Abstract
Increasing apoA-I synthesis may improve HDL functionality and lower CVD risk. As theobromine and fat increase fasting apoA-I concentrations, and the intestine is involved in apoA-I production, the acute effects of both were studied on duodenal gene transcription to better understand underlying mechanisms. In this crossover study, 8 healthy men received once a low fat (LF) meal, a LF meal plus theobromine (850 mg), or a high fat (HF) meal. Five hours after meal intake duodenal biopsies were taken for microarray analysis. Theobromine and HF consumption did not change duodenal apoA-I expression. Theobromine did not change gene expression related to lipid and cholesterol metabolism, whereas those related to glycogen/glucose breakdown were downregulated. HF consumption increased gene expression related to lipid and cholesterol uptake and transport, and to glucose storage, while it decreased those related to glucose uptake. Furthermore, genes related to inflammation were upregulated, but inflammation markers in plasma were not changed. In healthy men, acute theobromine and fat consumption did not change duodenal apoA-I mRNA, but inhibited expression of genes related to glucose metabolism. Furthermore, HF intake activated in the duodenum expression of genes related to lipid and cholesterol metabolism and to inflammation.
Introduction
Over the past decade, interventions aiming to increase serum high-density lipoprotein (HDL) cholesterol (HDL-C) concentrations to reduce the risk of cardiovascular disease (CVD) have not been successful. Recent insights, however, suggest that improving HDL functionality will more likely lower CVD risk than simply elevating circulating serum HDL-C concentrations1. In this respect, increasing serum apolipoprotein A-I (apoA-I) synthesis may be a promising approach, as serum apoA-I concentrations correlate with in vitro cholesterol efflux, a measure of HDL functionality2–4. For this, various pharmaceutical approaches are currently explored5. However, as a preventive strategy at a population level, dietary strategies are more useful. Unfortunately, the number of (novel) dietary components that increase apoA-I production is limited. Theobromine, a component from cocoa, has been reported to increase fasting serum apoA-I concentrations6. Mechanisms explaining the effects of theobromine on fasting serum apoA-I concentrations are however not clear. Since apoA-I is produced in enterocytes and hepatocytes7, and theobromine is absorbed in the small intestine8,9, we were interested in exploring the effects of acute theobromine consumption on gene expression in postprandial human duodenal biopsies, following theobromine intake. Duodenal biopsies were used, because apoA-I secretion is higher in the duodenum as compared with other parts of the intestine10.
Except theobromine, also exchanging carbohydrates for fatty acids increases fasting serum apoA-I concentrations11,12. Although the intestine is strongly involved in dietary lipid handling, the effects of fat intake on duodenal gene expression profiles have not been studied. Only a limited number of studies compared the effects of a low-fat/high-carbohydrate (LF) with a high-fat/low-carbohydrate (HF) meal on gene expression profiles, and so far only in human muscle biopsies13–15 and peripheral blood mononuclear cells (PBMCs)16. We therefore used a nutrigenomic approach to analyze differences in gene expression in human duodenal biopsies after adding 850 mg of theobromine to a LF meal, and after comparing HF with LF meal consumption to better understand effects of dietary theobromine and fat on duodenal apoA-I transcription and related pathways.
Results
Subject characteristics
All ten men completed the study. However, results of one man were excluded due to protocol violation, as he appeared not to be in fasting condition at start of one of the test days. Samples from a second subject were excluded due to technical issues during microarray analysis. Baseline characteristics of the final eight subjects are shown in Table 1.
Table 1.
Baseline characteristics of the participants who completed the study1.
| Mean ± SD | |
|---|---|
| Age (years) | 38 ± 15 |
| BMI (kg/m2) | 24.3 ± 2.0 |
| Serum total cholesterol (mmol/L) | 5.2 ± 0.9 |
| Plasma glucose (mmol/L) | 5.1 ± 0.3 |
| Systolic blood pressure (mmHg) | 130 ± 18 |
| Diastolic blood pressure (mmHg) | 79 ± 14 |
1Values are mean ± SD. n = 8.
Microarray analysis
From the 19697 genes present on the microarray, 10506 genes were expressed in the duodenum (expression value > 20 and > 5 probes per gene on the array). In comparison to the LF shake, 113 and 286 genes were differentially expressed after adding theobromine to the LF diet (LF-TB) and comparing it with the HF shake, respectively (Supplementary data Fig. 1). Twenty-three of these differentially expressed genes overlapped, i.e. the expression was significantly changed into the same direction after both the LF-TB and the HF interventions as compared with the LF diet. These 23 genes were SCN3B, PCDH11Y, NELL2, LYPD6B, ZNF485, FBXL16, PLA1A, ZBTB16, UPK2, DKK4, TIAM2, CKLF, SPRR1A, C12orf74, MTNR1A, KCNS3, MBOAT2, VSIG8, TEN1-CDK3, PDE10A, LRP12, TAS2R3 and RAD51AP1. So far, none of these 23 genes was described in relation to apoA-I transcriptional regulation. Unfortunately, apoA-I gene expression did not change when comparing the LF-TB meal vs. the LF meal (FC = 1.02, P = 0.758) or the HF meal vs. the LF meal (FC = 1.07, P = 0.310). Still, these 23 genes are potentially interesting targets to consider in the context of apoA-I transcription.
Next, gene expression profiles were further explored, to better understand the effects of adding theobromine to a LF diet or comparing a HF with a LF diet on duodenal gene expression. First, IPA was used to look at significantly changed pathways. Then, two more in depth analytical procedures were conducted, i.e. the Upstream Regulator Analysis and GSEA.
Adding TB to a LF meal
Ingenuity Pathway Analysis
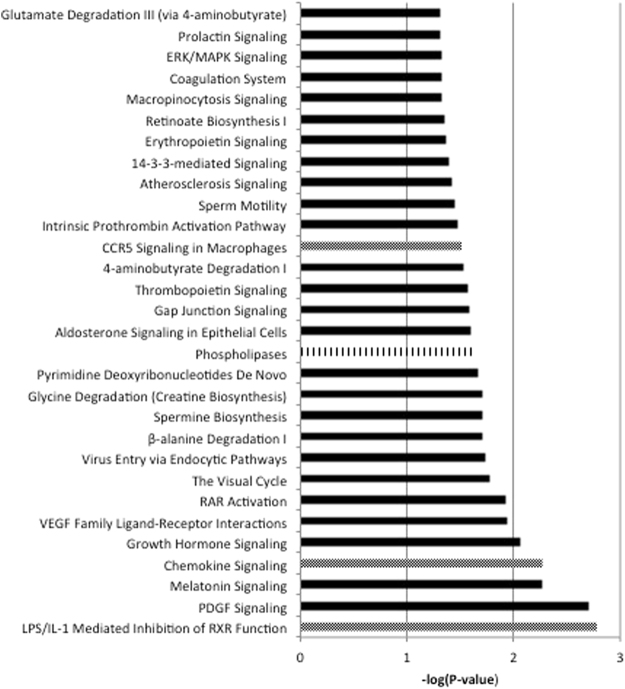
Thirty pathways were differentially regulated by theobromine consumption (Fig. 1). None of the pathways was related to cholesterol, lipid or glucose metabolism.
Figure 1.
Significantly different pathways after adding 850 mg of theobromine (TB) to the low-fat/high-carbohydrate (LF) diet. Dotted bars are pathways involved in the immune system, lined bars are pathways involved in cholesterol, lipid or glucose metabolism (n = 8).
Upstream Regulator Analysis
Adding TB to the LF meal changed the activation of 29 transcriptional regulators (Supplementary data, Table 1). Three of these upstream regulators, which were related to glucose metabolism, were inhibited. Further, the other identified upstream regulators were not linked to the lipid and cholesterol metabolism (Table 2).
Table 2.
Inhibited or activated upstream regulators after adding 850 mg of theobromine (TB) to a low-fat/high-carbohydrate (LF) meal or after comparing high-fat/low-carbohydrate (HF) with LF consumption functioning in lipid, cholesterol or glucose metabolism or the immune system (n = 8).
| Upstream regulator | Activation Z-score | Function (gene card ref) | Comparison |
|---|---|---|---|
| Ins1 | −3.02 | Decreases blood glucose | LF-TB vs. LF |
| Insulin | −2.08 | Decreases blood glucose | LF-TB vs. LF |
| INS | −1.95 | Decreases blood glucose | LF-TB vs. LF |
| INSIG2 | −1.95 | Feedback control of cholesterol synthesis | HF vs. LF |
| APOE | −1.73 | Main lipoprotein on chylomicrons | HF vs. LF |
| ACOX1 | −1.63 | Fatty acid β-oxidation pathway | HF vs. LF |
| SREBF2 | 1.93 | Lipid homeostasis | HF vs. LF |
| PPARα | 1.97 | Transcription factor in lipid and cholesterol metabolism | HF vs. LF |
| FABP2 | 2.00 | Uptake and transport of long chain fatty acids involved in TAG-rich lipoprotein synthesis | HF vs. LF |
| SREBF1 | 2.27 | Transcription factor which regulates lipid homeostasis | HF vs. LF |
| GCG | −2.42 | Proprotein for glucagon | HF vs. LF |
| GCK | −1.98 | Enzyme functioning in glucose utilization | HF vs. LF |
| Gsk3 | 1.98 | Glycogen synthesis | HF vs. LF |
| TNFSF12 | 1.65 | Activation of NFκB, inducer of proinflammatory cytokines | HF vs. LF |
| IL12 | 1.90 | Proinflammatory cytokine | HF vs. LF |
| IL2 | 1.92 | Proinflammatory cytokine | HF vs. LF |
| CD5 | 1.98 | Receptor in the regulation of T-lymphocyte proliferation | HF vs. LF |
| CCL5 | 1.98 | Chemoattractant for monocytes, T-lymphocytes, eosinophils | HF vs. LF |
| TNF | 1.99 | Survival, proliferation and differentiation of monocytes and macrophages | HF vs. LF |
| RELA | 2.02 | Subunit NFκB | HF vs. LF |
| MYD88 | 2.24 | Innate immune response | HF vs. LF |
| IL1A | 2.31 | Proinflammatory cytokine | HF vs. LF |
| TNFSF11 | 2.73 | T-lymphocyte dependent immune response | HF vs. LF |
| IL1B | 2.82 | Proinflammatory cytokine | HF vs. LF |
Gene Set Enrichment Analysis
In the GSEA, 6 gene sets were upregulated and 2 gene sets were downregulated (Supplementary data, Table 2). One of the downregulated set was glucose metabolism (Table 3).
Table 3.
Significantly inhibited or activated gene sets after adding 850 mg of theobromine (TB) to a low-fat/high-carbohydrate (LF) meal or after comparing high-fat/low-carbohydrate (HF) with LF meal consumption functioning in lipid, cholesterol or glucose metabolism or the immune system (n = 8).
| Name gene set | NES | FDR q-val | Comparison |
|---|---|---|---|
| Glucose metabolism | −1.96 | 0.137 | LF-TB vs. LF |
| Regulation of beta cell development | 1.96 | 0.119 | LF-TB vs. LF |
| Fatty acid beta oxidation | 2.09 | 0.025 | HF vs. LF |
| Regulation of lipid metabolism by PPARα | 1.93 | 0.057 | HF vs. LF |
| PPARα activates gene expression | 1.92 | 0.058 | HF vs. LF |
| Mitochondrial long chain fatty acid β oxidation | 1.88 | 0.075 | HF vs. LF |
| TAG synthesis | 1.82 | 0.106 | HF vs. LF |
| Regulation of lipid metabolism by PPARα | 1.81 | 0.104 | HF vs. LF |
| PPARα signaling pathway | 1.80 | 0.104 | HF vs. LF |
| PPARα targets | 1.80 | 0.101 | HF vs. LF |
| Statin pathway | 1.67 | 0.144 | HF vs. LF |
| SREBP signaling | 1.62 | 0.159 | HF vs. LF |
| Lipid digestion, mobilization and transport | 1.60 | 0.171 | HF vs. LF |
| Fatty acid TAG and ketone body metabolism | 1.60 | 0.167 | HF vs. LF |
| Lipid digestion, mobilization and transport | 1.56 | 0.184 | HF vs. LF |
| Metabolism of steroid hormones and Vit D | 1.58 | 0.175 | HF vs. LF |
| Steroid hormones | 1.57 | 0.178 | HF vs. LF |
| Regulation of cholesterol synthesis by SREBP SREBF | 1.56 | 0.181 | HF vs. LF |
| Lipid digestion, mobilization and transport | 1.55 | 0.183 | HF vs. LF |
| Cholesterol biosynthesis | 1.53 | 0.199 | HF vs. LF |
| Glucose metabolism | −2.29 | 0.001 | HF vs. LF |
| Metabolism of carbohydrates | −2.17 | 0.006 | HF vs. LF |
| Gluconeogenesis | −1.84 | 0.074 | HF vs. LF |
| Hexose transport | −1.83 | 0.073 | HF vs. LF |
| Carbohydrates digestion and absorption | −1.83 | 0.070 | HF vs. LF |
| Glycogen metabolism | −1.78 | 0.064 | HF vs. LF |
| Starch and sucrose metabolism | −1.77 | 0.058 | HF vs. LF |
| Hexose transport | −1.67 | 0.088 | HF vs. LF |
| Pancreatic secretion | −1.61 | 0.129 | HF vs. LF |
| Insulin signalling pathway | −1.59 | 0.136 | HF vs. LF |
| Glycolysis and gluconeogenesis | −1.47 | 0.193 | HF vs. LF |
| Type II diabetes mellitus | −1.47 | 0.196 | HF vs. LF |
| Glycogen storage diseases | −1.46 | 0.198 | HF vs. LF |
| Glucose transport | −1.45 | 0.196 | HF vs. LF |
| Chemokine receptors bind chemokines | 2.08 | 0.020 | HF vs. LF |
| Cell adhesion molecules CAMS | 1.82 | 0.113 | HF vs. LF |
| Cytokine cytokine receptor interaction | 1.82 | 0.110 | HF vs. LF |
| Cytokine and inflammatory response | 1.80 | 0.103 | HF vs. LF |
| Intestinal immune network for IGA production | 1.79 | 0.106 | HF vs. LF |
| RIP mediated NFκB activation via ZBP1 | 1.78 | 0.106 | HF vs. LF |
| ZBP1 DAI mediated induction of type I IFNS | 1.78 | 0.101 | HF vs. LF |
| IL1R pathway | 1.78 | 0.089 | HF vs. LF |
| Rheumatoid arthritis | 1.77 | 0.101 | HF vs. LF |
| NKT pathway | 1.74 | 0.110 | HF vs. LF |
| TNF signaling pathway | 1.73 | 0.112 | HF vs. LF |
| Staphylococcus aureus infection | 1.71 | 0.126 | HF vs. LF |
| NFκB signaling pathway | 1.65 | 0.176 | HF vs. LF |
| TOPB1 pathway | 1.60 | 0.168 | HF vs. LF |
| NTHI pathway | 1.59 | 0.173 | HF vs. LF |
| Inflammatory response pathway | 1.59 | 0.167 | HF vs. LF |
| TNFR2 pathway | 1.57 | 0.180 | HF vs. LF |
| Human complement system | 1.55 | 0.184 | HF vs. LF |
Overall gene expression pattern after adding TB to a LF meal
Overall, adding TB to a LF meal did not change the expression of genes related to lipid and cholesterol metabolism, whereas, expression of a number of genes related to glycogen and glucose breakdown were downregulated.
Comparing HF with LF meal consumption
Ingenuity Pathway Analysis
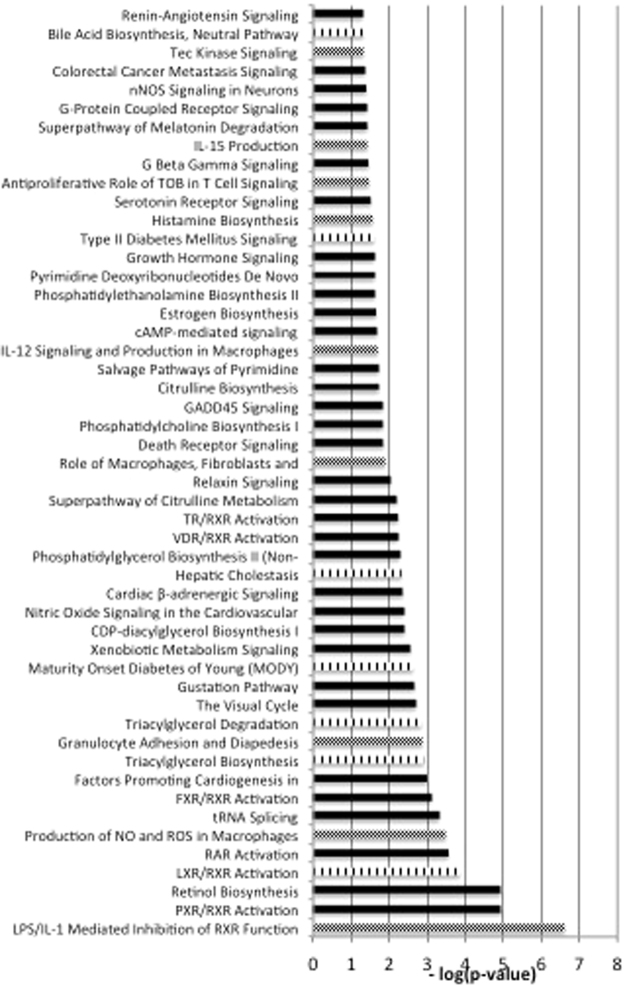
Fifty pathways were differentially regulated after intake of the HF meal as compared with the LF meal. Seven of these pathways were linked to cholesterol, lipid or glucose metabolism, such as LXR/RXR activation and triacylglycerol degradation/biosynthesis. Also 9 changed pathways were related to the immune response, including the production of IL12 and IL15 and the production of NO and ROS in macrophages (Fig. 2).
Figure 2.
Significantly different pathways comparing a high-fat/low-carbohydrate (HF) with a low-fat/high-carbohydrate (LF) diet. Dotted bars are pathways involved in the immune system, lined bars are pathways involved in cholesterol, lipid or glucose metabolism (n = 8).
Upstream Regulator Analysis
The activation of 113 transcriptional regulators was changed (Supplementary data, Table 3). After HF consumption regulators involved in lipid uptake and transport such as SREBF2, PPARα and FABP2 were activated, while regulators involved in fatty acid breakdown such as ACOX1 were inhibited. Furthermore, regulators involved in glucose uptake, including GCG and GCK were inhibited and regulators involved in glucose storage, such as Gsk3 were activated. Finally, HF meal consumption activated 11 proinflammatory regulators involved in the immune response: TNFSF12, IL12, IL2, CCL5, TNF, CD5, RELA, MYD88, TNFSF11, IL1A and IL1B (Table 2).
Gene Set Enrichment Analysis
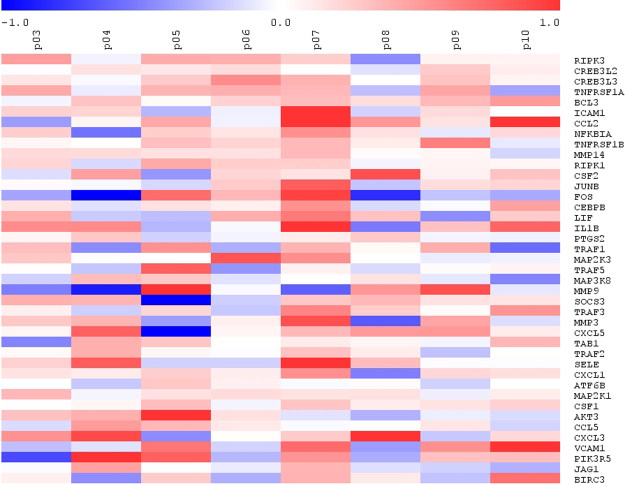
GSEA showed 83 upregulated gene sets and 176 downregulated gene sets after HF meal consumption (Supplementary data, Table 4). Seventeen gene sets involved in lipid and cholesterol metabolism were upregulated. These gene sets suggested an increased activity of PPARα, fatty acid oxidation, fatty acid, triacylglycerol (TAG) and lipoprotein metabolism, lipid digestion, mobilization and transport, cholesterol synthesis, steroid hormone metabolism and SREBP signaling. Furthermore, the 13 gene sets gene sets that were downregulated were involved in glucose and carbohydrate metabolism, suggesting an inhibited glucose and carbohydrate metabolism, gluconeogenesis and insulin signaling. Finally, 19 upregulated gene sets were involved in the immune system, including chemokine and cytokine activities, NFκB pathway, complement system and TNF signaling pathway (Table 3 and Fig. 3).
Figure 3.
Heatmap of the of the TNF signaling pathway from the GSEA results after the consumption of an acute high-fat/low carbohydrate (HF) vs. low-fat/high-carbohydrate (LF) meal (n = 8).
Overall gene expression pattern comparing HF with LF consumption
HF consumption increased the expression of genes related to lipid and cholesterol uptake and transport and glucose storage, while it decreased the expression of genes related glucose uptake. Furthermore, all three approaches showed upregulated expression of genes related to the immune response and inflammation.
Markers for inflammation, the acute phase response, endothelial function and intestinal damage
Since the three analytical approaches consistently showed differences in the expression profiles of genes involved in the cytokine and inflammatory responses after HF compared with LF meal consumption, a panel of plasma markers for inflammation and acute phase responses were analyzed to evaluate whether this acute change in inflammatory gene expression in the duodenum was also transferred into the circulation.
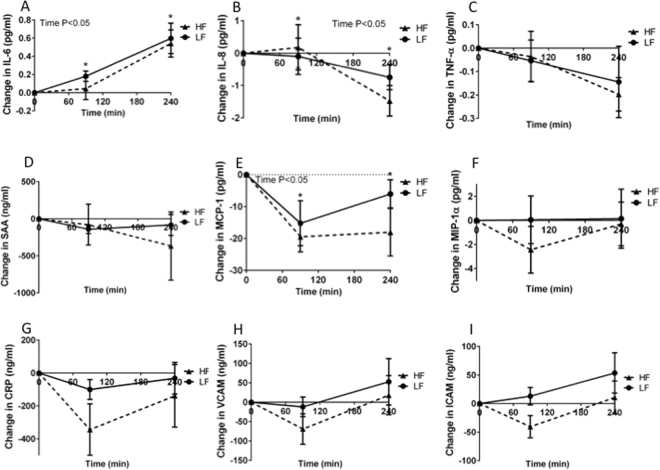
During the postprandial phase, IL-6 concentrations significantly increased over time (P < 0.001), while IL-8, MCP-1 and IFABP concentrations significantly decreased over time (P = 0.006, P < 0.001 and P < 0.001, respectively). Concentrations of all other markers for inflammation and the acute phase response, as well as endothelial function did not change over time (Fig. 4). Moreover, no differences in postprandial changes in markers for inflammation, acute phase response and endothelial function between the HF and LF meals were observed (Fig. 4). Finally, circulating levels of IFABP were measured to evaluate the potential effects of the HF and LF meal on intestinal damage. However, also IFABP concentrations were not different between HF and LF meal consumption (Fig. 5).
Figure 4.
Change in (A), interleukin 6 (IL-6) (B), interleukin 8 (IL-8) (C), tumor necrosis factor alpha (TNF-α) (D), serum amyloid A (SAA) (E), monocyte chemoattractant protein-1 (MCP-1) (F), macrophage inflammatory protein 1 a (MIP-1a) (G), high sensitive C-reactive protein (CRP) (H), vascular cell adhesion protein (VCAM) and (I), intercellular adhesion molecule (ICAM) after acute low-fat/high-carbohydrate (LF, black line, squares) and acute high-fat/low carbohydrate (HF, dotted line, triangles) meal consumption1. 1Values are mean ± SD. n = 8.
Figure 5.
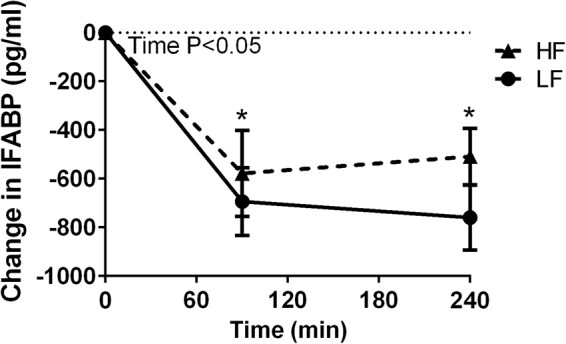
Change in intestinal fatty acid-binding protein (IFABP) after acute low-fat/high-carbohydrate (LF, black line, squares) and acute high-fat/low-carbohydrate (HF, dotted line, triangles) meal consumption. 1Values are mean ± SD. n = 8.
Discussion
It has been shown that consuming either theobromine6 or fat12 increased fasting serum apoA-I concentrations. To better understand the potential role of the intestine in the mechanisms underlying these effects on apoA-I concentrations, as well as to explore effects on gene expression profiles in general, we examined in the present randomized, double-blind, controlled study the acute effects of theobromine and of HF consumption on duodenal gene expression. Unfortunately, both theobromine and HF consumption did not change duodenal apoA-I gene expression. For this apparently discrepant result as compared to the assumed effects on serum apoA-I concentrations, there are at least four explanations. First, theobromine and fat only change duodenal apoA-I mRNA expression after longer-term consumption. Second, postprandial effects on apoA-I transcription are not evident within a time period of 5 hours. Third, theobromine and fat regulate apoA-I metabolism not at a transcriptional level in the duodenum, but only in the liver. However, this explanation is less likely, as postprandial serum apoA-I concentrations did also not change after theobromine and HF intake17. Fourth, theobromine and fat do not change apoA-I transcription but increases serum apoA-I concentrations via other mechanisms, e.g. a reduction in apoA-I clearance. Indeed, replacement of 13 energy % of carbohydrates for MUFA, which elevated apoA-I concentrations, decreased apoA-I fractional catabolic rate, while it did not change apoA-I production rate18. It should also be noted, that unexpectedly we could recently not confirm in a long-term study the effects of theobromine on apoA-I19, as reported by Neufingerl et al.6.
Except for evaluating effects on duodenal apoA-I mRNA expression, we also explored changes in gene expression profiles in general. In this context, acute theobromine consumption did not change the expression of genes related to lipid and cholesterol metabolism. This observation is in agreement with the earlier reported lack of acute effects in changing postprandial serum concentrations of apoB100, apolipoprotein B48, TAG and free fatty acids17, but not with the results of 4-weeks of theobromine consumption, which improved fasting lipid concentrations6,19. Except for apoB48, which is only synthesized in the intestine, the same considerations as those for apoA-I can be used to explain the lack of effects. Moreover, GSE analysis showed that theobromine inhibited duodenal expression of gene sets involved in glucose metabolism, suggesting a lower glycogen and glucose breakdown. Moreover, using the upstream regulator analysis, we identified downregulation of three factors (Ins1, Insulin and INS) that have a functional role in lowering plasma glucose concentrations. Thus, these changes are expected to translate into higher plasma glucose concentrations. Indeed, we have earlier reported increased postprandial glucose responses after 4-week TB-consumption19. Though suggestive, it is however unclear how and to what extent these decreases in gene expression relate to changes in glucose metabolism in the duodenum and to changes in plasma glucose and insulin responses. Besides these effects of theobromine on expression of genes involved in glucose metabolism, there were no other consistent clusters of metabolic processes that seemed to be sensitive to theobromine consumption. However, it should be noted that only effects of theobromine on a LF background diet were studied. Therefore, it cannot be excluded that effects of theobromine on a HF background would have been different.
Besides theobromine, also the acute effects of switching from a LF to HF diet on duodenal gene expression profiles were examined. Surprisingly, in humans these type of analysis during the postprandial phase has been done in muscle biopsies and circulating leukocyte fractions [13–16], but hardly in the small intestine, which plays a major role in TAG and cholesterol absorption and transport20. As expected, the HF meal increased postprandial serum TAG concentrations17, and upregulated genes related to lipid and cholesterol uptake and transport when compared with the LF meal. The upregulation in LXR/RXR pathway expression, triacylglycerol degradation and biosynthesis, as well as bile acid biosynthesis pathways, as observed in the GSE analysis, are illustrative. The changes in the expression of transcription factors, such as downregulation in INSIG2 (cholesterol biosynthesis), and upregulation in SREBF2 & SREBF1 (lipid homeostasis), and PPARα (lipid and cholesterol metabolism), are generally in line with these pathways. This can be explained by the higher amount of cholesterol and lipids that are present in the intestine after HF meal consumption, as these lipids need to be taken up and must be transported to other tissue. In agreement, in the intestines of mice, genes related to lipid metabolism were activated when long-term HF intake was compared with LF intake21. Also in human muscle biopsies, expression of genes functioning in the lipid metabolism increased after a HF meal13,15. In the present study, the expression of genes related to glucose uptake were decreased and those of glucose storage were increased comparing HF with LF consumption, which can be linked to the higher amount of fat in the HF meal and the increased amount of carbohydrates in the LF meal. In line with our results, in human muscle biopsies a shift in glucose metabolism from oxidation to storage was observed, when comparing HF with LF meal consumption14.
Besides the changes in expression of genes related to cholesterol ands lipid metabolism, which can easily be explained from a substrate point of view, the most apparent observation after consuming the HF meal was the upregulation of numerous gene sets and pathways associated with inflammation and immune function. Increased expression of cell adhesion molecules, cytokine and inflammatory responses as well as NF-κB and TNFα signaling were predominant. A number of studies looking in other tissues and species support our results. For example, in human PBMCs, the consumption of a HF breakfast increased the expression of IL-8 as compared with a LF breakfast22. Furthermore, inflammation was one of the most modulated biological processes after HF consumption in the intestines21 and adipose tissue23 of mice. In addition, in rat peripheral leukocytes a HF diet increased expression of genes related to leukocyte activation24. To visualize our findings, we presented the changes in all individual genes being part of the TNF signaling pathway (Fig. 3). This shows that a wide variety of inflammatory mediators are all upregulated during the HF meal as compared to the LF meal. This illustrates that it is not a single gene effect, but a general activation of inflammation processes activating cytokines, chemokines and adhesion molecules, which can all be linked to activated NF-kB signaling. This suggests that there is an ongoing activation of genes, already very early in the signaling cascade, which is confirmed by the activation of early upstream regulators like RELA (a subunit of NF-kB), MYD88, and the alarm cytokines IL1α and IL1β involved in inflammasome activation. The question is why HF diets activate these inflammatory systems. One explanation could be that saturated fatty acids, and particularly palmitic acid, which was present in significant amounts in the HF diets, is a potent substrate that is recognized by the pattern recognition receptor (PRR), Toll like receptor 4 (TLR4). Normally, TLR4 responds to lipopolysaccharide (LPS) molecules secreted by gram-negative bacteria. However, the LPS tail, which is the actual recognition site, is highly enriched in C16:0 molecules. Therefore, it has been suggested that other types of fat sources, such as long-chain polyunsaturated fatty acids, do not activate the NF-kB axis. Future studies are needed to confirm whether a HF-meal with a different fatty acid composition would have given different results.
Our results raise at least two additional questions. First, is the effect on inflammation a primary response to the intake of dietary fatty acids or is it a secondary response caused by enterocyte damage? Therefore, serum IFABP concentrations, a marker for enterocyte damage25, were determined, which were not different after the LF and HF meals, suggesting that the enterocytes were not damaged acutely after the HF meal. The second question is, whether the apparent increase in duodenal pro-inflammatory gene expression profiles translated into an inflammatory signature in postprandial serum samples. Our data clearly shows that none of the measured plasma markers differed after acute HF and LF consumption. This may indicate that the duodenal inflammatory signal needs longer than a few hours to translate into a systemic inflammatory response, or is not translated at all. Our results on plasma biomarkers are partly contradictory with other studies. Esser et al. found no changes in plasma CRP, ICAM-1, IL-6 and TNF-α, decreases in VCAM-1 and SAA, and an increase in IL-8 after an acute HF breakfast compared with an LF breakfast16. Nappo et al. found increased plasma TNF-α, IL6, ICAM-1 and VCAM-1 concentrations comparing HF with LF meal consumption26. It should be noted, however, that Esser et al. used a higher amount of fat16, while Nappo et al. studied diabetic patients26. Also, reported changes in plasma inflammation markers in response to HF and LF feeding have been very variable between studies27.
In conclusion, in healthy men, acute theobromine and fat consumption did not change apoA-I expression in the duodenum. Theobromine consumption inhibited intestinal gene expression related to glycogen and glucose breakdown, but did not change those related to lipid and cholesterol metabolism. Furthermore, HF intake activated expression of genes related to lipid and cholesterol uptake and transport and glucose storage, while it decreased those related to glucose uptake. Finally, microarray analyses suggested upregulation of inflammation in the duodenum after HF meal consumption, which was not translated into a systemic inflammatory response directly following a HF meal.
Methods
Study population and design
The design and results of the metabolic parameters of this double-blind crossover study have already been reported17. Briefly, ten apparently healthy men participated. During the screening visit, body weight, height and blood pressure were measured and a fasting blood sample was taken. Subjects were excluded when fasting serum total cholesterol concentrations ≥8.0 mmol/L or plasma glucose concentrations ≥7.0 mmol/L. After inclusion, all subjects participated in three test days, each separated by a one-week washout period. Two weeks before the start of the study, subjects were instructed to avoid products containing cocoa till the end of the study period. Also, the consumption of caffeine containing drinks was restricted to a maximum of 4 cups a day, since theobromine is a metabolite of caffeine. The study was conducted according the guidelines laid down in the Declaration of Helsinki. The study protocol was approved by the Medical Ethical Committee of the University Hospital Maastricht. All participants gave their written informed consent before entering the study. The study was registered on clinicaltrials.gov under study number NCT02085109, date of registration: 17-02-2014.
Test days
To minimize differences in dietary intake before the three test days, all subjects were provided with a standard low fat dinner the evening before each test day, which consisted of a commercially available macaroni, 3 crackers, and a dairy drink. The next morning, subjects came to the University in fasting condition, which means that after dinner the preceding evening, they had not consumed any foods or drinks, except for water. To reduce physical activity as much as possible, participants arrived by public transport or car on the morning of the test day. After a 15 min rest, the first fasting blood sample was collected (T0) via an intravenous cannula inserted into the antecubital vein. Subjects were then asked to consume a shake within 10 min. Three different shakes were provided in random order. One shake was low-fat/high-carbohydrate (LF), one shake was LF enriched with 850 mg of theobromine (LF-TB), and one shake was high-fat/low-carbohydrate (HF) (Table 4). The theobromine powder (Fagron, Uitgeest, the Netherlands) was added as the final ingredient to the blender jar before it was thoroughly mixed with the LF shake. The volumes of the shakes were standardized with water. The difference between the HF and the LF shakes was an exchange between fat and carbohydrates, while the amount of proteins between the three shakes was comparable (Table 4). Shakes were prepared by the research dietician to blind both researchers and participants. After consumption of the shakes, the volunteers were not allowed to eat or drink anything except water for the next 5 hours. Other blood samples were taken at T = 30 (T30), T = 60 (T60), T = 90 (T90), T = 120 (T120), T = 180 (T180) and T = 240 (T240) min. Five hours after meal intake, duodenal biopsies were taken at the Department of Endoscopy. During duodenoscopy, no sedatives were given. Four duodenal mucosal tissue samples, just proximal of the ampulla of Vater, were taken using standard biopsy forceps. The diameter of the biopsies varied between 2.0 mm and 2.2 mm. After sampling, biopsies were immediately frozen in liquid nitrogen, stored at −80 °C and analyzed at the end of the study for gene expression profiles.
Table 4.
Nutrient composition of the low-fat/high-carbohydrate (LF), LF with 850 mg theobromine (LF-TB) and high-fat/low-carbohydrate (HF) shakes.
| Nutrient | LF | LF-TB | HF |
|---|---|---|---|
| Energy (kcal) | 956 | 956 | 965 |
| Protein (g) | 19.4 | 19.4 | 17.9 |
| (E%)1 | 8 | 8 | 7 |
| Carbohydrates (g) | 193.7 | 193.7 | 85.7 |
| (E%) | 81 | 81 | 35 |
| Mono- and disaccharides (g) | 144.9 | 144.9 | 45.6 |
| Polysaccharides (g) | 48.8 | 48.8 | 40.1 |
| Total fat (g) | 10.5 | 10.5 | 60.6 |
| (E%) | 10 | 10 | 56 |
| Saturated fatty acids (g) | 3.2 | 3.2 | 36.0 |
| Monounsaturated fatty acids (g) | 4.0 | 4.0 | 18.7 |
| Polyunsaturated fatty acids (g) | 1.1 | 1.1 | 4.1 |
| Cholesterol (mg) | 334 | 334 | 341 |
| Theobromine (mg) | 0 | 850 | 0 |
1E%: energy percent.
Blood sampling and analysis
Blood was sampled in serum and EDTA-containing vacutainer tubes. Serum tubes were allowed to clot for 1 hour at 20 °C, followed by centrifugation at 1300 × g for 15 min at 20 °C. The EDTA tubes were placed on ice directly after sampling and centrifuged at 1300 × g for 15 min at 4 °C within 60 min after sampling. Serum and plasma aliquots were stored at −80 °C until analyses. All samples from one subject were analyzed within the same analytical run at the end of the study.
Because gene expression profiles clearly showed a more pro-inflammatory pattern after HF compared with LF consumption, concentrations of the inflammation and acute phase response markers: interleukin-6 (IL-6), interleukin-8 (IL-8), tumor necrosis factor-alpha (TNF-α), serum amyloid A (SAA), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein 1 alpha (MIP-1α), and high sensitive C-reactive protein (hsCRP), were measured in plasma samples of the HF and LF test days (T0, T90 and T240). In addition we analyzed plasma concentrations of soluble intercellular adhesion molecule-1 (sICAM-1) and soluble vascular cell adhesion molecule-1 (sVCAM-1) to evaluate a possible cross-talk between an “inflamed” intestine and the vascular wall. For these analyses, a commercially available Multi Spot ELISA kit (Meso Scale Discovery, Rockville, MD, USA) was used. Finally, in the same plasma samples, intestinal fatty acid-binding protein (IFABP) concentrations, which is a marker for damaged enterocytes25,28, were measured using a sandwich ELISA (R&D, Oxon, United Kingdom) to evaluate whether one acute HF meal can already damage the enterocytes.
Microarray processing and data analysis
Total RNA was extracted from one frozen duodenal mucosal biopsy using TRIzol reagent (Invitrogen, Breda, the Netherlands) and purified on columns using Qiagen RNeasy Micro Kit (Qiagen, Venlo, the Netherlands). Total RNA (100 ng) was labeled by Whole Transcript Sense Target Assay and hybridized to human whole-genome Affymetrix Gene 1.1 ST arrays targeting 19697 unique genes (Affymetrix, Santa Clara, CA). Microarray analyses were performed using MADMAX pipeline for statistical analysis of microarray data29. In short, microarrays were normalized with the robust multichip average method and probes were annotated as described30,31. This gene set was filtered on an expression of >10 on at least 5 arrays and measured with ≥5 probes. This filtered data set consisted of 10506 genes. Comparisons were made between the LF-TB and the LF meal and between the HF and the LF meal. Individual genes were defined as changed when comparison of the normalized signal intensities showed a p ≤ 0.05 in a 2-tailed paired intensity-based moderated t-statistics (IBMT) and a fold change of >1.2 or <−1.2 between the diets32. Further data analysis was performed on the filtered dataset with three different approaches i.e. Ingenuity Pathway Analysis (IPA), Upstream Regulator Analysis and Gene Set Enrichment Analysis (GSEA)33. Pathways were selected on a –log p-values of ≤1.3, which indicates a significant change of p ≥ 0.05 in that specific pathway comparing the LF-TB with LF diet or the HF with the LF diet. In the Upstream Regulator Analysis, Ingenuity software uses a curated database of interactions on the basis of the literature to link significant gene sets with upstream regulators. Significant linked gene sets were selected using a p-value of <0.05 for gene expression and a p-value of overlap of <0.05. A z-score above 1.5 indicates activation, whereas a z-score below -1.5 indicates inhibition of this upstream regulator. GSEA was performed on the unfiltered data set; gene sets were selected on a False Discovery Rate (FDR) q-value of <0.2 and were ranked on the Normalized Enrichment Score (NES). During microarray analysis we were especially interested in expression changes in apoA-I transcription and related pathways including the lipid, cholesterol and glucose metabolism, inflammation and the immune system.
Statistical analysis
All data are presented as mean ± SD unless otherwise indicated. Differences in changes of inflammatory markers, endothelial function markers and IFABP between the HF and LF meal were evaluated with general mixed models with subject as random factor, diet and time as fixed factors and a diet*time interaction. If this diet*time interaction was not significant, it was omitted from the model. If the factor time was significant, time points were compared to baseline concentrations, using Bonferroni’s corrections for multiple comparisons. Results were considered to be statistically significant if p ≤ 0.05. All statistical analyses were performed using SPSS 20.0 for Mac (SPSS Inc., Chicago, IL, USA).
Electronic supplementary material
Acknowledgements
We would like to thank M. Beckers and D. Luiten for their technical and dietetic assistance throughout the study.
Author Contributions
Designed the study: L.S., R.P.M., J.P. Conducted the study: L.S., R.P.M., R.J.J.R., J.P. Performed statistical analysis: L.S., R.P.M., M.V.B., J.P. Interpreted data: L.S., R.P.M., M.V.B., J.P. Wrote manuscript: L.S., R.P.M., J.P. Primary responsibility for final content: JP. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20068-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gadi R, Amanullah A, Figueredo VM. HDL-C: does it matter? An update on novel HDL-directed pharmaco-therapeutic strategies. Int J Cardiol. 2013;167:646–655. doi: 10.1016/j.ijcard.2012.05.052. [DOI] [PubMed] [Google Scholar]
- 2.Franceschini G, et al. Increased cholesterol efflux potential of sera from ApoA-IMilano carriers and transgenic mice. Arterioscler, throm, vasc biol. 1999;19:1257–1262. doi: 10.1161/01.ATV.19.5.1257. [DOI] [PubMed] [Google Scholar]
- 3.Kosmas CE, Christodoulidis G, Cheng JW, Vittorio TJ, Lerakis S. High-density lipoprotein functionality in coronary artery disease. Am J Med Sci. 2014;347:504–508. doi: 10.1097/MAJ.0000000000000231. [DOI] [PubMed] [Google Scholar]
- 4.Saleheen D, et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: a prospective case-control study. The lancet Diabetes Endocrinol. 2015;3:507–513. doi: 10.1016/S2213-8587(15)00126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikenaga M, Higaki Y, Saku K, Uehara Y. High-Density Lipoprotein Mimetics: a Therapeutic Tool for Atherosclerotic Diseases. J Atheroscler Thromb. 2016;23:385–394. doi: 10.5551/jat.33720. [DOI] [PubMed] [Google Scholar]
- 6.Neufingerl N, Zebregs YE, Schuring EA, Trautwein EA. Effect of cocoa and theobromine consumption on serum HDL-cholesterol concentrations: a randomized controlled trial. Am J Clin Nutr. 2013;97:1201–1209. doi: 10.3945/ajcn.112.047373. [DOI] [PubMed] [Google Scholar]
- 7.Oldoni F, Sinke RJ, Kuivenhoven JA. Mendelian disorders of high-density lipoprotein metabolism. Circulation research. 2014;114:124–142. doi: 10.1161/CIRCRESAHA.113.300634. [DOI] [PubMed] [Google Scholar]
- 8.Ellam S, Williamson G. Cocoa and Human Health. Annu Rev Nutr. 2013;33:105–128. doi: 10.1146/annurev-nutr-071811-150642. [DOI] [PubMed] [Google Scholar]
- 9.Rodopoulos N, Hojvall L, Norman A. Elimination of theobromine metabolites in healthy adults. Scan J Clin Lab Invest. 1996;56:373–383. doi: 10.3109/00365519609090590. [DOI] [PubMed] [Google Scholar]
- 10.Carlson TL, Kottke BA. ApoA-I secretion by rabbit intestinal mucosa cell cultures. Lipids. 1991;26:684–688. doi: 10.1007/BF02535614. [DOI] [PubMed] [Google Scholar]
- 11.Brouwer, I. Effect of trans-fatty acid intake on blood lipids and lipoproteins: a systematic review and meta-regression analysis. Geneva: World Health Organization. (2016).
- 12.Mensink, R. Effects of saturated fatty acids on serum lipids and lipoproteins: a systematic review and regression analysis. Geneva: World Health Organization. (2016).
- 13.Cameron-Smith D, et al. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. Am J Clin Nutr. 2003;77:313–318. doi: 10.1093/ajcn/77.2.313. [DOI] [PubMed] [Google Scholar]
- 14.Chokkalingam K, et al. High-fat/low-carbohydrate diet reduces insulin-stimulated carbohydrate oxidation but stimulates nonoxidative glucose disposal in humans: An important role for skeletal muscle pyruvate dehydrogenase kinase 4. J Clin Endocrinol Metab. 2007;92:284–292. doi: 10.1210/jc.2006-1592. [DOI] [PubMed] [Google Scholar]
- 15.Kakehi S, et al. Increased intramyocellular lipid/impaired insulin sensitivity is associated with altered lipid metabolic genes in muscle of high responders to a high-fat diet. Am J Physiol Endocrinol Metab. 2016;310:E32–40. doi: 10.1152/ajpendo.00220.2015. [DOI] [PubMed] [Google Scholar]
- 16.Esser D, van Dijk SJ, Oosterink E, Muller M, Afman LA. A high-fat SFA, MUFA, or n3 PUFA challenge affects the vascular response and initiates an activated state of cellular adherence in lean and obese middle-aged men. J Nutr. 2013;143:843–851. doi: 10.3945/jn.113.174540. [DOI] [PubMed] [Google Scholar]
- 17.Smolders L, Mensink RM, Plat J. An acute intake of theobromine does not change postprandial lipid metabolism, whereas a high-fat meal lowers chylomicron particle number. Nutr Res. 2017;40:85–94. doi: 10.1016/j.nutres.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Labonte ME, et al. Adding MUFA to a dietary portfolio of cholesterol-lowering foods reduces apoAI fractional catabolic rate in subjects with dyslipidaemia. Br J Nutr. 2013;110:426–436. doi: 10.1017/S000711451200534X. [DOI] [PubMed] [Google Scholar]
- 19.Smolders, L., Mensink, R. M., Boekschoten, M. V., de Ridder, R. J. J. & Plat, J. Theobromine does not affect postprandial lipid metabolism and duodenal gene expression, but has unfavorable effects on postprandial glucose and insulin responses in humans. Clinical Nutrition. epub ahead of print (2017). [DOI] [PubMed]
- 20.Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92:1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit NJ, et al. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC medical genomics. 2008;1:14–30. doi: 10.1186/1755-8794-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esser D, et al. Vascular and inflammatory high fat meal responses in young healthy men; a discriminative role of IL-8 observed in a randomized trial. PLoS One. 2013;8:e53474. doi: 10.1371/journal.pone.0053474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi MS, et al. High-fat diet decreases energy expenditure and expression of genes controlling lipid metabolism, mitochondrial function and skeletal system development in the adipose tissue, along with increased expression of extracellular matrix remodelling- and inflammation-related genes. Br J Nutr. 2015;113:867–877. doi: 10.1017/S0007114515000100. [DOI] [PubMed] [Google Scholar]
- 24.Fujimoto S, et al. Insulin resistance induced by a high-fat diet is associated with the induction of genes related to leukocyte activation in rat peripheral leukocytes. Life sciences. 2010;87:679–685. doi: 10.1016/j.lfs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Derikx JP, et al. A pilot study on the noninvasive evaluation of intestinal damage in celiac disease using I-FABP and L-FABP. J Clin Gastroenterol. 2009;43:727–733. doi: 10.1097/MCG.0b013e31819194b0. [DOI] [PubMed] [Google Scholar]
- 26.Nappo F, et al. Postprandial endothelial activation in healthy subjects and in type 2 diabetic patients: role of fat and carbohydrate meals. J Am Coll Cardiol. 2002;39:1145–1150. doi: 10.1016/S0735-1097(02)01741-2. [DOI] [PubMed] [Google Scholar]
- 27.Herieka M, Erridge C. High-fat meal induced postprandial inflammation. Mol Nutr Food Res. 2014;58:136–146. doi: 10.1002/mnfr.201300104. [DOI] [PubMed] [Google Scholar]
- 28.Adriaanse MP, et al. Serum I-FABP Detects Gluten Responsiveness in Adult Celiac Disease Patients on a Short-Term Gluten Challenge. Am J Gastroenterol. 2016;111:1014–1022. doi: 10.1038/ajg.2016.162. [DOI] [PubMed] [Google Scholar]
- 29.Lin K, et al. MADMAX - Management and analysis database for multiple ~omics experiments. J Integr Bioinform. 2011;8:160. doi: 10.1515/jib-2011-160. [DOI] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 31.Dai MH, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33:E175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartor MA, et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Academ Scienc USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.