ABSTRACT
Cancer is characterized by multiple genetic and epigenetic alterations, including a higher prevalence of mutations of oncogenes and/or tumor suppressors. Mounting evidences have shown that noncoding RNAs (ncRNAs) are involved in the epigenetic regulation of cancer genes and their associated pathways. The clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (CRISPR/Cas9) system, a revolutionary genome-editing technology, has shed light on ncRNA-based cancer therapy. Here, we briefly introduce the classifications and mechanisms of CRISPR/Cas9 system. Importantly, we mainly focused on the applications of CRISPR/Cas9 system as a molecular tool for ncRNA (microRNA, long noncoding RNA and circular RNA, etc.) editing in human cancers, and the novel techniques that are based on CRISPR/Cas9 system. Additionally, the off-target effects and the corresponding solutions as well as the challenges toward CRISPR/Cas9 were also evaluated and discussed. Long- and short-ncRNAs have been employed as targets in precision oncology, and CRISPR/Cas9-mediated ncRNA editing may provide an excellent way to cure cancer.
KEYWORDS: CRISPR-Cas9, genome editing, microRNA, long noncoding RNA, circular RNA, RNA editing, cancer
Introduction
Currently, cancer is one of the leading causes of death and is a refractory disease around the world which challenges the life and health of human beings.1 Although a multitude of exciting achievements have been made in the area of cancer therapy, including surgery, chemotherapy, radiation therapy, targeted biological therapy and new integrated combination therapy, the high possibility of relapse and chemo-/radiation- resistance and the detrimental side effects and toxicity are hurdles to ensure the quality of life.2 Therefore, further technical advances and novel therapeutic approaches for cancer therapy are urgently needed. Fortunately, the advent of the clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (CRISPR/Cas9) system has shed light on cancer therapy.
To date, CRISPR/Cas9 system is known as a molecular scissor and is widely used in various kinds of studies, including cancer research, drug discovery, treatment of mental disease, applications in plants, etc. CRISPR is a heritable and adaptive antiviral immune system of prokaryote, which primarily works against infectious invading viruses and phages and can be used to specifically cleave DNA in vitro.3-6 It is of importance and well established that cancer is a genetic disease characterized by mutations of multiple DNA and RNA in the cellular genome, and these characteristics of cancer confer cancer cells some distinct biological capabilities, including progressive growth, invasiveness, chemoresistance, and so forth.7,8 Correcting the mutations in the cancer cell genome is an appealing way to conquer cancer.9 A wealth of scientific evidence has shown that CRISPR/Cas9 system can correct mutations that cause cancer and have potential to be harnessed as a promising therapeutic technique to protect patients at the genetic level.10 Accumulating data have shown that CRISPR/Cas9-based system can target not only protein-coding genome but also the noncoding RNAs (ncRNAs) in human beings.10-12 In this review, we summarized the recent applications of CRISPR/Cas9 system in ncRNA-related genome editing, the off-target effects and CRISPR/Cas9-based novel techniques as well as the limitations and hurdles.
CRISPR system and mechanism
CRISPR was firstly discovered in Escherichia coli by Ishino and colleagues in late 1987.13 They found that the clustered repeat segments were interrupted by a series of spacer sequences and this phenomenon was later termed as CRISPR.14 Till now, researchers have discovered more than a dozen of different CRISPR/Cas systems that are categorized into three major groups (type I, II and III) and many subtypes according to their diverse mechanisms.15,16 CRISPR/Cas9 is one of the type II CRISPR systems in Streptococcus pyogenes (SpyCas9), and is the most globally used system in mammalian due to its high efficiency and accuracy.
The CRISPR/Cas9 system is also the first engineered CRISPR/Cas system for genome editing, partly because it contains an associated easily programmable single guide RNA (sgRNA) with a short recognition sequence which is only 20 nucleotides in length.17-19 The sgRNA consists of both CRISPR RNA (crRNA), which has a sequence complementary to the targeted sites, and trans-activating crRNA (tracrRNA), which is separately transcribed and partly complementary to the crRNA.19-21 Likewise, to exert the genomic editing function, the CRISPR/Cas9 system also needs a key enzyme component, Cas9 nuclease,3 which is closely related to these two RNA components. These RNAs are required to guide the Cas9 protein to the targeted sites and to activate the Cas9 nuclease. sgRNA combines with Cas9 protein to form a complex and strictly recognize the targeted complementary DNA sequence flanked at its 3′ end near the protospacer adjacent motif (PAM), which is commonly composed of NGG or NAG (N = A, T, G or C), and then initiate the DNA double strand breaks (DSBs) (Fig. 1).22
Figure 1.
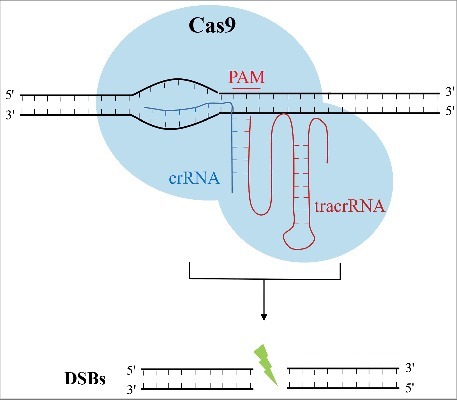
Schematic illustration of double strand breaks (DSBs) created by CRISPR-Cas9. Combined with crRNA and tracrRNA, Cas9 can specifically cut DNA double strands and cause DSBs.
dCas9, CRISPRa, CRISPRi, CRISPR-X and base-editing in cancer
dCas9, CRISPRa and CRISPRi
Apart from CRISPR knock in (CRISPR KI) and knock out (CRISPR KO), the CRISPR-Cas9 system has other diverse applications in cancer research area (Fig. 2). The catalytically inactive/dead Cas9 mutant (dCas9) is engineered to act as the transcriptional activator or suppressor of the target gene by blocking both the enzymatic activities of RuvC and HNH domains of the Cas9 nuclease, without altering the sequence.23-25 Many studies have indicated that these special reconstructive Cas9 can be fused to different domains to exert its stimulative or suppressive function of genetic transcription, and these strategies are now named as CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi), respectively.24-27
Figure 2.
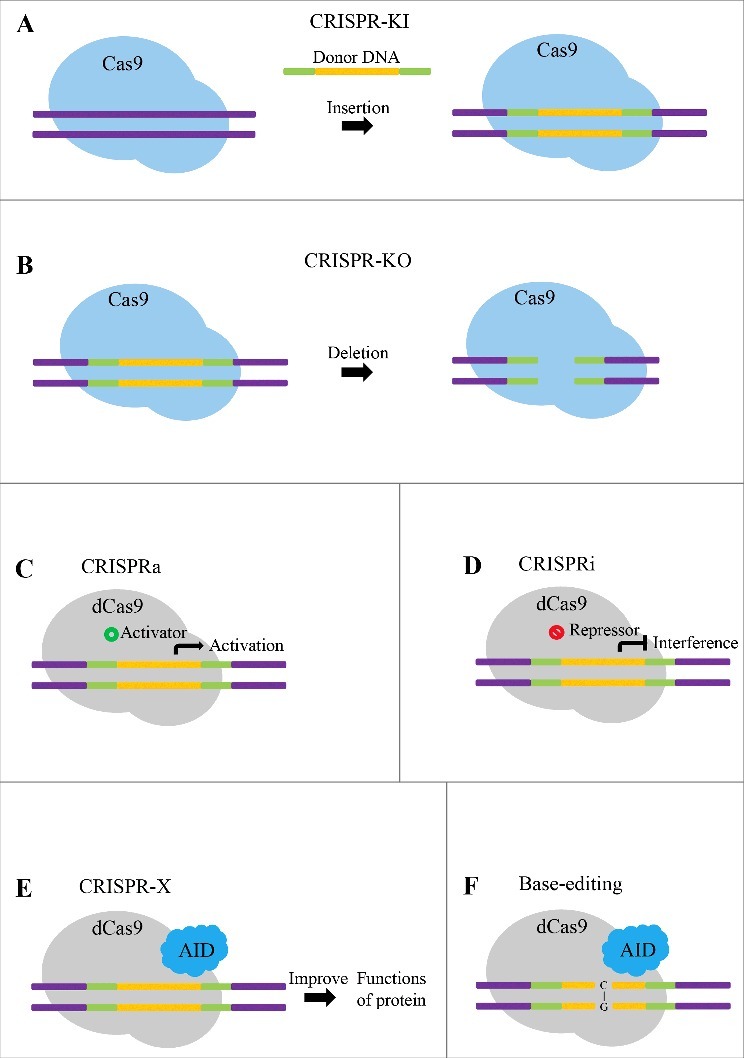
Techniques based on CRISPR-Cas9 system. (A) CRISPR-KI. Guided by the sgRNA, Cas9 can specifically knock the donor DNA into the targeted sequence. (B) CRISPR-KO. Guided by the sgRNA, Cas9 can specifically knock out the gene of interest. (C) CRISPRa. Guided by one or more sgRNAs, dCas9 can significantly activate the expression of endogenous genes. (D) CRISPRi. Co-expressed with the sgRNA, dCas9 can selectively and efficaciously repress transcription of targeted genes. (E) CRISPR-X. Combined with cytidine deaminase (AID), catalytically inactive dCas9 can be used to investigate and improve the functions of protein. (F) Base editing. Combined with diverse AIDs, dCas9 can mediate direct transition from C to T or from G to A without causing DSBs.
CRISPRa now is generally used to activate the expression of endogenous human genes. In one study, for example, Morgan L et al. demonstrated that a variant of dCas9, dCas9-VP64, can be directed by one or more guide RNAs (gRNAs) to up-regulate the expression of some specific genes in human cells, like, VEGFA and NTF3.28 Except from its use in human beings, CRISPRa is also used in other species. In another study, Cheng et al. used a nuclease-deficient dCas9, dCas9VP48, which can efficiently activate not only some exogenous reporter genes both in human and mouse cells, but also a number of endogenous genes, including IL1RN, SOX2, and OCT4.29
RNA interference (RNAi) is a universally used technique to suppress the expression of genes at the RNA level. Since dCas9 loses the endonuclease activity and acts as a DNA recognition protein-RNA complex when co-expressed with the sgRNA, this RNA-guided DNA recognition complex adds a new tool for researchers to control the expression of interested genes. Lei S and colleagues successfully demonstrated that CRISPRi can selectively and efficaciously repress transcription of target genes through silencing the transcription initiation and elongation. Interestingly, the off-target effects, which are commonly found when using CRISPR KI and CRISPR KO, are rarely detected.25 In another study, researchers used CRISPRi system to stably and effectively suppress the endogenous genes, like, CD71 and CXCR4, in HeLa cells.24
CRISPR-X
Recently, a powerful approach was based on dCas9 and was introduced to engineer and study the protein functions. Gaelen T et al. employed a system, named CRISPR-X, which includes catalytically inactive dCas9 and cytidine deaminase (AID) recruited by dCas9, to create specific point mutations in situ with limited off-target effects in the mammalian genome. Furthermore, the researchers mutated PSMB5 that was targeted by the therapeutic cancer drug bortezomib, and found CRISPR-X could identify the mutations that are closely related to bortezomib resistance.30 In short, this neoteric strategy can be used to investigate and improve the functions of proteins, and can be also applied to reveal the unknown mechanisms of drug-resistance.
Base-editing
The base-editing technology, which is based on the combination of diverse cytidine deaminases with dCas9, is another novel application of CRISPR/Cas9 system.31,32 Paired this engineered fusion with a gRNA, this complex can find cytosine within a targeted desire range of five nucleotides (nt) and then transform it into uracil, finally, facilitating a base substitution of C-to-T or G-to-A without introducing DSBs.31 This technology was firstly used by Komor and colleagues to mediate direct transition from cytidine to uridine thus generating single-base-pair substitutions in human and murine cell lines.31 With the help of electroporation and microinjection, this technology was used by Kim et al. to edit mouse embryos on a single base pair level efficiently and precisely.33 Aside from using in mammalian cells and embryos, CRISPR/Cas9 derived base-editing is likewise used in yeast and plants,34-36 indicating the huge potential of this novel technology in cancer research and other areas in the coming future.
In summary, CRISPR KI and CRISPR KO are globally and effectively used for creating mutations both in vivo and in vitro while CRISPRa and CRISPRi are employed to modulate transcriptional expression without causing any mutations. As a neoteric technology, CRISPR-X paves a path to investigate protein-protein interaction and provides a new skill for scientists to uncover the mystery of resistance to drugs. Another brand-new technology, base-editing, can result in specific point mutation without causing the cleavage of targeted sequence, which is different from CRISPR KI and CRISPR KO. These technologies may open various bright therapeutic avenues for the cure of cancer and other genetic diseases as long as we efficiently handle them and overcome limitations brought by them.
CRISPR/Cas9 as a tool for ncRNA editing
Prior to the emergence of CRISPR/Case9, RNAi was widely used to suppress the expression of protein coding genes.37 However, RNAi has been proven to be an inefficient approach when using for the interference of ncRNAs.38 ncRNAs, primarily including microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circularRNAs (circRNAs), have been proven to play a critical role in cancers (reviewed in39,40). As the majority of (about 99%) human genome, ncRNA-related region lacks protein-coding capability,41,42 and most of the genetic alterations occur in this vast region, targeting non-coding area with CRISPR/Cas9 system, therefore, maybe a feasible approach toward cancer therapy. Here, we mainly discuss the applications of CRISPR/Cas9 system in the areas of miRNAs and lncRNAs.
CRISPR/Cas9 for miRNA editing in cancer
So far, many studies have indicated that CRISPR/Cas9 system is an excellent option for ncRNA-related genome editing/regulating (Summarized in Table 1). As one of the primary ncRNAs discussed in this review, miRNAs have been widely investigated in CRISPR/Cas9-related area. Cloning CRISPR/Cas9 constructs with sgRNAs which specifically target the biogenesis processing sites of interested miRNAs, Chang et al. demonstrated that alterations generated by CRISPR/Cas9 on the structure of primary miRNAs can lead to the downregulation of these mature miRNAs both in vivo and in vitro.43 Moreover, this study also demonstrated that properly designed the sgRNAs, CRISPR/Cas9 can obviously minimize the off-target effects for the miRNAs with highly conserved sequences across the same family. In another study, Zhou and colleagues used CRISPR/Cas9 system to successfully knock out miR-3188 in hepatocellular carcinoma (HCC) cell lines, and found that miR-3188 KO effectively suppressed cell growth, invasion and migration, and inhibited xenografts tumor growth in nude mice.44 Recently, Yue's group reported that lentiviral CRISPR/Cas9 vectors were highly efficient in introducing indels (insertions and deletions) into the precursor miRNA sequences. Using this method, they successfully disrupted the expression of miR-21 and found that disruption of pre-miR-21 sequences results in decreased cell proliferation, migration and invasion in ovarian cancer cells.45
Table 1.
Applications of CRISPR-Cas9 in ncRNA study.
| ncRNA | Object | Effect | Efficiency | Reference |
|---|---|---|---|---|
| miR-21 | HEK-293 cells | Knockout | >1000-fold | 54 |
| miR-29a | HEK-293 cells | Knockout | High | 54 |
| miR-17 | In vitro and in vivo | Knockdown | 96% | 43 |
| miR-200c | In vitro and in vivo | Knockdown | 96% | 43 |
| miR-141 | In vitro and in vivo | Knockdown | 96% | 43 |
| miR-3188 | HCC cells | Knockout | High | 43 |
| miR-21 | SKOV3 and OVCAR3 cells | Disruption | High | 45 |
| miR-137 | A2780 cells | Disruption | High | 46 |
| miR-309 | Mosquito embryos | Disruption | 51.5% | 48 |
| miR-93 | HeLa cells | Disruption | 16% | 47 |
| miR-126a | Zebrafish embryos | Knockdown | High | 49 |
| miR-126b | Zebrafish embryos | Knockdown | High | 49 |
| Several miRNAs | Zebrafish/MV4-11 cells | Disruption | 90% | 50,52 |
| Rian | In vivo | Knockout | 33.3% | 93 |
| UCA1 | HCT-116 cells | Knockout | High | 54 |
| lncRNA-21A | HCT-116 cells | Knockout | High | 54 |
| AK023948 | MCF-7 cells | Knockout | High | 54 |
Except for editing/regulating these miRNAs discussed above, CRISPR/Cas9 has also shown vast applications toward other miRNAs, like, miR-137, miR-93, miR-309, miR-126a/b etc., in various cancer cells or organisms.46-52
CRISPR/Cas9 for lncRNA editing in cancer
In addition to miRNAs, lncRNAs have been reported to be successfully edited/regulated by CRISPR/Cas9 system. UCA1 (urothelial carcinoma-associated 1), an upregulated lncRNA in bladder cancer, could be targeted by specifically designed gRNAs of CRISPR/Cas9, and efficaciously inhibited both in vivo and in vitro,53 suggesting that CRISPR/Cas9 system can be used to modulate the expression of lncRNAs and further employed as a therapeutic approach towards clinical cancer therapy.
One of the limitations to apply CRISPR/Cas9 system to noncoding genes is that tiny indels may not necessarily generate functional loss of a certain noncoding gene. To overcome this obstacle, Ho and colleagues adopted a specific selection system, HR (homologous recombination), to integrate marker genes into the genome, and with the help of CRISPR-Cas9 system, UCA1 and lncRNA-21A as well as AK023948 were successfully knockout in HCT-116 and MCF-7 cells, respectively.54 Additionally, Shechner and colleague developed CRISPR-Display (CRISP-Disp), a targeted localization method that uses Cas9 to deploy large RNA cargos to DNA loci. They found that functional RNA domains up to at least 4.8 kb long could be inserted in CRISPR gRNA at multiple points, allowing the construction of Cas9 complexes with natural lncRNAs.55 This method may open an avenue for lncRNA study in the field of cancer research.
CRISPR/Cas9 for lncRNA editing in other diseases
Except for the application in lncRNA-related cancer research, CRISPR/Cas9 is also used for studying other diseases. LncRNA PEAT (Pax1 enhancer antisense transcript) is located near the upstream of the Pax1 gene and lacks an ORF and consensus TSS (transcription start site). In order to delete its transcribed unit completely via CRISPR/Cas9, researchers employed the existing annotation (mm10) and mapped RNA-seq reads to forecast the transcribed region in a mouse line.56 They also found that the deletion of PEAT mildly increased the expression of bone morphogenetic protein target gene as well as a number of ribosomal protein mRNAs. Another group showed that combining CRISPR/Cas9 with siRNAs or GapmeRs to delete or silence MANTIS (lncRNA n342419), a downregulated lncRNA in patients with idiopathic pulmonary arterial hypertension (IPAH), can efficiently inhibit the angiogenic sprouting of endothelial cells.57
CRISPR/Cas9-based screening
To facilitate the applications of CRISPR/Cas9 in both miRNA and lncRNA-related studies, several CRISPR/Cas9 based screening technologies have been invented.
With the help of CRISPR/Cas9 technology, Wallace et al. performed an unbiased global LOF screening to examine miRNAs which are involved in MV4-11 cell lines. Using this approach, they found that a subset of (27/197) evolutionarily conserved miRNAs can promote or inhibit the cellular proliferation and survival.52 This approach has the potential to identify functional non-coding components in mammalian genomes and provides a resource to guide coming work. In another earlier study,58 researchers focused their concentration on a functional screening to identify the cis-regions that control the processing of a certain miRNA, miR-142. Using a CRISPR/Cas9 based molecular chipper technology, they carried out a functional screen to identify cis-regulatory elements that control the biogenesis of miR-142 and verified it as an economical and easily customizable way, compared with microarray-based oligonucleotide synthesis, for sgRNA library construction.
Despite its limitations, this Molecular Chipper technology can be used not only for identifying functional non-coding regions in mammalian genomes but also for performing gene-expression-based screening of protein-coding gene regulation in the future.
So far, to our knowledge, more screenings for lncRNAs have been developed compared to miRNAs. In a recent study, Mo's group used a CRISPR/Cas9-based synergistic activation mediator (SAM) system to identify the underlying lncRNAs that have the ability to regulate AKT activity, and determined lncRNA AK023948 as a positive regulator for AKT in breast cancer cells,59 indicating that CRISPR/Cas9-based SAM library screening is a promising strategy for lncRNA investigation. Using a lentivirally delivered paired-guide RNA (pgRNA) CRISPR/Cas9 library, Zhu et al. invented a high-throughput genome-wide screening for human lncRNAs. With this method, they found more than fifty lncRNAs that can either positively or negatively regulate human cancer cell growth.60
CRISPRi, an important CRISPR/Cas9 based technology, is also developed to screen lncRNAs. Since CRISPRi exerts its function only within a small range (1 kb) around the targeted desire TSS,26 and dCas9 blocks only 23 bp of the targeted sequence,61 CRISPRi can generate precise interference of any lncRNA gene. Enlightened by this idea, Liu et al. developed a CRISPRi based platform for large-scale systematic screening.62 Targeting 16,401 lncRNA loci with CRISPRi libraries in diverse cell lines, including transformed cell lines and iPSCs (human induced pluripotent stem cells), they identified 499 lncRNAs which are needed for robust cell growth.62 This study indicated that the CRISPRi based platform may help to delineate the biological characteristics of lncRNA genome, and add a skill for large-scale screening of functional lncRNAs.
Besides, there are many investigations about non-coding genome which may not be transcribed into ncRNAs to expand the application of CRISPR/Cas9 in non-coding area. Zhang's group,63 for example, developed a high-throughput strategy using pooled CRISPR/Cas9 sgRNA system targeting 715 kb of sequence which surrounds three different genes NF1, NF2, and CUL3. Using this unbiased mutagenesis method, they showed how a Cas9-mediated systematic separation of non-coding sites can identify functional elements that are involved in cancer drug resistance and gene regulation.
Studies about ncRNA-related pathways also attract scientists' interest. Lately, Golden et al. used CRISPR/Cas9 genome-wide loss-of-function (LOF) screening combined with a fluorescent reporter of miRNA activity to identify new regulators that are involved in miRNA pathways in human cells.64 This strategy expands the use of CRISPR/Cas9 in the area of non-coding genome and provides a novel insight into the investigation of ncRNAs-related genome.
The emergence of CRISPR/Cas9 is opening a new way for scientists to edit genome. With the development CRISPR/Cas9 technology, ncRNA-based gene editing will become more efficient and the significant roles of ncRNAs in diseases will be fully investigated in the near future.
Off-target effect and solutions
Off-target effect
While the CRISPR/Cas9 system has many advantages over other genome editing technologies, like, ZFNs and TALENs, there are still some knotty problems that need to be solved, such as off-target effect. Sometimes, Cas9 can improperly bind to targets and generate mutations within the sequence which is outside the targeted sites.10
As off-target sites are determined by the nuclease and the sequence of sgRNA, therefore, some algorithms and methods including Genome Engineering, CCTop, Cas-OFFinder, E-CRISP, CRISPOR and CHOPCHOP v2 have been exploited to predict and assess the off-target sites and the efficiency of sgRNA22,23,65-78 (for detailed information, see Table 2). Off-target effect can generate some unnecessary mutations outside the desired target site and may cause some serious consequences.70-73 However, Schaefer et al. found that two CRISPR-treated mice (F03 and F05) shared the identical 117 indels and 1397 SNVs (single-nucleotide variants) by using whole-genome sequencing (WGS). The unexpectedly high number of mutations were induced after CRISPR/Cas9 editing in vivo.77 This work manifested that off-target effect is an extensively existed phenomenon in contrast to former studies. However, there are controversies about Schaefer et al's standpoint. Lareau and colleagues argued that Schaefer et al's experimental design lacked control, and the alleged “unexpected mutations” were merely due to the mice used in their experiment, which shared the common SNPs and indels prior to the Cas9-induced mutations. Therefore, data presented in Schaefer et al's paper were insufficient to support their conclusions.79 In addition to the viewpoint from Lareau et al., there are some other researchers questioning Schaefer et al's point of view. Kim et al.80 and Wilson et al.,81 for instance, both think that the observed variants in Schaefer et al's experiments are much more proper to be explained by the differences in the genetic background of the mice rather than mutations caused by Cas9. Thus, this issue is currently under drastic controversy and more efforts are needed to clarify it.
Table 2.
Common tools for off-target prediction and assessment.
| Tool name | Application | Classification | Reference |
|---|---|---|---|
| Genome Engineering | Prediction | Web-based | 22 |
| CCTop | Prediction | Web-based | 65 |
| Cas-OFFinder | Prediction | Web-based | 66 |
| E-CRISP | Prediction | Web-based | 67 |
| CRISPOR | Prediction | Web-based | 68 |
| CHOPCHOP v2 | Prediction | Web-based | 69 |
| CROP-IT | Prediction | Web-based | 74 |
| GUIDE-Seq | Detection | Web-based | 78 |
| WGS | Assessment/Detection | Assay | 70,73,77 |
| SURVEYOR | Assessment/Detection | Assay | 71,72 |
| ChIP-Seq | Assessment/Detection | Assay | 23,75,76 |
| Deep-targeted sequencing | Assessment/Detection | in silico | 73 |
CCTop, CRISPR/Cas9 target online predictor; CROP-IT, CRISPR/Cas9 off-target prediction and identification tool; GUIDE-seq, Genome-wide unbiased identification of DSBs enabled by sequencing; WGS, Whole-genome sequencing; ChIP-seq, chromatin immunoprecipitation sequencing.
Solutions toward off-target
To further reduce and finally, if possible, eliminate off-target effect, several approaches have been implemented. First, in one study, Ran et al. described an approach, which used the mutant variant of Cas9 with paired guide RNAs to cause DSBs at the desired target site, leading to dramatically minimized off-target activity.82 Additionally, this strategy has also been generally applied by many other laboratories.83-86 Second, Slaymaker and colleagues showed that enhanced specificity SpCas9 (eSpCas9) variants can essentially reduce off-target effects and retain powerful on-target cleavage.87 Another high-fidelity variant, named SpCas9-HF1, was designed to retain on-target activities and reduce non-specific DNA contacts. Due to its exceptional precision, SpCas9-HF1 can avoid most of the off-target mutations that are generally induced by wild-type SpCas9.88 Third, truncating sgRNA by 2–3nt was demonstrated to minimize undesired mutagenesis at off-target sites.86 Fourth, the delivery system is also an important factor which can affect the off-target phenomenon. Ramakrishna et al. showed that conjugating the Cas9 protein to cell-penetrating peptide (CPP), and then combing the gRNA to form nanoparticles as a delivery system resulted in efficient genome editing with distinctly reduced off-target effect compared to plasmid transfections.89 Finally, combining Cas9 with other nuclease may be a good choice. A study by Tsai et al. identified that fusing Cas9 together with RNA-guided FokI nucleases (RFNs) immensely improved the on-target specificity compared to the monomeric CRISPR-Cas.90
Briefly, in addition to taking off-target effect into consideration when we use this powerful tool, we should keep in mind that CRISPR/Cas9 system is not originally present in mammalian genome, and thus it may cause immune response and weaken treatment effectiveness when we push this system into clinical application.
Challenges and perspectives
Given the easy operability and economical nature of CRISPR/Cas9 system, scientists gain a better choice in their tool kit to easily and economically edit the genome of interest. Although techniques based on CRISPR/Cas9 system have many advantages over ZFNs, TALENs and RNAi in gene editing and in altering gene expression at the transcriptional level, there are still several limitations for scientists to overcome. Here, we mainly discussed the limitations in ncRNA area, off-target effect, immune response and the ethical issues.
Although the CRISPR/Cas9 system has been widely used to edit protein-coding genes or noncoding genes in human and other creatures, many limitations exist, especially when it is applied in non-coding RNA area. For one thing, due to the lack of distinct open reading frame (ORF), small indels generated by CRISPR/Cas9 system may fail to cause functional knockout of a given non-coding gene. For another, since many lncRNAs are derived from sense/antisense genes or bidirectional promoters, there is a possibility to affect the overlapping or adjacent genes in loci that harbor multiple genes when CRISPR/Cas9 system is applied in lncRNA editing. This phenomenon occurred to the lncRNAs NOP14-AS1, LOC389641, MNX1-AS1 etc when they were edited by CRISPR/Cas9 system.91 And this problem with lncRNA editing by CRISPR/Cas9 system needs to be solved prior to the use of CRISPR/Cas9 systems for editing lncRNAs on a genome wide scale.
The off-target effect, which could result in undesired genetic alterations that may lead to cancer or other knotty problems, is the main bottleneck of CRISPR/Cas9 technology. As discussed above, although many efforts have been invested the improvement of predicting and detecting the off-target effect, it is far from enough. To more precisely and effectively edit the targeted sequences, researchers should exploit more techniques and algorithms besides the approaches aforementioned.
Previous studies70-73 hold the view that off-target effect is an uncommon phenomenon, however, Schaefer and colleagues raised doubts about this conclusion. They believe that the off-target effect extensively exists.77 Although their viewpoint challenges the former conclusion, this is not a crisis for the use of CRISPR/Cas9, it is, on the contrary, a “pushing hand” for researchers to carefully detect the off-target effect when they use this novel technology. And their work will undoubtedly drive this technology toward a better orientation.
Another obstacle is the unfavorable immune response, which is brought by bacterial Cas9. Generally, bacterial Cas9 is delivered by viral vectors, especially by adenoviral vectors, which are well-known to cause an immune response. In a previous study,92 for example, researchers delivered Streptococcus pyogenes–derived Cas9 (SpCas9) into mice by the adenoviral vector to edit Pten. They found remarkable increase of antibodies, including IgG1, IgG2a/b, as well as IL-2, in the mice which had received SpCas9. These results raised some questions: How can we reduce the immune response? What can we do to manage immune response in the patients when using CRISPR/Cas9 in the clinic in the future? Fortunately, with the huge number and variety of bacteria, we have enough time to find and engineer both new and less immunogenic variants of Cas9 before putting CRISPR/Cas9 technology into the clinical area.
Ethical issues are problems that researchers cannot bypass before using CRISPR/Cas9-related technique in real applications. Thus far, most of countries across the world permit researchers editing genome for the sake of improving the grain yield in agriculture and for other biological usage. However, manipulating human eggs, sperms and embryos is still very controversial. There are two issues that may cause the ethicists' concern. First, CRISPR/Cas9 technology is an immature and less developed gene-editing technology. Scientists know little about the long-term impact of genetic alteration on the future generations. Once mistakes or off-target effect takes place, it will result in serious unexpected consequence. In addition, CRISPR/Cas9 technology is enabled to edit any genome they want. This will bring about huge social conflicts if CRISPR/Cas9 technology is applied for anti-evolution and anti-human use. Although there is divergence, we optimistically believe that scientists will reach a consensus with ethicists and laws towards this issue.
Albeit there are still various limitations and obstacles toward CRISPR/Cas9 system, we hopefully hope that this budding and gradually mature technology could contribute to drug discovery, cancer therapy as well as the cure of other previously supposed incurable genetic diseases in the coming future.
Funding Statement
This work was supported by research grants from the Natural Science Foundation of Zhejiang (LY15C060003, LQ18H200001), the Non-profit Technology Research Program of Zhejiang (LGF18H160006), the Natural Science Foundation of Ningbo (2017A610247), the Scientific Innovation Team Project of Ningbo (2017C110019), the National Undergraduate Training Program for Innovation and Entrepreneurship (201711646018) and the K.C.Wong Magna Fund at Ningbo University.
Disclosure of potential conflicts of interest
No potential conflicts of interest are disclosed.
Acknowledgments
The authors thank their respective laboratory members and collaborators for critical review of this article. The authors apologize that space constraints prevent them from citing all relevant publications.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al.. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459-66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709-12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579-86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234-44. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget. 2016;7:12305-17. doi: 10.18632/oncotarget.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16:89-100. doi: 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuo C, Hou W, Hu L, Lin C, Chen C, Lin X. Genomic Editing of Non-Coding RNA Genes with CRISPR/Cas9 Ushers in a Potential Novel Approach to Study and Treat Schizophrenia. Front Mol Neurosci. 2017;10:28. doi: 10.3389/fnmol.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canver MC, Bauer DE, Orkin SH. Functional interrogation of non-coding DNA through CRISPR genome editing. Methods. 2017;121–22:118-29. doi: 10.1016/j.ymeth.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429-33. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565-75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 15.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al.. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467-77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602-7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960-4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827-32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al.. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670-6. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al.. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442-51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al.. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647-61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al.. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583-8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977-9. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163-71. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036-42. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420-4. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305):aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 33.Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35:435-37. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 34.Satomura A, Nishioka R, Mori H, Sato K, Kuroda K, Ueda M. Precise genome-wide base editing by the CRISPR Nickase system in yeast. Sci Rep. 2017;7:2095. doi: 10.1038/s41598-017-02013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al.. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35:441-43. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 36.Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438-40. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 37.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25-33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 38.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7-21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 39.Tian H, Zhou C, Yang J, Li J, Gong Z. Long and short noncoding RNAs in lung cancer precision medicine: Opportunities and challenges. Tumour Biol. 2017;39:1010428317697578. doi: 10.1177/1010428317697578. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472-80. [PMC free article] [PubMed] [Google Scholar]
- 41.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al.. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al.. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131-8. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep. 2016;6:22312. doi: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou SJ, Deng YL, Liang HF, Jaoude JC, Liu FY. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24(9):1577-1587. doi: 10.1038/cdd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huo W, Zhao G, Yin J, Ouyang X, Wang Y, Yang C, Wang B, Dong P, Wang Z, Watari H, et al.. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer. 2017;8:57-64. doi: 10.7150/jca.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Chen W, Zeng W, Wan C, Duan S, Jiang S. microRNA-137 promotes apoptosis in ovarian cancer cells via the regulation of XIAP. Br J Cancer. 2017;116:66-76. doi: 10.1038/bjc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Q, Meng X, Meng L, Chang N, Xiong J, Cao H, Liang Z. Small indels induced by CRISPR/Cas9 in the 5′ region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol. 2014;11:1243-9. doi: 10.1080/15476286.2014.996067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Zhao B, Roy S, Saha TT, Kokoza VA, Li M, Raikhel AS. microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2016;113:E4828-36. doi: 10.1073/pnas.1609792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Zhu RF, Li FF, Liang YL, Wang C, Qin YW, Huang S, Zhao XX, Jing Q. MicroRNA-126a Directs Lymphangiogenesis Through Interacting With Chemokine and Flt4 Signaling in Zebrafish. Arterioscler Thromb Vasc Biol. 2016;36:2381-93. doi: 10.1161/ATVBAHA.116.308120. [DOI] [PubMed] [Google Scholar]
- 50.Narayanan A, Hill-Teran G, Moro A, Ristori E, Kasper DM, C AR, Lu J, Nicoli S. In vivo mutagenesis of miRNA gene families using a scalable multiplexed CRISPR/Cas9 nuclease system. Sci Rep. 2016;6:32386. doi: 10.1038/srep32386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Dai Z, Liang Y, Yin M, Ma K, He M, Ouyang H, Teng CB. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep. 2014;4:3943. doi: 10.1038/srep03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace J, Hu R, Mosbruger TL, Dahlem TJ, Stephens WZ, Rao DS, Round JL, O'Connell RM. Genome-Wide CRISPR-Cas9 Screen Identifies MicroRNAs That Regulate Myeloid Leukemia Cell Growth. PLoS One. 2016;11:e0153689. doi: 10.1371/journal.pone.0153689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhen S, Hua L, Liu YH, Sun XM, Jiang MM, Chen W, Zhao L, Li X. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget. 2017;8:9634-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho TT, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, Mo YY. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015;43:e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664-70. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stafford DA, Dichmann DS, Chang JK, Harland RM. Deletion of the sclerotome-enriched lncRNA PEAT augments ribosomal protein expression. Proc Natl Acad Sci U S A. 2017;114:101-06. doi: 10.1073/pnas.1612069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Günther S, et al.. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017;136:65-79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng J, Roden CA, Pan W, Zhu S, Baccei A, Pan X, Jiang T, Kluger Y, Weissman SM, Guo S, et al.. A Molecular Chipper technology for CRISPR sgRNA library generation and functional mapping of noncoding regions. Nat Commun. 2016;7:11178. doi: 10.1038/ncomms11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun. 2017;8:14422. doi: 10.1038/ncomms14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al.. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279-86. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113-26. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al.. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355(6320):aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545-49. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang TC, Kopp F, et al.. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542:197-202. doi: 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473-5. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122-3. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 68.Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al.. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272-6. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27-30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, et al.. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111:11461-6. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. CRISPR/Cas9 System as an Agent for Eliminating Polyomavirus JC Infection. PLoS One. 2015;10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L, Grishin D, Wang G, Aach J, Zhang CZ, Chari R, Homsy J, Cai X, Zhao Y, Fan JB, et al.. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43:e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677-83. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 76.Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, Zhang Y. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014;24:1009-12. doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 2017;14:547-48. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al.. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187-97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lareau C, Clement K, Hsu JY, Pattanayak V, Joung JK, Aryee MJ, Pinello L. “Unexpected mutations after CRISPR-Cas9 editing in vivo” are most likely pre-existing sequence variants and not nuclease-induced mutations. 2017; https://doi.org/ 10.1101/159707. [DOI]
- 80.Kim ST, Park J, Kim D, Kim K, Bae S, Schlesner M, Kim JS. Questioning unexpected CRISPR off-target mutations in vivo. 2017; https://doi.org/ 10.1101/157925. [DOI]
- 81.Wilson CJ, Fennell T, Bothmer A, Maeder ML, Reyon D, Cotta-Ramusino C, Fernandez CA, Marco E, Barrera LA, Jayaram H, et al.. The experimental design and data interpretation in ‘Unexpected mutations after CRISPR Cas9 editing in vivo’ by Schaefer et al. are insufficient to support the conclusions drawn by the authors. 2017; https://doi.org/ 10.1101/153338. [DOI]
- 82.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380-9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132-41. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281-308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, et al.. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399-402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 86.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279-84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84-8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490-5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020-7. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569-76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, et al.. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum Gene Ther. 2015;26:432-42. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, Du Y, Tate PH, Huang X, Zhang W. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014;11(7):829-35. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]


