Abstract
INTRODUCTION
Circadian alterations are prevalent in Alzheimer’s disease (AD) and may contribute to cognitive impairment, behavioral symptoms and neurodegeneration. Epigenetic mechanisms regulate the circadian clock, and changes in DNA methylation have been reported in AD brains, but the pathways that mediate circadian deregulation in AD are incompletely understood. We hypothesized that aberrant DNA methylation may affect circadian rhythms in AD.
METHODS
We investigated DNA methylation, transcription, and expression of BMAL1, a positive regulator of the circadian clock, in cultured fibroblasts and brain samples from two independent cohorts of aging and AD.
RESULTS
DNA methylation modulated rhythmic expression of clock genes in cultured fibroblasts. Moreover, rhythmic methylation of BMAL1 was altered in AD brains and fibroblasts and correlated with transcription cycles.
DISCUSSION
Our results indicate that cycles of DNA methylation contribute to the regulation BMAL1 rhythms in the brain. Hence, aberrant epigenetic patterns may be linked to circadian alterations in AD.
Keywords: DNA methylation, circadian rhythms, methylation cycles, Alzheimer’s disease, BMAL1, neurodegeneration, epigenetics, fibroblasts, circadian clock, brain
1. INTRODUCTION
Alzheimer’s disease (AD) is the most prevalent neurodegenerative disorder of aging worldwide. AD is characterized by accumulation of neuritic plaques and neurofibrillary tangles in the brain, accompanied by cognitive impairment and memory loss [1]. Disturbances of circadian rhythms affect almost all AD patients, evidenced by altered sleep/wake cycles, thermoregulation [2] and exacerbations of cognitive impairment and confusion during the evening (“sundowning”) [3, 4], and represent a major cause of hospitalization and morbidity in AD [5]. Recent basic and clinical studies have revealed correlations in circadian regulation with amyloid-β production and clearance [6], therefore disturbance of this pathway may contribute to AD pathogenesis [6] and provide a novel therapeutic target in slowing or reversing the progression of AD.
In mammalian cells, circadian rhythms are generated by the transcriptional/translational oscillation of the core components of the biological clock, including the positive regulators BMAL1, CLOCK and NPAS2, and the negative regulators CRY1/2 and PER1/2/3 [7]. These rhythms subsequently regulate the expression of >10% of the transcriptome, synchronizing metabolic and physiologic functions to predictable changes in the environment [8]. At the organism level, circadian rhythms are coordinated by the main pacemaker located in the suprachiasmatic nucleus (SCN) that signals to peripheral oscillators throughout the body [9]. Recently, transcriptome-wide analysis in post-mortem human brains revealed rhythmic patterns of gene expression in regions outside the SCN, including prefrontal cortex and hippocampus [10, 11].
Epigenetic mechanisms contribute to the regulation of the circadian clock. Transient changes in histone modifications modulate clock gene transcription [12], and rhythmic methylation cycles have been uncovered in the SCN master clock in rodents [13] and in secondary oscillators in the human frontal cortex [14]. Although significant changes in methylation have been reported in AD brains [15] and altered circadian methylation rhythms have been noted in the human frontal cortex in the context of AD [14], the pathways that mediate circadian deregulation in AD are yet not fully elucidated, and nothing is known about epigenetic mechanisms as they pertain to the clock in AD.
We investigated whether aberrant DNA methylation contributes to circadian deregulation. Using fibroblasts and postmortem brain samples from two large AD cohorts and matched controls, we provide strong evidence for a role of DNA methylation cycles in the regulation of rhythmic BMAL1 transcription in the brain and gene expression rhythms in cells; providing a link between epigenetic and circadian alterations associated with AD.
2. METHODS
2.1-Study Populations
We evaluated 66 postmortem frontal cortex samples (Brodmann’s area 9) from the Shiley-Marcos Alzheimer’s Disease Research Center at UCSD (Supp. Table 1). The parent study was reviewed and approved by the UCSD Human Research Protections Program. All subjects provided written informed consent prior to participating and donating tissue. We analyzed DNA 5mC methylation and RNAseq data from 396 participants from the Religious Orders Study and Rush Memory and Aging Project (ROS/MAP) (Supp. Table 1). All participants provided written informed consent and signed an Anatomic Gift Act. The studies were approved by the Institutional Review Board of Rush University Medical Center. Recruitment and assessment of these studies are reported elsewhere [16, 17].
2.2-BMAL1 gene expression
RNA was extracted from frozen brain samples or from 1×106 fibroblasts using the RNeasy Lipid Tissue Mini kit (Qiagen). Total RNA (1 µg) was reversed-transcribed with SuperScript VILO cDNA synthesis kit (Life Technologies). Quantitative real-time PCR (qPCR) was run in duplicate samples with a probe that detects three human BMAL1 isoforms (Taqman Hs00154147, Life Technologies) or the corresponding mouse Bmal1 transcripts (Taqman Mm00500226, Life Technologies). Probes detecting species specific β-actin transcripts, known to be stably expressed across the 24 hr day [18] were used as internal control for both, human and mouse assays. Relative transcript abundance was calculated as the inverse ratio to threshold cycle (1/dCt) and normalized using the lowest and highest levels as 0 and 100% respectively.
Detailed RNAseq methods and analysis for ROS/MAP cases are reported elsewhere [14].
2.3-BMAL1 Protein levels
Protein analysis of brain BMAL1 was performed on n=10 subject per group, selected according to their time of death to include representative samples distributed across the 24 h of the day. Extracts were obtained from 200 mg cortical tissue according to the nuclear fractionation protocol (Abcam). Western blotting was performed as described [19] using anti-BMAL1 antibody (Abcam, ab140646,1:1000) and anti β-Actin (Abcam, ab 8227, 1:1000). Quantity One (v.4.6.9, BioRad) was used for densitometry analysis. Data were expressed as the normalized ratio of BMAL1/ACTIN densitometry levels (normalization was performed using the lowest and highest values per group as 0 and 100% respectively). Detection of BMAL1 on fixed human fibroblast cultures was performed as described previously [19] using anti-BMAL1 antibody (Abcam, ab93806, 1:250) and FITC-anti rabbit secondary (Vector, 1:75).
2.4-DNA methylation
DNA methylation in brain samples was assessed in the ADRC cohort using the Illumina Infinium Human Methylation 450k BeadChip (Illumina) as described [20]. Briefly, genomic DNA extracted from frozen cortical tissue (DNeasy Blood & Tissue Mini kit, Qiagen) was bisulfite converted (EZ DNA Methylation kit, Zymo) and used for genome-wide methylation profiling at the UCSD IGM Genomics Core, under standard protocols (Illumina). The methylation status of a specific CpG site was expressed as β values, calculated as the ratio of the fluorescence intensity signals of the methylated (M) and unmethylated (U) alleles, using GenomeStudio Software (Illumina). For each individual subject, we calculated average β values for each of the 24 probes interrogated by the array that map onto the BMAL1 gene. Methods of DNA isolation, array processing and data analysis on the ROS/MAP cohort are described in detail elsewhere [14].
2.5-Fibroblast analysis
Primary human fibroblast cultures were established as reported [21] from dermal punch biopsies taken from healthy control individuals and patients diagnosed with probable AD participating in a longitudinal study at the UCSD ADRC (approved by UCSD IRB). Biopsies (4 mm punch) were taken from the back of the forearm. After collection, biopsy material was immediately processed by a technician to establish cell lines. Human fibroblasts were cultured in DMEM containing 15% fetal bovine serum (FBS), L-glutamine, NAA and penicillin/streptomycin. At the time of experiments, cell lines had been passaged 3–10 times. NIH3T3 cells stablyexpressing the Per2::luc reporter were developed in our laboratory as reported previously [22] using a construct reported in detail elsewhere [23]. NIH3T3 cells were cultured under similar conditions as the human cells, but with the addition of Hygromicyn B (200 µg/mL) as selection marker for the Per2::luc reporter. In order to make lumicycle experiments comparable to PCR time course assays, cells were synchronized in both experiments by changing the growth media to serum free DMEM, conditions that produce similar results to those employing serum shock or other stimuli [24].
2.6-Luminometry and circadian analysis
Case-control matching was maintained during cell culture procedures and experiments. Human fibroblast cells were split and grown in 35 mm plates to 70% confluence. Cells were then transduced with a Per2::luc lentiviral reporter construct (generously provided by Andrew Liu, University of Memphis). Viral particles were produced by the UCSD viral vector core facility. Additional details of the lentiviral construct have been published previously [24]. Infections were performed using 1×107 infectious units/plate and polybrene 0.1% for the 48 h prior to starting rhythm measurements. At the time of luminometery, cells were fully confluent (approximately 1.2 × 106 cells/plate). To synchronize rhythms, growth medium was replaced with HEPES-buffered, serum-free recording media containing 1 mM luciferin (Biosyth) immediately before luminometry [24]. Rhythm measurements were conducted in a luminometer (Actimetrics) as described [25]. Photoemissions from each cell line were measured in duplicate for 70 s, every 10 min over 5 days at 35°C. For drug experiments employing 5-Aza-2’-deoxycytidine (5-Aza-dC, Cayman Chemical;) or S-Adenosyl methionine (SAM from Cayman Chemical), drug or vehicle (PBS) was added to the recording media, and administered through a single media change at the onset of luminometer recording and remained present in the growth medium over the course of the experiment. None of the drug concentrations used significantly decreased viability. At the conclusion of the experiment, background subtracted luminometry data were fit to a damped sine wave by the least squares method using commercial software (Lumicycle Analysis). To reduce signal artifacts, the first 14 h (human cells) or 24 h (mouse cells) were excluded. Rhythm parameters (amplitude, phase, period) were then estimated for each trace and averaged across replicates.
2.7-Quantification of DNA methylation in fibroblasts
Global methylation levels were determined by ELISA (MethylFlash methylated DNA quantification kit, Epigentek) on 100 ng genomic DNA isolated from NIH3T3 fibroblasts treated with 50 µM 5-Aza-dC; 1 µM SAM or vehicle for 48 h, as per manufacturer’s instructions. Methylation at BMAL1 cg13286116 was determined on synchronized human fibroblast cultures from 2 control and 4 AD lines, sampled at the indicated times. Bisulfite-converted DNA was amplified using primers 5’-atggagaagtattatagaaagatttgg-3’ (sense) and 5’-acctatatccctaactcccaac-3’ (antisense). PCR products were subcloned into TOPO-TA vector (Life Technologies). Finally, 6 clones per cell line were sequenced. Analysis was performed using the Quantification Tool for Methylation Analysis (QUMA [26]) and expressed at each time point as the percentage of clones in each line that were fully methylated at cg13286116.
2.8-Statistic analysis
Statistical analysis of fibroblast-based assays was performed using one-way ANOVA adjusted for multiple observations and followed by Tukey’s post-test with a significance of p<0.05 or non-parametric Mann Whitney t test (unpaired; two-tailed) with a significance of p<0.05. Pearson’s correlation was used to analyze the association between DNA methylation levels and transcription at BMAL1. Fisher r-to-z transformation was used to investigate the significance of the difference between correlation coefficients across groups. Analyses were performed in Prism 6 (GraphPad, San Diego). Results are shown as average with S.E.M. as indicated. To investigate the oscillatory pattern of methylation, average β values were plotted as a function of time of death and non-linear fitting to sinusoidal curves with a fixed period of 24 h was performed as indicated.
We characterized 24-hour rhythms of DNA methylation separately in each of the diagnostic subgroups of the ROS/MAP cohort (Supp. Table 1) using cosine functions; described in detail in Supp. Methods.
3. RESULTS
3.1-Circadian cycle abnormalities in fibroblasts from individuals with AD correlate with aberrant BMAL1 expression
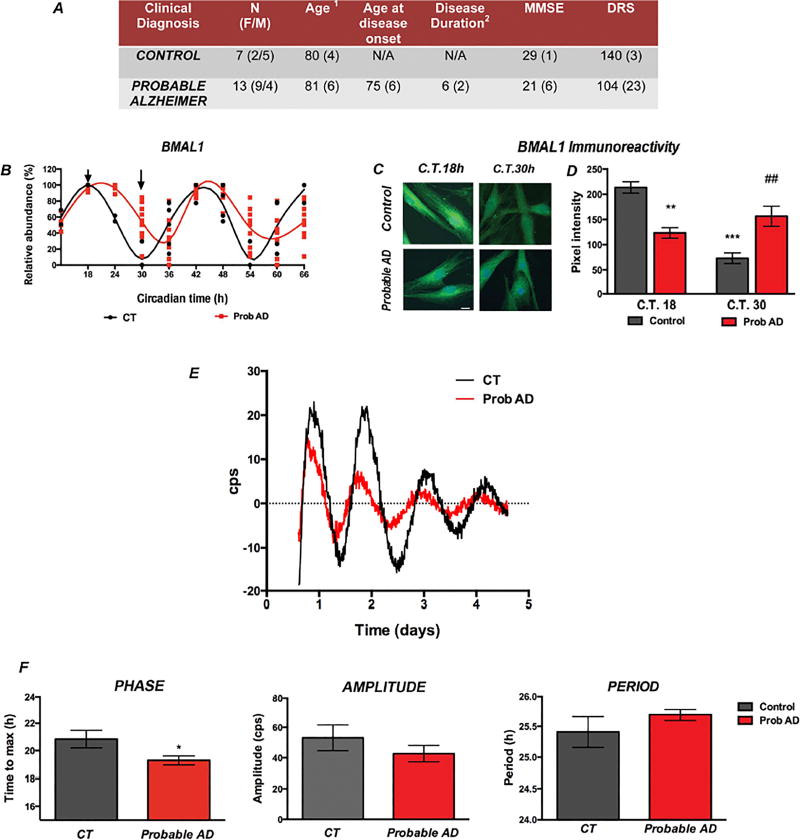
To understand the circadian alterations associated with AD pathology, we started by studying human fibroblasts cultures derived from 13 patients diagnosed with probable AD and 7 age-matched healthy controls (Fig. 1A). Time-course analysis in synchronized cultures showed a delayed expression peak (acrophase) of BMAL1 transcription in AD compared to control cells (Fig. 1B). Quantification of nuclear BMAL1 content at the times of transcript peak and nadir also showed significant differences between AD fibroblasts and control cells, even when the effect of transcriptional changes on protein abundance may appear later and may also be affected by protein stability and turnover (Fig. 1C–D).
Figure 1. Altered circadian rhythms in AD patients’ fibroblasts associate with aberrant BMAL1 transcription.
A. Clinical and demographic characterization of cell line donors. (1Age at skin biopsy. 2Calculated in years from clinical diagnosis to biopsy date). B. Lowess curves representing BMAL1 transcript abundance at different circadian times. Symbols represent duplicate determinations per cell line tested. C. Immunohistochemical detection of BMAL1 comparing protein abundance in control and AD fibroblasts at the times when BMAL1 transcripts reached peak and nadir values in control cells (indicated by arrows in Figure 1B). Imaged at 60 ×. Scale bar = 50 µm. D. Quantification of pixel intensity of BMAL1 immunoreactivity. ** p<0.01 and ***p<0.001 in comparison to control cells at peak time; or nadir time ## p<0.01 as per unpaired non-parametric Mann-Whitney t test. E–F. Rhythmic Per2::luc expression was measured from transduced control and AD-derived fibroblasts to determine circadian parameters. E. Representative data for Per2::luc rhythms obtained from a control subject and a patient with diagnosis of probable AD. F. Mean circadian rhythm parameters in AD case-control group analyses. * p<0.05 probable AD (n=13) vs. control fibroblasts (n=7) as determined by unpaired, non-parametric Mann-Whitney t test.
To evaluate whether these changes were associated with the core transcriptional clock, we used a bioluminescent reporter (Per2::Luc) to record circadian oscillations longitudinally in real time [24, 25]. We transduced cultured fibroblasts with the Per::Luc reporter construct and recorded emissions during five days in culture. Exogenous reporter constructs usually contain a small fragment of the promoter region and may not undergo epigenetic changes resulting from AD. Therefore, in order to study the effects of epigenetic modification of BMAL1, it is necessary to use a reporter of BMAL1 output, like Per2::Luc. Since these clock genes are tightly linked, epigenetic modifications of BMAL1 that impact transcription, should be reflected in Per2 expression. Notably, AD cells showed a significant phase advance and changes in cycle amplitude and period compared to controls (Fig. 1E–F), suggesting an association between molecular circadian alterations with AD. Period length, amplitude and phase were not significantly correlated with gender or age Supp. Fig. 1). However, we observed a significant correlation between circadian phase (Tmax) measured in fibroblasts and the morningness scores determined in a subset of participants who completed the Basic Language Morningness Scale (BALM) [27] questionnaire, suggesting that individual differences in donor chronotype may be reflected in cell culture (Supp. Fig. 2).
3.2-DNA methylation changes modulate circadian cycles in cultured fibroblasts
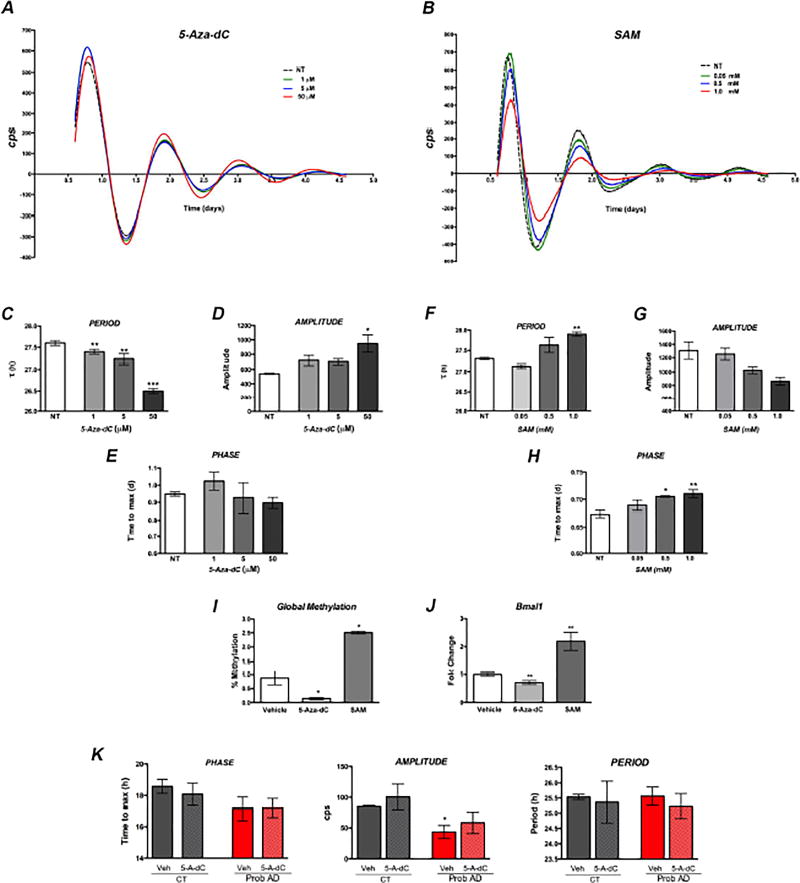
Recently, 24 h methylation rhythms were described in the human frontal cortex, cycles that appear to be correlated with age and dementia [14]. Moreover, changes in DNA methylation were also reported to mediate the effects of day length on clock plasticity [13]. To directly test whether DNA methylation modulates circadian rhythms in cells, we treated NIH3T3 fibroblasts with 5-Aza-2’ deoxycytidine (5-AZA-dC), a cytidine analog that inhibits DNA methyl-transferases, to decrease global DNA methylation; or exposed fibroblasts to the methyl donor S-adenosyl-methionine (SAM), to increase methylation (Fig. 2I). We observed concentration-dependent alterations in circadian rhythms in response to both treatments (Fig. 2A–B). Reduced methylation resulted in shorter period length, advanced phase and increased amplitude (Fig. 2C–E). Higher methylation elicited the opposite effects, with increased period length, delayed phase and decreased rhythm amplitude (Fig. 2F–H). Importantly, these drug-induced changes in methylation had specific effects on Bmal1 transcription (Fig. 2J). To determine if methylation changes may also be involved in the alteration of circadian cycles in AD fibroblasts, we performed preliminary studies on a subset of the human fibroblasts lines (n=3 control and 3 Prob. AD). Treatment with 50 µM 5-Aza-dC induced similar changes in phase, amplitude and period as those observed for NIH3T3 cells, although the magnitude of the changes was lower (Fig. 2K). These results support a role for DNA methylation in modulating the circadian clock, likely involving transcriptional changes in Bmal1, and provide a link between epigenetic disruption and circadian alterations in AD.
Figure 2. Pharmacological modulation of DNA methylation induces changes in circadian rhythms in cultured fibroblasts.
Rhythmic Per2::luc expression in NIH 3T3 mouse fibroblasts treated with 5-Aza-dC; SAM or vehicle (NT). A–B. Concentration-dependent changes in circadian rhythms were observed in response to both compounds. C–E. Reduced global methylation shortens period; increases amplitude and advances phase. F–H. Increased methylation levels lengthen period; decreases amplitude and delays phase. I–J. Both treatments significantly changed global DNA methylation (I) and expression level of Bmal1 transcripts (J). K. Circadian rhythms are modulated by changes in methylation in human fibroblasts treated with 50 M 5-Aza-dC. Error bars represent S.E.M. * p<0.05; ** p<0.01 and *** p<0.001 in comparison to vehicle treated cells as per One Way ANOVA (C–I) or paired parametric t test (J–K).
3.3-Rhythmic DNA methylation patterns correlate with transcription of BMAL1 and are significantly dysregulated in the brains from patients with early AD
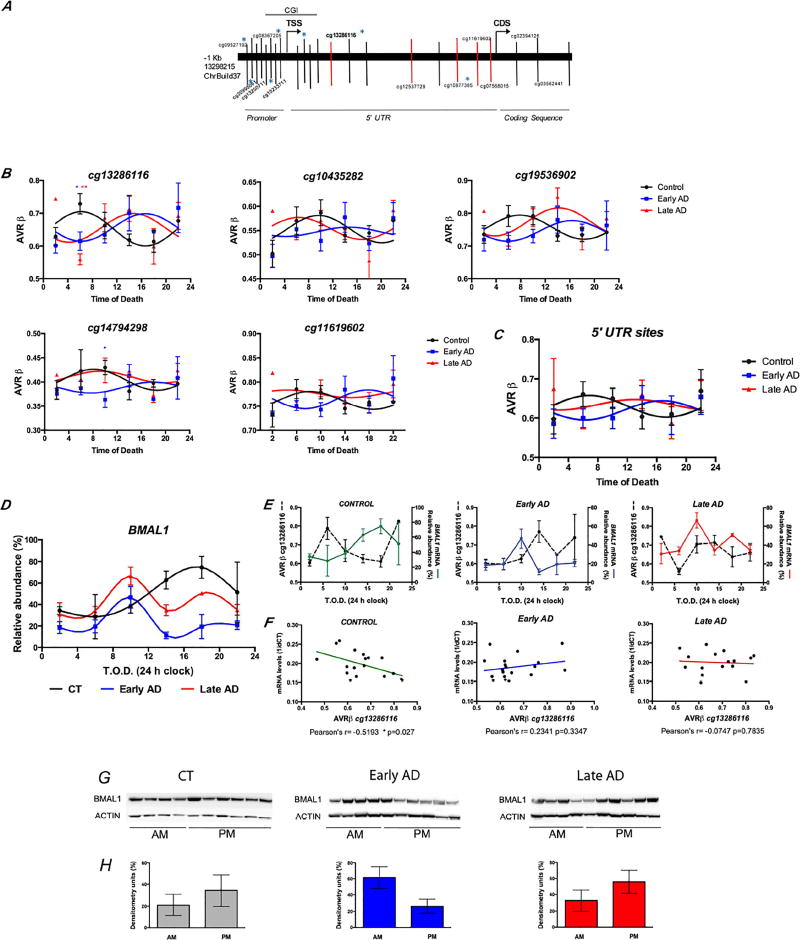
Based on these in vitro results, we performed a cross-sectional analysis in midfrontal cortex human postmortem samples. We included 66 samples from the Shiley-Marcos Alzheimer’s Disease Research Center (ADRC) that were stratified according to ante-mortem cognitive evaluation and postmortem neuropathological burden (Supp. Table 1). To investigate the role of DNA methylation on BMAL1 expression we interrogated a genomic region of >90 Kb covered by the Illumina 450k Methylation array, that corresponded to the promoter; 5’-untranslated region (5’UTR), and a portion of the coding sequence (CDS) of BMAL1 (Fig. 3A). Overall, CpG sites at the promoter, including those at the CG island, appear unmethylated, while CpGs located at the 5’-UTR and CDS show intermediate and high methylation levels respectively. This pattern is preserved in control and AD brains (not shown). To evaluate rhythmic patterns of variation at specific sites, methylation levels from each subject were examined as a function of time of death (T.O.D.) and fit to cosinor models as previously reported [14]. We centered the analysis on the 5’UTR sequences of BMAL1, which we hypothesized were most likely to be involved in the circadian modulation of transcription given their intermediate levels of methylation. We observed oscillatory patterns with significant differences in methylation at peak times between AD cases (early and late) compared to controls. These alterations may reflect underlying changes in phase and amplitude (Fig. 3B). Notably, the five CGs showing the highest methylation amplitude reached peak values at similar times, suggesting coordinated epigenetic activity in this region. Indeed, combined analysis of cg13286116; cg10435282; cg19536902; cg14794298 and cg11619602 replicated their individual patterns of methylation, with rhythms from early AD patients being nearly completely antiphase to brains from controls, whereas brains from late AD cases showed lower amplitude and dampened rhythms (Fig. 3C).
Figure 3. Rhythmic oscillation of DNA methylation correlate with BMAL1 transcription in AD brains.
A. Schematic representation of BMAL1 genomic region screened. Vertical bars indicate the location of probes in the Illumina 450k array. Blue * denotes probes showing significant rhythms with 24 h period. Red bars depict 5’ UTR probes that showed altered methylation rhythms in AD brains. Gray bar indicates the position of CG island Chr11:13298796–13300735 in ChrBuild37. B. Comparison of methylation rhythms at 5’UTR CpGs between control, early and late AD cases. Panels show non-linear fit of methylation values to sinusoidal curves. Blue * p<0.05; red ** p<0.01 in comparison to controls at the same time (one way ANOVA, with Tukey’s posthoc test). C. Representation of fit-sinusoidal curves for the combined 5’UTR sites from (B). D. Locally weighted scatterplot smoothing (Lowess) curves representing BMAL1 transcript abundance as a function of time of death as 4 h bins (T.O.D.). Symbols represent mean values ± S.E.M. E–F. Analysis of cg13286116 methylation and BMAL1 transcript abundance. Lower panel shows significant correlation (Pearson’s R) for control cases, which is lost in AD brains. G. Immunohistochemical detection of BMAL1 protein in frontal cortex samples at different T.O.D. ACTIN immunoreactivity is showed as loading control. H. Densitometric analysis of BMAL1 protein comparing abundance during AM vs. PM hours (as indicated in G). Data represents normalized values of BMAL1/ACTIN ratios.
We next profiled the cycles of BMAL1 transcription in the brain by qPCR (Fig. 3D). Circadian oscillation of BMAL1 was evident in control subjects, with peak expression in the early evening (16:00–20:00), in agreement with previous reports [11]. By contrast, BMAL1 transcription was significantly altered in both early and late AD brains, with lower amplitude and advanced phase. BMAL1 transcript abundance significantly correlated with cg13286116 methylation in the control group, but this association was not found in AD brains (Fig. 3E–F). Pearson’s correlation coefficients were significantly weaker in the AD samples compared to controls (p=0.028). Finally, to evaluate the impact of epigenetic and transcriptional alterations on the circadian clock machinery, we assessed the oscillation of nuclear BMAL1 protein levels in cortical tissue. Although individual variability was high, BMAL1 protein was elevated during PM hours in control cases (Fig. 3G–H). In contrast, early AD cases showed the highest BMAL1 levels in the morning, while BMAL1 accumulation shifted towards night hours in late AD cases (Fig. 3G–H). Taken together, these results show that aberrant methylation rhythms correlate with abnormal patterns of BMAL1 transcription and protein expression in AD. These alterations differ across disease stages, and may represent a molecular marker of progression.
Next, we validated these results on an independent cohort. We performed secondary analysis using the comprehensive brain epigenomic and transcriptomic data from two cohort studies of aging and AD, the Religious Order Study and the Rush Memory and Aging Project (ROS/MAP) [14], including 396 cases from the methylome analysis and 171 from the RNAseq that matched our criteria (Supp. Table 1).
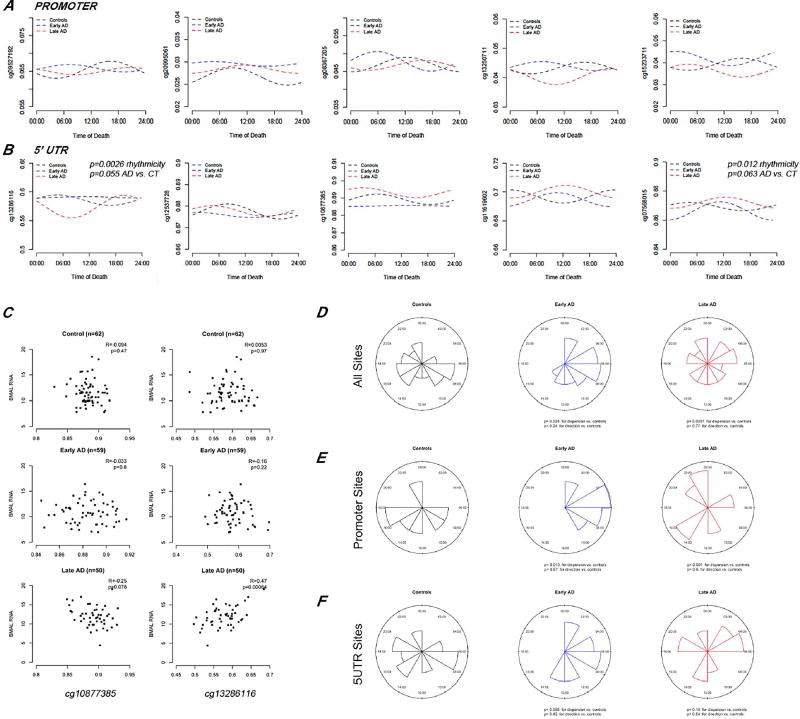
Visual inspection of the best fit cosine curves showed quantitative differences in methylation at individual sites within the BMAL1 promoter and the 5’ UTR in early and late AD cases, similar to what we observed in the initial study (Fig. 4A–B). Cosinor analyses determined that 24 h methylation rhythms were present in the 5’UTRs of control subjects at cg22322535 (p=0.042); in early AD at cg07568015 (p=0.012) and in late AD at cg13286116 (p=0.0026). Moreover, there was a trend toward different methylation rhythms between early AD and controls for cg07568015 (p=0.063) and between late AD and controls for cg13286616 (p=0.055). BMAL1 transcription correlated with methylation of 5’UTR CGs, including cg13286116, a site that showed significant alterations in AD, validating the results from the discovery cohort (Fig. 4C). Evaluation of the distribution of acrophase methylation timing across all CGs interrogated for BMAL1, showed significant changes in dispersion in early and late AD cases in comparison to controls, also illustrating changes in the acrophase timing across groups (Fig. 4D–F). We therefore hypothesize that dynamic methylation cycles at the 5’UTR have a role in the regulation of rhythmic transcription of BMAL1.
Figure 4. Validation of rhythmic DNA methylation, BMAL1 transcription correlations and acrophase alterations in the ROS/MAP cohorts.
A–B. Individual CpG site analysis on 396 ROS/MAP participants adjusting for age at death and sex. Lines represents best-fit cosine curve. Selected panels showing comparison of fitted cosinor curves per group for probes located at BMAL1 promoter (A) and 5’ UTR (B) regions. Top p value refers to rhythmicity in the group. Bottom p value refers to rhythmicity differences in comparison to the control group. C. Selected scatterplots showing association of the combined signal of all BMAL1 isoforms detected by RNAseq with methylation levels at individual CpGs. Pearson’s R coefficients and p values for the correlation are indicated. D–F. Distribution of methylation acrophase times for all 22 BMAL1 probes detected (D); or for probes located at BMAL1 promoter (E) or 5’UTR regions (F) in controls, early and late AD brains.
3.4-Similar methylation abnormalities at BMAL1 are observed in AD cells and brains
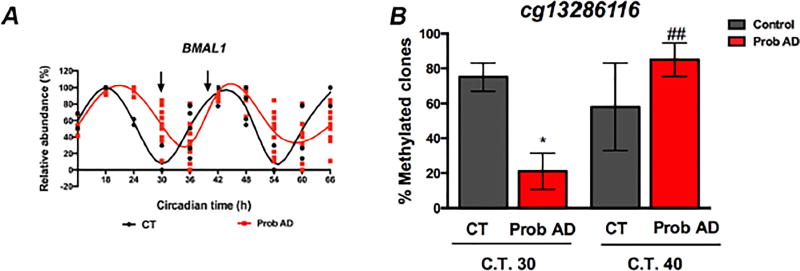
Finally, we examined whether AD patient fibroblasts and brain samples presented similar alterations in BMAL1 methylation. We quantified the methylation levels the site that showed most significant changes in AD brains, cg13286116, using bisulfite sequencing. DNA was obtained from control and AD fibroblasts at the circadian times when BMAL1 mRNA reached its lowest abundance in control cells (C.T.30) and again at a later time when BMAL1 transcripts peak (C.T.40; Fig. 5A). About 80% of the control clones were fully methylated at C.T.30, compared to 20 % of AD clones at the same time point. Ten hours later, 50% of control clones were fully methylated, while 80% of AD clones were methylated (Fig. 5B). These results, although limited by the small sample size, suggest that the differences observed in BMAL1 methylation patterns in the brains of AD patients are indeed reflected in fibroblast cell lines, thereby supporting the validity of our cell-based functional studies.
Figure 5. Differences in BMAL1 methylation between AD patients and controls are also evident in fibroblast cells.
Bisulfite sequencing was used to compare the levels of methylation at site cg13286116 between control subjects and AD patients’ cells at the times when BMAL1 transcription is close to nadir (C.T.30) and peak (C.T.40) in control fibroblasts. A. Panel B from Fig. 1 is provided as a reference of BMAL1 transcript levels at the time points tested. B. The graph shows the percentage of sequenced clones that showed fully methylated cg13286116 at each time point. * p<0.05 in comparison to control cells, ## p<0.01 in comparison to Probable AD cells at C.T.30 as per unpaired Mann-Whitney t test.
4. DISCUSSION
In the present study we provide evidence that DNA methylation regulates BMAL1 expression rhythms in human brain samples and fibroblasts from AD patients and controls. To the best of our knowledge, this is the first report describing the association between rhythmic methylation and cyclic clock gene expression, and its potential role in circadian alterations in the AD brain. Characterization of circadian rhythms in real-time using AD patient fibroblasts enabled us to directly compare BMAL1 methylation changes in AD brains with functional differences in cultured cell lines. Moreover, by showing that pharmacological manipulation of DNA methylation leads to changes in rhythms and BMAL1 expression, our data support a causal role for methylation as a modulator of circadian cycles. This suggests that the aberrant brain methylation patterns reported in AD could alter clock gene expression and neuronal circadian rhythms, thereby contributing in part to the sleep and behavior alterations associated with pathology.
Circadian disruption is recognized as an important clinical feature in the course of AD [28–30]. However, the underlying molecular alterations linked to such deregulation are yet to be discovered. Epigenetic mechanisms are intimately linked with the regulation of circadian clocks. Chromatin modifiers SIRT1 and HDAC3 are recruited to clock-regulated genes in a circadian pattern [31, 32]. CLOCK-BMAL1 dimers induce histone modifications at the promoters of clock-controlled genes to activate transcription [33] [34]. Still, the potential role of DNA methylation on this system in underexplored [35–37]. In model organisms, like Neurospora, methylation at the frq locus is associated with modulation of circadian period and phase [38, 39]. A recent study reported that the effects of day length on clock plasticity are likely modulated by DNA methylation changes in the mouse SCN [13]. Altered circadian light cycles affected behavior, global transcription and DNA methylation, and older mice failed to entrain to light changes [13]. Our results support the hypothesis that similar epigenetic mechanisms may regulate circadian clocks in the human brain, and may be affected by AD pathology.
AD postmortem brains show an acceleration of age-related changes in methylation [40] and genes associated with increased disease susceptibility show abnormal methylation and transcription levels, even early in AD [15]. These observations, in addition to the results reported here suggest that DNA methylation may represent an accessory feedback loop that helps synchronize the expression of clock genes and maintain rhythmicity. Therefore, aberrant methylation may weaken rhythms and impact downstream rhythmic processes. At the molecular level, circadian clock function is based on interlocking transcriptional/translational mechanisms where positive regulators modulate the expression of their own repressors [7]. Therefore, while we provided evidence that DNA methylation modulates BMAL1 transcription, BMAL1 in turn may directly regulate the enzymes that direct methylation, the DNA methyl transferases (DNMTs) and demethylases. Changes in Dnmt expression were observed in response to altered day length in mouse SCN [13], hence neuronal degeneration in the SCN reported in AD [29] may also initiate epigenetic alterations that will further impact circadian regulation in peripheral clocks. Additional mechanistic studies are needed to completely understand the potential complexity of this mechanism. With respect to other disease specific changes in AD, BMAL1 protein levels may be directly affected by accumulation of amyloid-β (Aβ) during the course of AD pathology. A recent study reported that Aβ enhances the degradation of Bmal1 and Cbp, another circadian clock regulator, reducing binding of transcription factors to the Per2 promoter, and altering the oscillation of Per2 mRNA and protein expression in cell culture models and in the SCN of the 5×FAD transgenic mice model of AD [41].
In humans, the phase of circadian behavior presents high variability among different individuals as a result of the interaction of different chronotypes, daily life schedules and metabolism that impact the clock function at a cellular level [42]. As our sample size increased in the validation cohort, we observed a higher dispersion in the methylation rhythms, evidenced by lower amplitudes and higher p-values for rhythmicity in controls than AD cases, an effect that may be associated to individual life-styles. In “late AD” cases with severe dementia, exposure to tight schedules of care may contribute to entrain the clock to external clues, reducing the inter-individual variability. These confounding external factors limit the present study.
Functional longitudinal studies are not feasible in the brain of living human subjects, but circadian rhythms can be reliably measured from skin fibroblasts, and likely reflect the activity of non-SCN brain oscillators [42], since period length (of about 24 h) is genetically determined. Although we observed alterations in BMAL1 transcription and protein levels in both, brain samples and fibroblast cultures, the changes are different. The post-mortem brain studies were conducted in subjects living in regular light-dark cycles that impose limits in the BMAL1 rhythm and affect the expression profile. Cell culture studies, on the other hand, were conducted under constant conditions and reflect the free running rhythm. Therefore, the two data sets are not directly comparable. Noteworthy, the ability of detecting alterations in fibroblasts that may resemble alterations in the human brain, beyond their expected differences in epigenotype, exemplifies the power of this cellular system for monitoring circadian rhythmicity in vivo.
The clinical continuum of AD includes three distinctive disease stages: preclinical; mild cognitive impairment (MCI); and dementia [43]. Circadian and sleep-wake dysfunction are well described in advanced AD, but less studied in individuals with MCI, although 60% of them report a prevalence of sleep disturbance [44] and are at higher risk to develop dementia [44, 45]. Circadian abnormalities have been associated with degeneration of SCN neurons [46] and decreased melatonin levels in the cerebrospinal fluid (CSF) of AD patients [47]. In our study, we detected the most striking alterations in the frontal cortex from “early AD” patients (i.e. a group intermediate between MCI and late AD), supporting the idea that epigenetic and circadian alterations may not only be a consequence of SCN cell loss, but deregulation of peripheral clocks in other brain regions at initial disease stages may also contribute to circadian impairment.
Sleep quality plays an important role in preserving memory and cognition, and reducing the risk of converting to dementia in susceptible individuals [48]. Here we show significant deregulation of BMAL1 in association with early AD. BMAL1 not only controls the transcription of core clock genes, but also regulates cerebral redox homeostasis. Targeted deletion of Bmal1 in mouse neurons and glia triggers degeneration of synaptic terminals and neuronal oxidative damage and impairs cortical functional connectivity [49]. Our findings suggest that epigenetic alterations early in AD could contribute to cognitive impairment and neurodegeneration via deregulation of circadian rhythms. If so, then early interventions, including epigenetic and chronotherapies, may prove effective in delaying disease progression.
Supplementary Material
HIGHLIGHTS.
DNA methylation cycles associate with rhythmic BMAL1 transcription in the brain.
This epigenetic mechanism is altered early in Alzheimer’s disease progression.
Aberrant BMAL1 methylation and transcription may contribute to circadian impairment observed in AD.
RESEARCH IN CONTEXT.
Systematic review: No previous study has investigated the role of dynamic cycles of DNA methylation on cellular circadian rhythms in AD.
Interpretation: Our findings show that cycles of DNA methylation may contribute to the regulation of circadian rhythms in the brain via modulation of BMAL1 transcription. This epigenetic mechanism is altered early in AD pathology, likely resulting in aberrant BMAL1 expression and possibly circadian impairment.
Future directions: Future studies will focus on elucidating the mechanisms of cyclic DNA methylation and identifying how rhythms interact with specific pathological mechanisms in AD.
Acknowledgments
We thank Anthony Adame and Taeyeon Kim for immunochemistry analysis. This work was support by the University of California Academic Senate grant RN127H; NIA P50 AG05131; a pilot grant from the UCSD Shiley-Marcos ADRC; New Investigator Research Grant to promote Diverstiy (NIRGD-13-284795) from the Alzheimer’s Association to P.D.; VA Career development award 1IK2BX001275 to M.J. M. National Institutes of Health grants P30AG10161 (DAB) RF1AG15819 (DAB) R01AG17917 (DAB) R01AG36042 (DAB) R01AG36836 (PLD). Canadian Institutes of Health Research grant MSH136642 (ASL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
P.C. performed brain and fibroblast methylation, gene transcription and protein analysis. M.M. carried out circadian analysis on fibroblasts and contributed with data analysis and interpretation. A.L. analyzed transcription and methylation data from ROS/MAP cohorts. D.S. contributed with clinical analysis and case selection criteria. E.M. performed neuropathalogical analysis and adviced on case selection. D.G. performed clinical studies. P.C., M.M, A.L., E.M., P.D.J., D.B. and P.D. contributed to manuscript writing. P.D. designed and supervised the study and performed data analysis and interpretation.
References
- 1.Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362:329–44. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 2.Van Someren EJ. Circadian and sleep disturbances in the elderly. Exp Gerontol. 2000;35:1229–37. doi: 10.1016/s0531-5565(00)00191-1. [DOI] [PubMed] [Google Scholar]
- 3.Gehrman P, Marler M, Martin JL, Shochat T, Corey-Bloom J, Ancoli-Israel S. The timing of activity rhythms in patients with dementia is related to survival. J Gerontol A Biol Sci Med Sci. 2004;59:1050–5. doi: 10.1093/gerona/59.10.m1050. [DOI] [PubMed] [Google Scholar]
- 4.Slats D, Claassen JA, Verbeek MM, Overeem S. Reciprocal interactions between sleep, circadian rhythms and Alzheimer's disease: focus on the role of hypocretin and melatonin. Ageing Res Rev. 2013;12:188–200. doi: 10.1016/j.arr.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Pollak CP, Perlick D. Sleep problems and institutionalization of the elderly. J Geriatr Psychiatry Neurol. 1991;4:204–10. doi: 10.1177/089198879100400405. [DOI] [PubMed] [Google Scholar]
- 6.Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, et al. Sleep drives metabolite clearance from the adult brain. Science. 2013;342:373–7. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhr ED, Takahashi JS. Molecular components of the Mammalian circadian clock. Handb Exp Pharmacol. 2013:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–7. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 9.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 10.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950–5. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim AS, Myers AJ, Yu L, Buchman AS, Duffy JF, De Jager PL, et al. Sex difference in daily rhythms of clock gene expression in the aged human cerebral cortex. J Biol Rhythms. 2013;28:117–29. doi: 10.1177/0748730413478552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahar S, Sassone-Corsi P. The epigenetic language of circadian clocks. Handb Exp Pharmacol. 2013:29–44. doi: 10.1007/978-3-642-25950-0_2. [DOI] [PubMed] [Google Scholar]
- 13.Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, et al. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17:377–82. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- 14.Lim AS, Srivastava GP, Yu L, Chibnik LB, Xu J, Buchman AS, et al. 24-hour rhythms of DNA methylation and their relation with rhythms of RNA expression in the human dorsolateral prefrontal cortex. PLoS genetics. 2014;10:e1004792. doi: 10.1371/journal.pgen.1004792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–63. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr Alzheimer Res. 2012;9:628–45. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res. 2012;9:646–63. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosir R, Acimovic J, Golicnik M, Perse M, Majdic G, Fink M, et al. Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol Biol. 2010;11:60. doi: 10.1186/1471-2199-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, et al. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286:9031–7. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8:1030–8. doi: 10.4161/epi.25865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy MJ, Le Roux MJ, Wei H, Beesley S, Kelsoe JR, Welsh DK. Calcium channel genes associated with bipolar disorder modulate lithium's amplification of circadian rhythms. Neuropharmacology. 2016;101:439–48. doi: 10.1016/j.neuropharm.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, Parks D, et al. Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci. 2008;121:3629–35. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–16. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy MJ, Wei H, Marnoy Z, Darvish RM, McPhie DL, Cohen BM, et al. Genetic and clinical factors predict lithium's effects on PER2 gene expression rhythms in cells from bipolar disorder patients. Transl Psychiatry. 2013;3:e318. doi: 10.1038/tp.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–5. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee MK, Lee HJ, Rex KM, Kripke DF. Evaluation of two circadian rhythm questionnaires for screening for the delayed sleep phase disorder. Psychiatry Investig. 2012;9:236–44. doi: 10.4306/pi.2012.9.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu RY, Zhou JN, Hoogendijk WJ, van Heerikhuize J, Kamphorst W, Unmehopa UA, et al. Decreased vasopressin gene expression in the biological clock of Alzheimer disease patients with and without depression. J Neuropathol Exp Neurol. 2000;59:314–22. doi: 10.1093/jnen/59.4.314. [DOI] [PubMed] [Google Scholar]
- 29.Stopa EG, Volicer L, Kuo-Leblanc V, Harper D, Lathi D, Tate B, et al. Pathologic evaluation of the human suprachiasmatic nucleus in severe dementia. J Neuropathol Exp Neurol. 1999;58:29–39. doi: 10.1097/00005072-199901000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Wang JL, Lim AS, Chiang WY, Hsieh WH, Lo MT, Schneider JA, et al. Suprachiasmatic neuron numbers and rest-activity circadian rhythms in older humans. Ann Neurol. 2015;78:317–22. doi: 10.1002/ana.24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 32.Sun Z, Feng D, Everett LJ, Bugge A, Lazar MA. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harbor symposia on quantitative biology. 2011;76:49–55. doi: 10.1101/sqb.2011.76.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature structural & molecular biology. 2010;17:1414–21. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 35.Liu HC, Hu CJ, Tang YC, Chang JG. A pilot study for circadian gene disturbance in dementia patients. Neurosci Lett. 2008;435:229–33. doi: 10.1016/j.neulet.2008.02.041. [DOI] [PubMed] [Google Scholar]
- 36.Nakatome M, Orii M, Hamajima M, Hirata Y, Uemura M, Hirayama S, et al. Methylation analysis of circadian clock gene promoters in forensic autopsy specimens. Leg Med (Tokyo) 2011;13:205–9. doi: 10.1016/j.legalmed.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Satou R, Sugihara N, Ishizuka Y, Matsukubo T, Onishi Y. DNA methylation of the BMAL1 promoter. Biochem Biophys Res Commun. 2013;440:449–53. doi: 10.1016/j.bbrc.2013.09.124. [DOI] [PubMed] [Google Scholar]
- 38.Belden WJ, Lewis ZA, Selker EU, Loros JJ, Dunlap JC. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet. 2011;7:e1002166. doi: 10.1371/journal.pgen.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciarleglio CM, Axley JC, Strauss BR, Gamble KL, McMahon DG. Perinatal photoperiod imprints the circadian clock. Nature neuroscience. 2011;14:25–7. doi: 10.1038/nn.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song H, Moon M, Choe HK, Han DH, Jang C, Kim A, et al. Abeta-induced degradation of BMAL1 and CBP leads to circadian rhythm disruption in Alzheimer's disease. Mol Neurodegener. 2015;10:13. doi: 10.1186/s13024-015-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown SA, Kunz D, Dumas A, Westermark PO, Vanselow K, Tilmann-Wahnschaffe A, et al. Molecular insights into human daily behavior. Proc Natl Acad Sci U S A. 2008;105:1602–7. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naismith SL, Hickie IB, Terpening Z, Rajaratnam SM, Hodges JR, Bolitho S, et al. Circadian misalignment and sleep disruption in mild cognitive impairment. J Alzheimers Dis. 2014;38:857–66. doi: 10.3233/JAD-131217. [DOI] [PubMed] [Google Scholar]
- 45.Schlosser Covell GE, Dhawan PS, Lee Iannotti JK, Hoffman-Snyder CR, Wellik KE, Caselli RJ, et al. Disrupted daytime activity and altered sleep-wake patterns may predict transition to mild cognitive impairment or dementia: a critically appraised topic. Neurologist. 2012;18:426–9. doi: 10.1097/NRL.0b013e318272f7ef. [DOI] [PubMed] [Google Scholar]
- 46.Stopa EG, Volicer L, Kuo-Leblanc V, Harper D, Lathi D, Tate B, et al. Pathologic evaluation of the human suprachiasmatic nucleus in severe dementia. J Neuropathol Exp Neurol. 1999;58:29–39. doi: 10.1097/00005072-199901000-00004. [DOI] [PubMed] [Google Scholar]
- 47.Zhou JN, Liu RY, Kamphorst W, Hofman MA, Swaab DF. Early neuropathological Alzheimer's changes in aged individuals are accompanied by decreased cerebrospinal fluid melatonin levels. J Pineal Res. 2003;35:125–30. doi: 10.1034/j.1600-079x.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 48.Lim AS, Yu L, Kowgier M, Schneider JA, Buchman AS, Bennett DA. Modification of the relationship of the apolipoprotein E epsilon4 allele to the risk of Alzheimer disease and neurofibrillary tangle density by sleep. JAMA Neurol. 2013;70:1544–51. doi: 10.1001/jamaneurol.2013.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. The Journal of clinical investigation. 2013;123:5389–400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.