Abstract
DNA sliding clamps are rings that tether certain enzymes to DNA. How clamp proteins slide on DNA has remained a mystery. A new crystal structure, together with molecular dynamics and NMR studies, has revealed how the human PCNA clamp slides on DNA.
Most of the central enzymes needed to duplicate DNA were found within a decade or two after the discovery of the double helix [1]. These were: the helicase to unwind duplex DNA strands; DNA polymerases that copy the separated strands into new duplexes; and RNA primase that makes RNA primers that give a start site for the DNA polymerases [2]. But it took a further 20 years before the functions of two other core proteins essential for replication in all cells were elucidated. One of these core protein is the Escherichia coli β clamp, the first protein known to encircle DNA [3,4]: its function is to bind DNA polymerase III, the E. coli replicase, and slide behind the polymerase, holding it to DNA for processive incorporation of thousands of nucleotides in one binding event. The human equivalent is the PCNA ring that is structurally superimposable with the E. coli β clamp [5]. The other core component is the clamp loader, a pentameric complex that uses ATP to open and close the clamp around DNA, and is ubiquitous in all cells [3,6]. Sliding clamps are now known to be used in many processes beyond replication, including repair, cell cycle control, and nucleosome assembly [7]. Because of the unusual nature of the clamp interaction with DNA, the actual mechanism by which these clamps slide along DNA has remained largely unknown.
A recent study [8] has illuminated the clamp sliding process along DNA and the findings are fascinating. A crystal structure of human PCNA encircling a 10 base pair duplex DNA has been solved to high resolution, and the architecture of the clamp–DNA interface is now clear (Figure 1). The inner diameter of the clamp is about 30 Angstroms in diameter, compared to 20 Angstroms for duplex DNA, and the DNA is tilted about 30° relative to the inner channel. This is similar to the tilt observed in the crystal structure of the E. coli β clamp bound to primed DNA, except the β clamp was not caught in a sliding mode because the primed site was held in the protein binding pocket which acts as a placeholder to keep the clamp at the 3′ terminus when the polymerase is not present [9]. In contrast, the PCNA–DNA complex shows a remarkable alignment of basic residues that line the central channel and they track along just one strand of the duplex. The PCNA trimer, like the β dimer, contains 12 α helices that line the central channel, four per subunit. The DNA tracking residues are lysines that lie on the surface of the α-helices and spatially define a pitch similar to B-form DNA (Figure 1). In β and PCNA clamps, the α helices are arranged perpendicular to the DNA backbone, and thus to track a DNA helix the six tracking lysines are strategically spread out over 5 different α helices.
Figure 1. Human PCNA–DNA crystal structure.
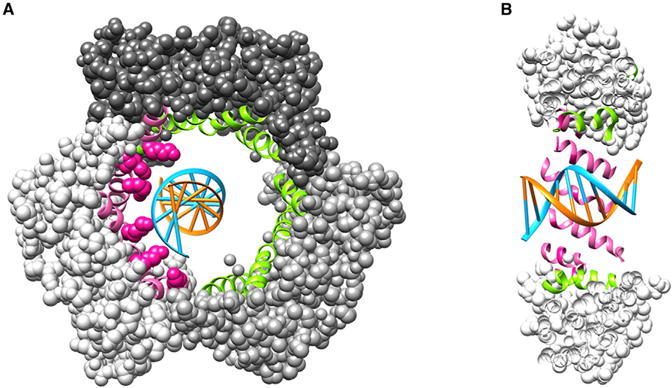
(A) Front view of the PCNA trimer–DNA (PDB ID 5L7C) in space-filling representation; each subunit is a different shade of grey. The α helices lining the central chamber are in ribbon representation. In pink are the helices that interact with DNA (the blue strand), and in green are the remaining α helices (the six lysine side chains that interact with DNA are in pink space-filling representation). DNA is in blue and orange. (B) Cut away side-view of PCNA–DNA. The representation and colors of the protein and DNA are the same as in (A).
NMR studies and molecular dynamics simulations revealed that the lysines walk along successive backbone phosphates of one DNA strand during sliding [8], and predict a sliding rate far faster than the DNA polymerase, consistent with single-molecule measurements of rapid clamp diffusion on DNA [10]. The fast rate of clamp diffusion on DNA ensures that it will not hinder the DNA polymerase rate. The molecular dynamics simulations also concluded that clamps mainly spiral when they diffuse along DNA [8,11]. The simulations show that the 30° tilt of DNA to the central axis of the clamp is maintained during sliding, such that when viewed from the side, a tilted clamp that runs along one DNA ‘rail’ propagates the tilt in a helical fashion and gives the appearance of alternating between forward and backward tilts every one-half turn of DNA (Figure 2A). This is the natural consequence of sliding on a helical substrate, and De March et al. [8] refer to this sliding process as ‘cogwheeling’ along the DNA. They point out that certain biochemical studies of mutant proteins suggest that polymerases and clamp loaders are specific for the particular tilt that PCNA has adopted at the point that it is positioned at a primer terminus [12,13].
Figure 2. The PCNA tilt on DNA may function in different ways.
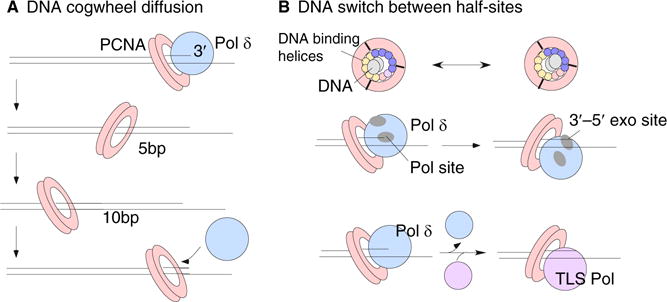
(A) The PCNA tilt is maintained during diffusion on DNA, causing it to spin during diffusion along DNA. As it spins, the tilt will alternate between a forward and then backward tilt every five base pairs. If polymerase δ requires a specific tilt of PCNA on DNA for active synthesis, this type of diffusion will enable PCNA to maintain the specific tilt during diffusion, such that when PCNA samples the 3′ terminus it will be in the correct tilt for activity with polymerase δ. (B) Top: the PCNA inner chamber is lined by 12 α helices; five α helices carry the charged residues that track one strand of DNA. Thus there are two sites in one circular clamp and this could enable a ‘tilt switch’ in one step without diffusion. The middle diagram illustrates the use of this type of switch for bringing the exonuclease site into the correct orientation for removal of a mismatched nucleotide at the primer 3′ terminus. The bottom diagram illustrates a speculative case in which a translesion DNA polymerase may require a different PCNA tilt for activity relative to polymerase δ. In this case, a direct switch in tilt could alter specificity of PCNA for a replicative polymerase δ or a translesion DNA polymerase.
The tilt of PCNA at the 3′ terminus may therefore define protein binding specificity of the clamp for different factors that interact with the clamp. Presumably the clamp loader, replication factor C (RFC), places the clamp on the DNA at the correct tilt for interacting with DNA polymerase δ, a polymerase that functions at a eukaryotic replication fork with PCNA. Interestingly, human DNA polymerase δ often dissociates from PCNA during synthesis and must reassociate to finish replicating the DNA, a finding that would seem to contradict the usual processive actions endowed on polymerases by a sliding clamp tether [14]. Polymerase δ requires PCNA for activity, and the way polymerase δ rapidly finds its way back to active status is explained by the cogwheel diffusion process. While the PCNA clamp is expected to diffuse away from the primer terminus whenever the polymerase dissociates from the clamp, the rapid cogwheel diffusion process enables the ‘active’ tilt to be maintained during clamp diffusion on DNA. During rapid clamp diffusion the clamp will frequently resample the primer terminus, and upon its arrival it will be in the proper tilt specified for activity with the DNA polymerase (Figure 2A). In other words, the cogwheeling of PCNA during sliding will maintain the correct tilt when it samples the 3′ terminus, and thereby facilitate repairing of a productive clamp–polymerase interaction for continued synthesis.
There are times when different proteins must act on the same clamp, either in succession or while bound to the clamp at the same time [15]. The homotrimeric structure of PCNA provides it with multiple DNA tracking sites per PCNA trimer. Furthermore, because the clamp tracks using only five of its 12 α helices, the clamp can conceivably ‘switch’ from one tracking site to another and thereby change the tilt relative to a 3′ terminus (Figure 2B). This tilt switching process could alter the specificity of PCNA for an interacting partner. Studies of PCNA diffusion show that this type of switching does occur, but not as frequently as cogwheel diffusion [11]. It is speculated that this switch may be useful for proofreading by DNA polymerase δ, which has a 3′–5′ exonuclease site in a separate location from the polymerase site that removes nucleotides that are incorrectly placed into DNA by the polymerase [8]. Therefore, if the polymerase accidentally incorporates a wrong nucleotide into DNA, the polymerase–PCNA may switch between different DNA tracking sites and reposition the primer terminus from the polymerase site to the exonuclease site. Indeed, a particular tilt of PCNA with DNA has been documented by Morikawa’s group [16,17] for Pyrococcus furiosus (Pfu) PCNA on DNA with polymerase B or DNA ligase, and computational studies suggest the switch from polymerase to exonuclease sites may occur by a direct switch of this tilt as indicated in Figure 2B [18]. Although speculative, another useful discrimination between active and inactive tilts could occur during the switch between a high fidelity DNA polymerase that encounters a DNA lesion and low fidelity specialized translesion DNA polymerases that are specially constructed to traverse lesions [19,20]. Thus, we speculate in Figure 2B that the translesion DNA polymerase might function with PCNA when it is in a tilt that is not active with a high fidelity polymerase, and this may facilitate and regulate the switching between them. Alternatively, the two polymerases could interact with the same tilt and trade on and off the clamp in two separate events.
It is quite gratifying to see the inner workings of this beautifully symmetrical PCNA ring, and how nature has been able to sculpt it to satisfy the requirements of the many different enzymes that use DNA sliding clamps. Through this recent study [8] we can now finally visualize how PCNA slides along DNA and the manifestations of the movements and protein dynamics that occur as it does so.
References
- 1.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg A, Baker TA. DNA Replication. 2nd W.H. Freeman; New York: 1992. [Google Scholar]
- 3.Stukenberg PT, Studwell-Vaughan PS, O’Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 4.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 5.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21 (WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 6.Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 7.De Biasio A, Blanco FJ. Proliferating cell nuclear antigen structure and interactions: too many partners for one dancer? Adv Protein Chem Struct Biol. 2013;91:1–36. doi: 10.1016/B978-0-12-411637-5.00001-9. [DOI] [PubMed] [Google Scholar]
- 8.De March M, Merino N, Barrera-Vilarmau S, Crehuet R, Onesti S, Blanco FJ, De Biasio A. Structural basis of human PCNA sliding on DNA. Nat Commun. 2017;8:13935. doi: 10.1038/ncomms13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurence TA, Kwon Y, Johnson A, Hollars CW, O’Donnell M, Camarero JA, Barsky D. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J Biol Chem. 2008;283:22895–22906. doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, van Oijen AM. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J Biol Chem. 2009;284:17700–17710. doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Hingorani MM. Impact of individual proliferating cell nuclear antigen-DNA contacts on clamp loading and function on DNA. J Biol Chem. 2012;287:35370–35381. doi: 10.1074/jbc.M112.399071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedglin M, Pandey B, Benkovic SJ. Stability of the human polymerase δ holoenzyme and its implications in lagging strand DNA synthesis. Proc Natl Acad Sci USA. 2016;113:E1777–E1786. doi: 10.1073/pnas.1523653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgescu RE, Langston LD, O’Donnell M. A proposal: Evolution of PCNA’s role as a marker of newly replicated DNA. DNA Repair. 2015;29:4–15. doi: 10.1016/j.dnarep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayanagi K, Kiyonari S, Saito M, Shirai T, Ishino Y, Morikawa K. Mechanism of replication machinery assembly as revealed by the DNA ligase-PCNA-DNA complex architecture. Proc Natl Acad Sci USA. 2009;106:4647–4652. doi: 10.1073/pnas.0811196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayanagi K, Kiyonari S, Nishida H, Saito M, Kohda D, Ishino Y, Shirai T, Morikawa K. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc Natl Acad Sci USA. 2011;108:1845–1849. doi: 10.1073/pnas.1010933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Yan C, Kossmann BR, Ivanov I. Secondary interaction interfaces with PCNA control conformational switching of DNA polymerase PolB from polymerization to editing. J Phys Chem B. 2016;120:8379–8388. doi: 10.1021/acs.jpcb.6b02082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kath JE, Chang S, Scotland MK, Wilbertz JH, Jergic S, Dixon NE, Sutton MD, Loparo JJ. Exchange between Escherichia coli polymerases II and III on a processivity clamp. Nucleic Acids Res. 2016;44:1681–1690. doi: 10.1093/nar/gkv1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase h. Biochemistry. 2014;53:2793–2803. doi: 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]


