 from IL1RL1 expression. (E) Association between IL1RL1 expression and the levels of its associated methylation probes and acetylation peaks. Colors indicate the genotype of rs13015714: minor allele homozygotes (yellow), heterozygotes (green), major allele homozygotes (blue).
Using Bonferroni correction with the CIT test (n=411, two-tailed), we observed that 9% of the association sets conform to the EM model, 3% conform to the TM model, 85% conform to IM, and the remaining 3% could not be classified (Figure 4C, Table S5). As an example, an xQTL SNP (rs13015714) associated with Celiac disease (GWAS p<10−8) was found to affect IL1RL1 gene expression (p<10−11), DNA methylation (p<10−30) and histone modification (p<10−12), but the impact of this SNP on gene expression appeared to be fully mediated by epigenetic modifications (Figure 4D–E), and thus this SNP conforms to the EM model. We additionally tested whether GWAS SNPs (downloaded from the GWAS catalog1) are preferentially enriched for any of these models but did not find any model-specific enrichment.
A large fraction of the shared xQTL SNPs appear to affect gene expression directly. This result could be explained by: 1) epigenetic modification playing a passive role21 where gene expression in fact lies upstream of epigenetic modification (3% based on the TM model), 2) regulation of gene expression being dependent on a more complex combination of epigenetic marks that are not measured in our subjects, and 3) artefactual decorrelation between the expression and epigenomic features due to technical or other factors. Thus, we should interpret the detected mediation as only a subset of true mediation, i.e. these may be the most robust subset of mediation events. Further work and additional data may be needed to assess this issue more comprehensively. Indeed, when we separately included only DNA methylation or histone modification into the model, we identified a smaller subset of association sets for which an effect on gene expression was fully explained by the epigenetic features: 3% for DNA methylation and 6% for histone modification. Thus, a complementary (non-redundant) combination of DNA methylation and histone acetylation seems to be required to capture the mediation effect, and adding other non-redundant epigenetic features would likely further enhance detection of this type of functional propagation.
Enrichment of disease susceptibility SNPs among xQTL SNPs
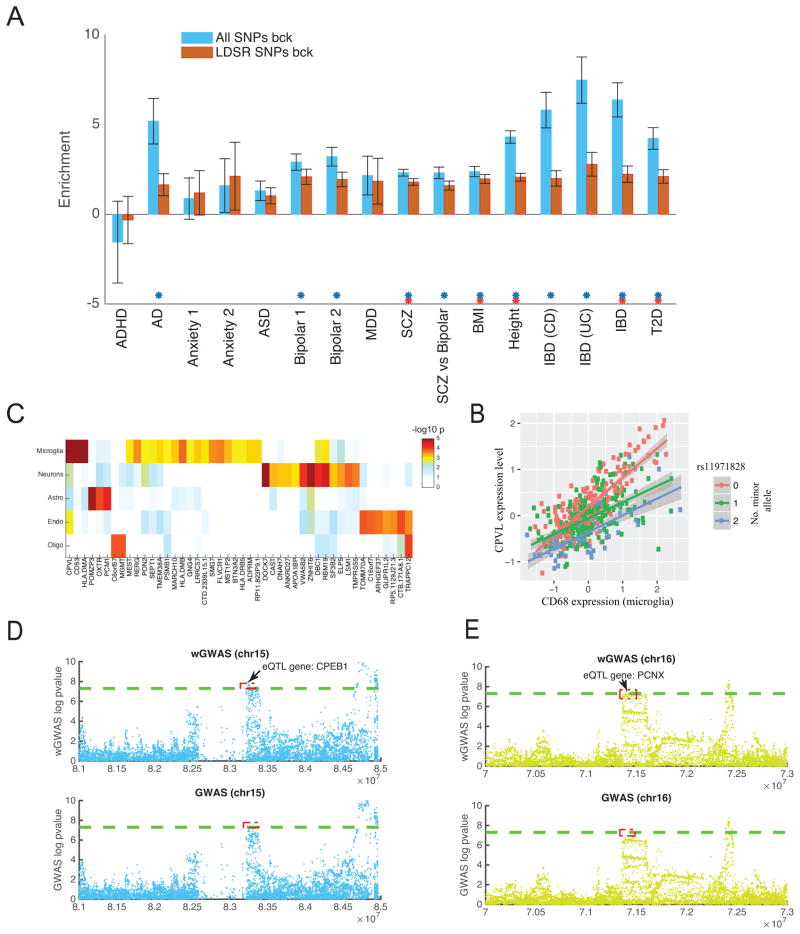
Studies have shown that SNPs associated with eQTLs are more likely to influence complex traits and disease susceptibility6,10. Here, we provide further support for this observation for eQTLs, mQTLs, and haQTLs by performing an enrichment analysis on reported p-values of 16 GWAS datasets, including large-scale GWAS meta-analyses of AD36, Schizophrenia37, and type II Diabetes38 (Supplementary Information). Enrichment was assessed using stratified linkage disequilibrium (LD) score regression (LDSR)39. For all 12 GWAS studies (out of 16) with over 20,000 samples (Table S6, Figure 5A), significant enrichment was observed for the xQTL SNPs. We also repeated this analysis using a more stringent background model, where we considered enrichment of our xQTLs against a background set of SNPs falling in “generic” annotation categories as provided in the LDSR software39. Again, significant enrichment, albeit with lower effect size, was observed for many of the GWAS studies (Figure 5A, Table S6). Next, we hypothesized that SNPs shared between xQTL types, which affect multiple molecular phenotypes, are more likely to impact downstream processes and could constitute a list of “high confidence” functional SNPs. We therefore compared all xQTL SNPs that are shared across at least two molecular traits, against those xQTLs that are only found for one molecular trait. Indeed, we observed enrichment for the shared xQTLs, but their enrichment was not always higher than the background xQTL SNPs, i.e. somewhat trait dependent (Table S6). To test the robustness of the results to window size, we repeated the analysis with 100Kb windows for all three xQTL types (Table S7). The overall trend remained the same with slightly higher enrichment observed.
Figure 5. Application of the xQTL Resource for translational studies.
(A) Enrichment of xQTL SNPs in published GWAS datasets based on the LDSR model39. Enrichments are with respect to two sets of background SNPs: 1) all genome-wide SNPs and 2) SNPs falling in generic functional sites previously defined by LDSR. The error bars reflect standard deviation. (B) −log10 p-value of interaction test in quantifying cell-specificity. 46 genes that survived FDR correction at a q-threshold of 0.2 shown. (C) Level of CPVL expression vs. a marker of microglial proportion (CD68 gene). CPVL expression is found to increase with increasing proportion of microglia, particularly among major allele homozygotes (pink dots). (D–E) Zoomed in Manhattan plot around the PCNX (D) and CPEB1 (E) loci, showing the results of the published standard GWAS (bottom panel) and our weighted GWAS (top panel). Each dot is one SNP. The dotted green line is the standard genome-wide significance threshold (p < 5x10−8).
The enrichment results are reassuring, and, as we describe later, we can use our list of xQTL SNPs to enhance susceptibility locus discovery in GWAS studies. Investigators can also confidently use our xQTL lists to annotate GWAS SNPs related to the brain or nervous system which will accelerate the transition to functional studies. For example, we used our eQTLs to map the 21 SNPs (and correlated SNPs in LD with r2 > 0.8) reported in the IGAP AD GWAS and identified four candidate AD genes that are absent from the reported gene list defined by proximity36 (MADD, MTCH2, PILRA, and POLR2E). The TSS of these eQTL mapped genes were >100Kb, on average, from their respective AD SNPs. MTCH2, PILRA, and POLR2E have also been found in recent eQTL mapping studies40, demonstrating the robustness of our results. MADD has not been previously reported in this context but is a good candidate given that its expression correlates with neuronal cell death in AD41 and that it has also been reported to modulate AD-related tau toxicity in a Drosophila model42.
Accelerating the transition to functional studies in specific cell types
Selection of the relevant cell type to target for in vitro or in vivo follow-up functional studies is challenging since our xQTLs, like those identified in many other studies, rely on tissue profiles generated from a complex mixture of cell types. To help prioritize cell types for such follow-up efforts, we repeated the analyses relating each SNP to a given molecular feature but additionally included a variable that estimates the proportion of a cell type in the profiled tissue and an interaction term to identify those SNPs whose effects depend on the proportion of a target cell (Supplementary Information). This approach was recently validated using whole-blood data43.
Using eQTL results as an example (n=494, two-tailed), we examined the potential specificity of each lead eQTL SNP for five cell types that are abundant in the cortex: neurons, microglia cells, astrocytes, oligodendrocytes, and endothelial cells. We found that assignment to a single cell type is ambiguous for most eQTLs (p-values available at http://mostafavilab.stat.ubc.ca/xQTLServe). In a minority of cases, our analysis returned an unambiguous cell type for the lead eQTL. For example, at an FDR<0.05, we identified 6 significant cell-specific eQTLs (1 astrocytic, 3 microglia, and 1 neuronal). An example is presented in Figure 5C. The CPVL locus harbors an eQTL effect (rs11971828) that is stronger in microglial cells. With a more lenient discovery strategy where we thresholded the interaction term at an FDR<0.2, we found putative cell-type specific effects in neurons (n=13) and microglia (n=22) (Figure 5B). Even though only a small number of cell-specific eQTLs were identified with multiple testing correction, our results can still be useful in prioritizing cell types for follow-up experiments based on the observation that suggestive cell-type specific eQTL genes show clear cell type preferences. Many of these “top” cell-specific eQTL genes tend to conform to the expected function of the implicated cell. For example, the MGMT locus harbors an eQTL that ranks among the top 3 for oligodendrocytes-specificity (p=1.5x10−4). MGMT is known to play a role in oligodendrocyte function, and its mutations are associated with oligodendrogliomas. These cell-specific results are intriguing but require molecular validation using purified cell populations from the cortex with matched genotypes to be confirmed.
xQTL-weighted GWAS for gene discovery efforts
Our large compendium of brain xQTLs can also be leveraged to accelerate gene discovery by boosting statistical power in GWAS. The simplest way of using our xQTL SNP list would be to restrict association analysis to our xQTL SNPs. However, such a strategy would miss other relevant SNPs that are not in our list (or were not tested in the cis xQTL analysis). Thus, we opted to use a weighted Bonferroni procedure44, which permits all SNPs to be analyzed but weights their p-values by their potential phenotypic relevance. We refer to this approach as an “xQTL-weighted GWAS”. Provided that the weights are non-negative and average to one, strong control on family-wise error rate is guaranteed44. We employed a binary weighting scheme, where p-values of xQTL SNPs were divided by w1 and all other SNPs were divided by w0 with s = w1/w0 > 1 (see Supplementary Information for s selection). Consistent with the standard GWAS convention, significance was declared at p < 5x10−8. To not over-count the number of significant hits due to correlations between SNPs, we applied PLINK1.945 on the 1000 Genomes phase 1 data19 to remove SNPs among the significant hits that are in linkage disequilibrium with one other (r2<0.2).
We compared five approaches: (1) no weighting, (2) weighting xQTL SNPs found for any of the molecular phenotypes, (3) weighting SNPs within predefined windows from the molecular features (1Mb, 5Kb, and 1Mb for eQTL, mQTL, and haQTL analyses) to account for distance bias, (4) weighting generic functional SNP in the LDSR baseline model39, and (5) weighting xQTL SNPs that are shared across any of the molecular phenotypes. Over the 19 GWAS datasets (Supplementary Information), weighting xQTL SNPs resulted in equal or more GWAS hits than no weighting, except for inflammatory bowel disease (Table S8). For 8 of the 19 studies, the xQTL-weighted GWAS approach found at least 2 new independent loci (Table S8). By contrast, weighting SNPs within predefined windows from the molecular features as well as weighting SNPs in the LDSR baseline model resulted in little change in detection sensitivity. Interestingly, the gain in sensitivity was not always the highest when we weighted the shared xQTL SNPs. Also, compared to weighting the DGN eQTL SNPs, weighting the union of all xQTL SNPs found in this study identified more additional independent susceptibility SNPs for a majority of the tested GWAS datasets, which demonstrates that additional signals are captured by mQTL and haQTL SNPs. In particular, weighting the xQTL SNPs found 22, 18, and 9 additional independent SNPs for schizophrenia, height, and inflammatory bowel disease, respectively, compared to no weighting. In contrast, weighting the DGN eQTL SNPs found only 9, 3, and 2 additional independent SNPs. In fact, weighting just the ROSMAP eQTL SNPs identified 17 additional independent SNPs for schizophrenia, which illustrates the presence of eQTLs in our data that are enriched in brain diseases and not observed in blood.
Among the brain diseases that we examined, the largest detection gain was obtained with the schizophrenia dataset37, where 18 additional loci met genome-wide significance (excluding those near the MHC region) and were not in linkage disequilibrium (r2<0.2) with the reported susceptibility SNPs37. 7 of these 18 SNPs were found to be associated with eQTLs (Table S8), including rs57709857, which influences LSM1, a gene previously found in a Han Chinese schizophrenia study46. However, the LSM1 locus had not reach genome-wide significance in individuals of European ancestry47. The list of eQTL genes also includes PCNX (associated with rs2189806), a gene encoding a member of the Notch signalling pathway that was reported to harbour a de novo copy number variant linked to Autism Spectrum Disorder48, and CPEB1 (associated with rs1864699), which was recently found to be implicated in experience-dependent neuronal development and circuit formation49 (Figure 5D–E). Thus, several of our new schizophrenia loci have some face validity, but additional replication efforts are required to ensure that these are robust findings. In terms of the percentage increase in detection sensitivity, the largest gain was observed for Bipolar disorder50, where the standard GWAS approach identified one significant hit, whereas xQTL-weighted GWAS identified 2 additional independent loci.
Conclusion
Using one of the largest multi-omic datasets for brain tissue, we generated a list of xQTLs as a Resource for the neuroscience community to further investigate the interplay between the genome, epigenome, and transcriptome in disease susceptibility. Our list of xQTLs replicates well in both brain and blood datasets, but it also contains xQTLs that appear unique to brain. Notable biological insights drawn from this Resource include significant sharing of xQTL SNPs across the measured molecular phenotypes. Also, the effects of some eQTL SNPs are fully mediated by our two epigenetic features, but further work and more data are needed to comprehensively assess the extent to which epigenomic features mediate eQTL effects. Overall, we created a large new reference with which investigators can functionally annotate their results, enhance their analyses as illustrated by our xQTL-weighted GWAS approach, and guide functional studies as with our cell type analysis. This Resource can be easily accessed through our portal, xQTL Serve.
Supplementary Material
Acknowledgments
We thank the participants of ROS and MAP for their essential contributions and gift to these projects. This work has been supported by many different National Institute of Health (NIH) grants: P330AG10161, U01 AG046152, R01AG16042, R01 AG036836, R01 AG015819, R01 AG017917, R01 AG036547. The U01 AG046152 grant (to PLD and DAB) is a component of the AMP-AD Target Discovery and Preclinical Validation Consortium, a program of the National Institute of Aging and the Foundation of the NIH. Data and samples used in this study are available at the RADC Research Resource Sharing Hub at www.radc.rush.edu.
Footnotes
Accession Codes
The molecular datasets used in this manuscript are available via the AMP-AD Knowledge Portal: RNA-seq data - doi:10.7303/syn3388564; ChIP-seq data - doi:syn4896408; DNA methylation data - doi:10.7303/syn3157275. The results of the study are available through the xQTL Serve website: http://mostafavilab.stat.ubc.ca/xQTLServe
Contributions
Study design: SM, BN, PLD. Sample collection: DAB. Data generation and quality control analyses: BN, CM, HK, EP, JX, SM, PLD. Analyses: BN, CW, CG, SM. Interpretation of results and critical review of the manuscript: BN, CM, HK, EP, JX, CG, DAB, SM, PLD.
References
- 1.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein DB. Common genetic variation and human traits. New England Journal of Medicine. 2009;360:1696. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 4.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannon E, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 14.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett DA, et al. Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jager PL, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging. 2012;33:1017 e1011–1015. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Jager PL, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nature Neuroscience. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez-Arcelus M, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do C, et al. Mechanisms and Disease Associations of Haplotype-Dependent Allele-Specific DNA Methylation. Am J Hum Genet. 2016;98:934–955. doi: 10.1016/j.ajhg.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome research. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh JG, Muruve DA, Power C. Inflammasomes in the CNS. Nat Rev Neurosci. 2014;15:84–97. doi: 10.1038/nrn3638. [DOI] [PubMed] [Google Scholar]
- 30.Pontillo A, Catamo E, Arosio B, Mari D, Crovella S. NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis Assoc Disord. 2012;26:277–281. doi: 10.1097/WAD.0b013e318231a8ac. [DOI] [PubMed] [Google Scholar]
- 31.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaffney DJ, et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson AD, et al. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet Genomics. 2008;18:781–791. doi: 10.1097/FPC.0b013e3283050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida H, et al. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 35.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karch CM, Ezerskiy LA, Bertelsen S, Goate AM. Alzheimer’s Disease Risk Polymorphisms Regulate Gene Expression in the ZCWPW1 and the CELF1 Loci. PLoS One. 2016;11:e0148717. doi: 10.1371/journal.pone.0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc Natl Acad Sci U S A. 2004;101:4210–4215. doi: 10.1073/pnas.0307349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dourlen P, et al. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westra HJ, et al. Cell Specific eQTL Analysis without Sorting Cells. PLoS Genet. 2015;11:e1005223. doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roeder K, Devlin B, Wasserman L. Improving power in genome-wide association studies: weights tip the scale. Genet Epidemiol. 2007;31:741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]
- 45.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang L, Hu F, Zeng X, Gan L, Luo XJ. Further evidence for the association between the LSM1 gene and schizophrenia. Schizophr Res. 2013;150:588–589. doi: 10.1016/j.schres.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 48.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruderfer DM, et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.