ABSTRACT
In most organisms, gene expression over the course of the day is under the control of the circadian clock. The canonical clock operates as a gene expression circuit that is controlled at the level of transcription, and transcriptional control is also a major clock output. However, rhythmic transcription cannot explain all the observed rhythms in protein accumulation. Although it is clear that rhythmic gene expression also involves RNA processing and protein turnover, until two years ago little was known in any eukaryote about diel dynamics of mRNA translation into protein. A recent series of studies in animals and plants demonstrated that diel cycles of translation efficiency are widespread across the tree of life and its transcriptomes. There are surprising parallels between the patterns of diel translation in mammals and plants. For example, ribosomal proteins and mitochondrial proteins are under translational control in mouse liver, human tissue culture, and Arabidopsis seedlings. In contrast, the way in which the circadian clock, light-dark changes, and other environmental factors such as nutritional signals interact to drive the cycles of translation may differ between organisms. Further investigation is needed to identify the signaling pathways, biochemical mechanisms, RNA sequence features, and the physiological implications of diel translation.
KEYWORDS: Protein synthesis; ribosome; RNA, circadian clock; diurnal cycle
Introduction
Translation, the synthesis of proteins from mRNA by cytoplasmic ribosomes, is known to be sensitive to environmental conditions, such as nutrient status and stresses in animals1,2 or light and darkness in plants.3,4 Because the cellular environment changes over the course of the diel day-night cycle, one would expect that translational efficiency (see glossary of terms) of individual mRNAs will fluctuate over the course of the day. But to what degree is this the case? And are cycles of translation driven by, enhanced by, or dampened by the circadian clock? Until recently, in most organisms very little was known about the diel control of translation.5-8
Most eukaryotes and several prokaryotic organisms possess a timekeeper, termed the circadian clock, which allows the organism to measure the time of day, anticipate daily changes and even sense the seasons of the year. Circadian clocks function as endogenous, quasi-autonomous cellular oscillators with a free-running period of approximately 24 hours. Typically, the clock is reset once a day by the lights-on signal at dawn, which is also known as the zeitgeber stimulus. However, after entrainment, the clock is able to run continuously for many days without any external input. In most organisms that have been studied, the heart of the circadian clock consists of a cell-autonomous central oscillator that is constructed from a circuit of gene expression events that are regulated at the level of transcription9,10 although clocks that function at the translational level11 and at the protein level12 have also been described. The clock orchestrates numerous cellular and higher-order functions, in part by regulating the transcription of other genes, an activity referred to as the clock output pathway. The transcriptional clocks of mammals and plants will be described briefly below.
Deciphering the biochemical and molecular basis of circadian clocks has been an intense area of research. Clocks are known to be exquisitely calibrated circuits of transcription, protein accumulation, and degradation. Phase delays in the accumulation of mRNA and protein are common. But to what degree does translational regulation play a role in clock function and clock output? And is the translational regulation that may occur cyclical in nature or not?
Early experiments in the sea slug Aplysia and the marine photosynthetic dinoflagellate, Gonyaulax polyedra (renamed Lingulodinium polyedrum) established that translation of new proteins must be critical for clock function, given that a pulse of a translation inhibitor shifted the phase of the clock in these organisms.13,14 In addition, research rooted in the 1960s established that the clock of the giant unicellular algae Acetabularia resides in the cytoplasm, rather than the nucleus, and involves cyclical protein synthesis.15 However, in the years following, it became evident that the genetically tractable clocks of multicellular eukaryotes are constructed around transcriptional feedback loops.10,16 Although routinely referred to as a ‘transcriptional-translational feedback loop’, the clock's translational arm was simply thought to be required to produce the transcription factors for a functional oscillator. In most organisms, the role of translation in clock function, and even in clock output, was neglected or dismissed, in part because few central clock proteins were revealed to be RNA binding proteins. Yet, work in Lingulodinium clearly indicated that central oscillators and clock outputs could operate at the level of translation.17,18 In addition, more recent studies have unveiled numerous linkages between RNA biology and circadian clocks.19-22 Specifically, cycles of gene expression at the level of mRNA are converted into dynamic patterns of protein levels through a series of biochemical steps, of which translation is only one, besides splicing,23,24 polyadenylation,25 RNA methylation,26 and cellular sequestration. At the protein level, new protein synthesis by translation is in a dynamic equilibrium with protein turnover.27 In short, a thorough investigation of the nexus between diurnal rhythms and mRNA translation was long overdue.
Each clock consists of interlocked feedback loops at the transcriptional level. In mammals, the master circadian pacemaker resides in the suprachiasmatic nucleus (SCN) of the brain.10 The clock in the SCN synchronizes peripheral oscillators that are located in many if not all organs. The central clock is reset at dawn by light signals perceived through the eye and communicated to the SCN through the optic nerve. At a cellular level, the clock consists of two transcriptional feedback loops. In the first loop, BMAL1 (Brain-Muscle-Arnt-like 1, also known as ARNTL) and CLOCK (Circadian Locomotor Output Cycles Kaput) are transcription factors that activate the transcription of Per (Period) and Cry (Cryptochrome). PER and CRY proteins dimerize, and interact with and inhibit the CLOCK/BMAL1 complex, forming a negative feedback loop. The ensuing drop in Per and Cry gene transcription together with degradation of PER and CRY proteins resets the cycle. A second loop is activated by retinoid-related orphan receptors (RORa, b, c) and repressed by REV-ERBα/REV-ERBβ. This loop induces a delay in Cry1 expression important for timing the clock10 (Fig. 1A).
Figure 1.
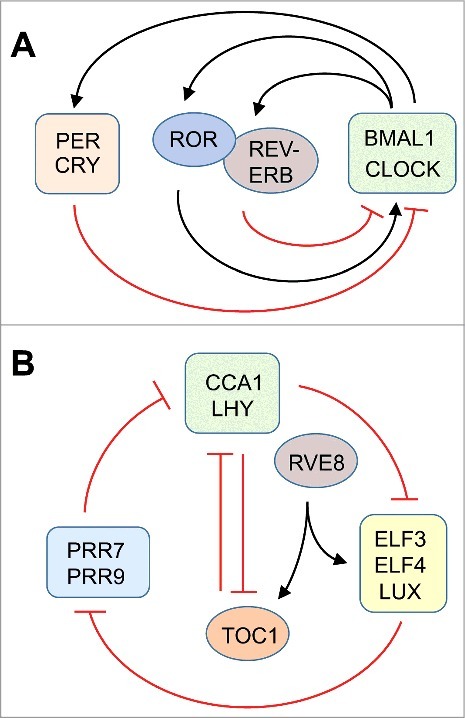
A schematic representation of the core elements of the circadian clock in the mouse (A) and Arabidopsis (B). Arrows and T-bars indicate positive and negative influences, respectively. The mouse model is adapted from87 and the plant model from.16,34
Nutritional signals such as glucose and fat interfere with clock function in mammals. For example, a high-fat diet impairs BMAL1 recruitment to its targets on chromatin.28 These findings suggest that the clock is influenced by nutritional inputs. In addition, the insulin-AKT-mTOR pathway, which responds to nutritional signals, rhythmically phosphorylates BMAL1 in the nucleus.29,30 Vice versa, there is also evidence that the clock affects mTOR signaling.31,32
While the mammalian clock is a positive/negative feedback loop, the plant circadian clock contains a central toggle switch as well as a three-way repressilator (Fig. 1B).16,33,34 The ‘dusk’ transcription factor TOC1 (TIMING OF CAB EXPRESSION1) represses the ‘dawn’ transcription factors, CCA1 (CIRCADIAN CLOCK ASSOCIATED1) and LHY (LATE ELONGATED HYPOCOTYL) and vice versa. Around dawn, CCA1 and LHY repress the evening complex, consisting of the transcription factors LUX, ELF3, and ELF4 (LUX ARRHYTHMO, EARLY FLOWERING3 and 4). The evening complex represses the day-time transcription factors, PRR9 (PSEUDORESPONSE REGULATOR9) and PRR7, which in turn repress CCA1 and LHY, closing the repressilator circuit. Balancing out the repressors of transcription are activators, for example RVE8 (REVEILLE8),35 which stimulates transcription of ELF4 and TOC1. Additional feedforward and feedback pathways define the period length of the circadian cycle and presumably add robustness to the clock. Light activates phytochromes and cryptochromes, photoreceptors, which among other targets, activate the genes for PRR9, CCA1 and LHY. Arabidopsis may also use translational regulation to perfect the timing and strength of circadian rhythms.36 As is the case in the mammalian clock, nutritional signals (sucrose in Arabidopsis) have been shown to help reset the plant clock via PRR7,37 GI (GIGANTEA),38 phytochrome-interacting transcription factors, and LHY and CCA1.39
While transcriptional regulation of and by the circadian clock has been extensively described, many rhythms at the protein level remain unexplained. Initial studies in the mouse liver and Arabidopsis found rhythmic proteins without rhythmic mRNA expression, pointing to post-transcriptional diurnal or circadian control.40-43 However, in Arabidopsis for instance, among hundreds of abundant proteins, nearly all are non-rhythmic, even though most are encoded by rhythmic mRNAs40,44,45 suggesting that translational regulation or turnover may silence transcript-level rhythms. These considerations focus the lime light on a fundamental but long neglected question: how the clock and diurnal light dark and nutrient cycles affect the translation of mRNAs into proteins. The purpose of characterizing translational regulation is twofold. It may explain discrepancies between transcript-level cycles and protein-level cycles and may explain if or how the clock and light-dark cycles cooperate to regulate translation.
Translation of specific groups of mRNAs cycles over the course of the day
The global percentage of RNA found in polysomes, i.e. actively translated, fluctuates over the course of day. It varies between approximately 40-60% at the end of night and 65-80% during the day in Arabidopsis6,7,46 and between 70% during the day and 80% during the night in the mouse liver.47 Six recent papers have now yielded an overview of diurnal or clock-dependent translational regulation on a transcriptome-wide scale. The studies include two in mouse liver, one comparing liver and kidney, one in human U2OS tissue culture cells, and one in Arabidopsis thaliana. An additional study of translation state in the Drosophila brain identified three genes with flat total mRNA levels but cycling ribosome-association,48 i.e. clear evidence of cycles in translation state.
Each of the mouse studies entrained the animals to cycles of 12h light / 12h dark (see Table 1). In the initial study,47 the animals were fed only at night, the time when mice are normally active, to reduce variation from erratic feeding behavior. Translation efficiency of mRNAs was scored genome-wide over two days every two hours. Two percent of mRNAs displayed a cycle in polysome loading, a measure of translation efficiency that is independent of the mRNA level.47 Many of the translationally regulated mRNAs in the mouse encoded ribosomal proteins. Their translation was stimulated shortly before dusk and dropped sharply right after dawn.47 This shows that rhythmic translation occurs in the mouse liver. The rhythm of translation of ribosomal proteins was accompanied by similar rhythms of nascent rRNA transcript abundance, unspliced ribosomal protein (RP) mRNAs (interpreted to reflect the transcription rate), and the abundance of unassembled ribosomal proteins (interpreted to reflect the protein synthesis rate). Thus, some combination of the diel feeding cycle, the light-dark cycle and the circadian clock coordinately orchestrates ribosome biogenesis. Two strains of clock-deficient mice seemed to be generally compromised in their cycles of transcription. However, the translation state of the ribosomal protein mRNAs was not measured in the clock-deficient strains.
Table 1.
Transcriptome-wide studies of diurnal translation.
| First author Year Organism | Light and other environmental conditions | Did the study address the role of the circadian clock? | Methodology to score translation state | Major findings |
|---|---|---|---|---|
| Jouffe 2013 Mouse liver47 | Light-dark (12/12 hr.) cycles with feedings only at night or starvation conditions | Bmal1 knockout and Cry1/Cry2 double knockout | Microarrays of polysomal and total mRNA | 1. Two percent of mRNAs are translated with a rhythm unexplained by mRNA abundance. |
| 2. Rhythmic translation of ribosome biogenesis mRNAs peaks at night. | ||||
| 3. Rhythmic phosphorylation of pre-initiation complex subunits. | ||||
| Huang 2013 Fly clock cells in the head 48 | Constant darkness after entrainment | Constant conditions | Translating ribosome affinity purification (total RNA-Seq for only a few genes) | 1. Translation of most mRNAs peaks around ZT8 or ZT12 (bimodal pattern), while published mRNA levels peak anytime. |
| 2. Certain mitochondrial proteins preferentially translated during the day, signaling and transcription at night. | ||||
| 3. Cycles of translation efficiency confirmed in a small number of mRNAs. | ||||
| Missra 2015 Arabidopsis 46 | Light-dark (16/8 hr.) cycles | CCA1 overexpressing strain | Microarrays of three fractions of (non-)polysomal and total mRNA | 1. ∼2000 mRNAs with translation cycles, peaks centered around noon or midnight. |
| 2. Ribosomal and mitochondrial proteins preferentially translated at night. | ||||
| 3. The clock broadly affects phase and amplitude of translation cycles. | ||||
| 4. Translational control of clock genes. | ||||
| Janich 2015 Mouse liver 50 | Light-dark (12/12 hr.) cycles; ad libitum feeding | No, only examined day-night timing effects | RPF and total RNA sequencing libraries | 1. Rhythmic biosynthesis of core clock proteins is determined by mRNA availability. |
| 2. 147 rhythmically translated mRNAs. Protein biosynthesis machinery up around dusk and early night. | ||||
| 3. Translation of uORFs as a novel regulatory mechanism of the clock. | ||||
| Atger 2015 Mouse liver49 | Light-dark (12/12 hr.) cycles; feedings only at night or ad libitum | Bmal1 knockout mice | RPF and total RNA sequencing | 1. Rhythmic ribosome footprints are mainly due to mRNA availability. |
| 2. Disruption of the clock affects translational rhythms for 16 genes. | ||||
| 3. Mitochondrial proteins preferentially translated at ZT10; translation machinery at ZT17. | ||||
| 4. Feeding restriction tightens or phase-shifts translation of selected cohorts. | ||||
| Jang 2015 Human U2OS cells53 | Synchronized with dexamethasone | siRNAs targeting ARNTL(BMAL1) | RPF and total RNA sequencing | 1. Circadian translational rhythms are bimodal. |
| 2. 40 genes had oscillations in translation efficiency. | ||||
| 3. Oscillation in P-body abundance associated with LSMI expression. | ||||
| Castelo-Szekely 2017 Mouse kidney51 | Light-dark (12/12 hr.) cycles; ad libitum feeding | No, only examined day-night timing effects | RPF and RNA sequencing libraries; compare with liver data | 1. Translational regulation is organ-specific. |
| 2. 92 rhythmically translated mRNAs with peaks at ZT4 and ZT16. | ||||
| 3. Ribosome occupancy is more similar between kidney and liver than mRNA level. |
RPF, ribosome protected fragments, ribosome footprints
Extending,49 Atger and coworkers49 compared cycles of ribosome footprints in mice fed either only at night or ad libitum to address the role of feeding cycles on diurnal translation. Translation state was estimated by comparing ribosome footprint density and exonic RNA sequence reads. Cycles of translation efficiency were inferred from discrepancies between transcript cycles and ribosome footprint cycles. The rhythmicity of the transcription rate was again estimated from intronic RNA-sequence reads. Of the genes with rhythmic translation, those translated preferentially at ZT10 were enriched for mitochondrial functions; genes translated with a peak at ZT17 tended to be part of the translation machinery. Bmal1 knockout mice were employed to demonstrate the clock's effect on rhythmic transcript levels and translation. Overall, the translation of few mRNAs was clearly clock-dependent, and this group did not include the ribosome biogenesis cluster.49
A similar, simultaneous study compared the diel cycle of mRNA transcripts with that of ribosome footprints and calculated translation efficiencies from the ratio of footprints to total mRNA.50 A total of 147 rhythmically translated mRNAs were identified with high confidence, among these mRNAs for iron metabolism and, again, ribosomal proteins. Aside from 147 mRNAs with robust TE cycles on top of flat mRNA levels, there were mRNAs with cyclical mRNAs but lacking a cycle of footprints. These mRNAs are also candidates for diel translational control, but were not analyzed in detail. By comparing data from liver with more recent data from the kidney,51 it became clear that translation cycles are strongly organ-specific, as few if any of the mRNAs cycled in both organs. These data may indicate that different organs respond differently to cyclical cellular signals. Although the role of the clock in driving cycles of translation efficiency was not explicitly addressed, it is notable that the rise in translation efficiency anticipates the light-dark transition, as it did in,50 a characteristic of clock-controlled events. For comparison, a study that monitored transcriptional activity of RNA polymerase III in the same organ also observed an anticipatory rise before dusk and was able to attribute it to the circadian clock.52
In human U2OS osteosarcoma cells53 the cell cycle was synchronized to the circadian cycle with dexamethasone, and the translation state of mRNAs was scored under free-running conditions by comparing ribosome footprints with total mRNA abundance. The amplitude of the cycles was generally small, which meant limited statistical power when mRNAs were screened for cycles in translational efficiency, yet 40 such mRNAs were identified. However, two classes of mRNAs yielded good evidence for diel translational control: mRNAs with footprint cycles on the basis of flat mRNA levels, and mRNAs with transcript cycles but lacking footprint cycles. The former were enriched for functions in RNA-biology including translation, while the latter tended to code for signaling proteins and transcriptional regulators. A third group, mRNAs that cycled at both transcript and footprint levels generally had no phase-shift between the two suggesting the absence of diel translational control for these mRNAs. Regarding possible mechanisms for translational control, the authors implicated the RNA binding protein RBM10, short 3′ UTRs, and uORFs. Interestingly, P-bodies, organelles where translationally silent mRNA are stored, oscillate in abundance in a diurnal fashion and in a clock-dependent manner.53
Finally, Arabidopsis seedlings were grown in a 16hr light/8hr dark cycle, and translation states of mRNAs were scored by microarray analysis of non-polysomal, small polysomal, large polysomal, and total mRNAs.46,54 The translation state of most mRNAs peaked around noon or midnight with around 2000 mRNAs (∼15% of the active transcriptome) having robust translational changes during a diel cycle. The cohort of genes translationally upregulated at night was enriched for cytosolic ribosomal and mitochondrial proteins. Photosystem I mRNAs were preferentially translated at night and dawn, while photosystem II mRNAs were upregulated toward dawn, and mRNAs for light harvesting antenna proteins were preferentially translated during the day. mRNAs for proteins involved in redox regulation, protein turnover, and the circadian clock were also strongly represented among translationally cycling mRNAs. A strain overexpressing the clock gene CCA1 was used to address the role of the circadian clock. In the clock-deficient CCA1-ox strain, the translation patterns were significantly altered, suggesting that fluctuations of ribosome loading in Arabidopsis are orchestrated in concert between a functional circadian clock and external cues.46
In summary, in both plant and animal kingdoms a fraction of the transcriptome experiences diel changes in ribosome loading. Strikingly, most of the studies identified ribosome biogenesis and mitochondrial proteins as a target of translational control. The role of the clock in driving the patterns of translation dynamics is not immediately clear in all cases. The clock may or may not drive, enhance, or even suppress cycles of translation.
Bimodal pattern of translation cycles
Strikingly, in all of the studies the times of peak translation are distributed over the day in a bimodal pattern. Arabidopsis transcripts with diel translation cycles fell into two major groups. One peaked in the morning and the other peaked during the night.46 While the time resolution of the study was not sufficient to precisely map peak times, a mathematical sine model revealed this bimodal pattern quite clearly. Human U2OS cells also revealed a bona fide bimodal distribution of peak translation times. RNA-only cyclers, which by inference must be subject to anti-phasic translational control, peaked in a bimodal fashion at CT (cycle time) 5±1 or CT17±1. In contrast, mRNAs that cycled only at the translation level tended to be translated at a single time of day (unimodal distribution around CT5±1).53 In the mouse, the bimodal pattern was particularly clear in the kidney, with peak translation at ZT4 or ZT16.51 In the liver, a small number of mRNAs with iron response elements (IREs) in their 5′UTR also had robust translation rhythms, which peaked at ZT0 (dawn).50 However, the majority of mRNAs with a translation cycle peaked in a broad period around dusk; of those, one subset peaking at ZT10 (evening) was enriched for mitochondrial electron transport functions while mRNAs peaking at ZT12-ZT17 (dusk and early night) trended towards protein synthesis functions.47,49,50
Thus, studies in four distinct model systems, two intact organs, a cell culture, and a plant, demonstrate that translational control is substantially bimodal, with one prominent peak each during the day and during the night. Moreover, the same pattern also appears in the fruit fly48 although in this study transcriptional and translational control were not separated. The physiological significance of the bimodal pattern remains unclear – why would it not be advantageous to tailor translation to a wider variety of peak times across the day? On a mechanistic level, one might hypothesize that the bimodal pattern of translation may result from a fairly simple signaling pathway, as little as a single pathway, to which individual mRNAs are coupled either positively or negatively or not at all. This pathway may be the circadian clock, but as we will see, the role of the clock in the phasing of translation efficiencies is complex.
Interaction of the endogenous clock and exogenous signals in regulating TL cycles
The clock affects translation cycles in complex ways. The impact of the clock on translation is arguably most clear and direct in Arabidopsis, where the translation cycles in the clock-deficient CCA1-overexpressor strain were for the most part phase-shifted or otherwise altered compared to the clock-entrained wild type.46 Clock-control of translation was also evident in the fly brain, albeit for only a small number of mRNAs.48 However, the role of the clock is not simply to impose translational cycles on otherwise translationally ‘flat’ mRNAs. First of all, robust translation cycles were observed even in clock-deficient conditions.46,49 These results suggest that translation cycles can be driven by external conditions, such as light-dark cycles in Arabidopsis46 or feeding cycles in the mouse,47,49 and feeding cycles are themselves heavily influenced by the clock (Fig. 2). Nutritional and hormonal signals such as sucrose and auxin in plants or sugars and insulin in animals regulate translation through the TOR pathway.47,55,56 Likewise, plants respond to light-dark shifts by altering polysome loading,6,57,58 while ribosome levels stay constant.7
Figure 2.
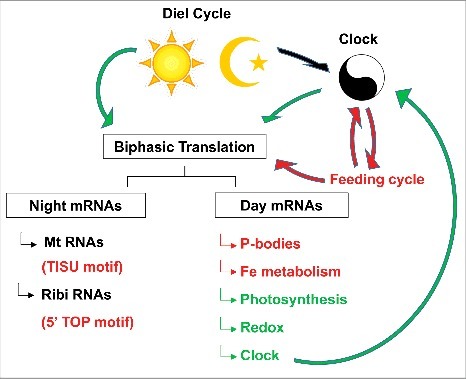
The cartoon summarizes how light dark-changes in the environment, the circadian clock, and feeding cycles influence translation. Arrows and text colored red are based on data from animals, while those in green stem from plants, and those in black apply in both kingdoms. Strikingly, a separation of translation into mRNAs that are preferentially translated at night or during the day is common in both kingdoms, and the functional annotations also partially overlap. Mt, mitochondrial protein; Ribi ribosome biogenesis proteins; Fe, iron. For details see text.
In the mouse liver, focusing again on mRNAs with flat transcript levels and cycling footprint density (or vice versa), BMAL1 knockout did not phase-shift or abolish the translation cycles in a uniform way.49 Instead, this experiment demonstrated that the clock has fairly mild, yet diverse effects on shaping the cycles of translation in the mouse liver. For example, translation of many mRNAs remained unchanged, some mRNAs lost their TL cycle, and others shifted their phase.49 In Arabidopsis, this complex effect of the clock was also striking. The clock triggered the translational cycling of some mRNAs and advanced the phase of translation of other mRNAs – effects in keeping with typical roles of the clock at the transcript level. Moreover, and quite clearly, the clock also suppressed the translational cycling of several hundred mRNAs, cycling that is presumably caused by the light-dark shifts over the day.46
Taken together, the clock modulates translation cycles imposed by other conditions, e.g. by integrating nutritional signals. Whether the clock alone can drive cycles of translation efficiency under otherwise constant conditions is less clear. This was addressed in Arabidopsis46 but only for a few mRNAs, and the results were ambiguous. Likewise, the tissue culture experiments by Jang and coworkers only revealed a fairly limited number of mRNAs with ARNTL/clock-dependent cycles of translational efficiency.53
A recent study in the mouse liver has extended what we know about the interplay of feeding rhythms, light-dark cycles, and the clock, specifically for ribosome biogenesis.59 Beyond the known cycle of translation state47,49,50 and TOR signaling,47 the level of polyadenylated ribosomal RNAs also cycles up and down. These poly(A)-rRNAs are most likely a degradation intermediate, and indeed, poly(A)-rRNA is highest during the day, about eight hours before the peak in ribosome loading of the ribosomal protein mRNAs. Strikingly, in the liver, not only the fraction of RNA found in polysomes, but the total amount of RNA per cell, and even the cell size all cycle with a peak at night.59 This cycle of ribosome biogenesis events is observed only when mice are fed during the night. When the mice are fed during the day instead, the cycle is masked.59 This raises the question how the feeding regimen interacts with the light-dark regimen and the circadian clock to regulate the cycle of ribosome biogenesis. Possibly it is the asynchrony between the feeding cycle and the circadian clock that masks the ribosome biogenesis cycle. This idea can now be tested.
The phasing of translation for functional classes of mRNAs
To understand the effects of diel translation, the functional classes of proteins that are affected must be examined. In both the mouse liver and in Arabidopsis mRNAs related to protein synthesis and mRNAs functioning in mitochondria, particularly during oxidative phosphorylation, are prominent among the translationally cycling mRNAs.47,49,50 Strikingly, the same classes were also the most robustly cycling in Arabidopsis.46 Similarly, in U2OS tissue culture cells the transcriptionally flat but translationally cycling mRNAs were enriched for functions in RNA-biology.53 Regarding the timing, while the protein synthesis-related mRNAs peaked at night in both organisms (ZT17 in Arabidopsis; ZT12–15 in mouse liver,47,50 the mitochondrial protein mRNAs peaked during the day in the mouse (ZT4–12, depending on the feeding regimen)49 but during the night in Arabidopsis.46 Although the dynamics of translation appear overtly similar, it should be understood that plants experience their peak energy status during the day while mice preferentially feed at night and rest during the day. In summary, in both mice and plants, translational control contributes to metabolic regulation by enhancing ribosomal and mitochondrial mRNA expression during the appropriate times for each organism.
In the mouse liver, two percent of mRNAs experience cycles in poly(A) tail length.60 The NOCTURNIN deadenylase targets mRNAs for ribosome biogenesis and for mitochondrial proteins.61 Strikingly, these are the same functional classes of mRNAs that experience elevated translation efficiency at night. However, the mechanistic link between diurnal deadenylation and translation has not been established, especially considering that short poly(A) tails are generally associated with translational repression rather than translational stimulation.
Are clock mRNAs under translational control?
In Arabidopsis, clock gene mRNAs and mRNAs for photoreceptor proteins, which constitute the input pathway for the clock, are prominent among translationally cycling mRNAs46 and fall into two groups. The first group (e.g. PRR9, ELF3, and phytochrome and cryptochrome photoreceptors) has translation peaks at dawn or noon (ZT0 or ZT6). The second group (e.g., the evening complex subunit LUX, TOC1, PRR5, and the photoreceptor UVR8) peaks at night (ZT18). Several of these mRNAs, including TOC1, LUX, GI, and PRR5, show a 6-hour delay between maximal transcript abundance and maximal translation. Consistent with the general pattern, when the clock was disrupted, the translation peaks of the ‘dawn-noon’ mRNAs were delayed until noon or later, while those of the ‘night’ mRNAs were typically delayed until dawn or noon. According to these data, Arabidopsis relies not only on transcriptional control but also on translational control at the level of ribosome loading to fine-tune expression of clock genes.46
In contrast to the situation in Arabidopsis, in mouse liver cells, the ribosomal footprint profiles of all central clock transcripts, such as the anti-phasic Bmal/Arntl and Nr1d1/REV-ERBα mRNAs closely matched their mRNA abundance rhythms.49,50 Likewise, in U2OS tissue culture cells, clock gene mRNAs such as Nr1d1 and Nr1d2 were not prime targets of translational control, and the known delay between peak mRNA level and peak protein levels of clock gene expression could not be attributed to translational control.53 Nevertheless, the central clock mRNAs of the mouse are subject to a form of translational control, which is independent of time of day, as evident from the distinct translational efficiencies of different clock mRNAs. For example, of the two paralogs REV-ERBα and REV-ERBβ the translational efficiency of the latter is five-fold lower, correctly predicting lower protein levels despite higher mRNA levels.50 Moreover, the translational efficiencies of the Nr1d1 and Nr1d2 mRNAs differ between liver and kidney in the mouse.50,51 Taken together, the extent of translational regulation of the clock mRNAs is organism-specific. This answers one of the major questions about the interaction between the clock and diel translation, yet still leaves the mechanism of translational rhythms largely unknown.
These data extend a small number of specific case studies of translational control for clock mRNAs, starting with Arabidopsis LHY mRNA.36 For example, in the mouse, translation of the Cryptochrome and Period mRNAs is inhibited and stimulated, respectively, by heterogeneous nuclear ribonucleoprotein Q, which binds to the 5′ untranslated region of Per1 mRNA and stimulates the activity of an internal ribosome entry site.62-64 In Drosophila, among other examples,65 the translation of a specific, alternatively spliced isoform of the Doubletime mRNA, dgt-RC, is regulated by the rhythmic RNA binding protein, LARK. Because Doubletime encodes CKIδ/ε, which is a determinant of period length in Drosophila, these data are evidence for translational control of the central oscillator itself.66
Mechanistic reasons for rhythms of translation
The mechanisms for diurnal regulation of translation are not understood. However, a number of studies have identified correlative evidence around phosphorylation of translation factors, mRNA sequence features particularly in 5′ untranslated regions, and the activation of specific signaling pathways.
Rhythmic phosphorylation events of translation initiation factors may contribute to TL cycles in mice. Robust rhythmic phosphorylations of proteins in the AKT-TOR pathway, including translation initiation factors eIF4E (peak phosphorylation at ZT6-12) and eIF4G, eIF4B, 4E-Binding Protein 1 (4E-BP1), and ribosomal protein RPS6 were observed.47,49 These proteins are components of the cap-binding complex and the ribosome and are phosphorylated preferentially at night, the period when the mice are active and feeding and when translation of ribosome biogenesis mRNAs is highest. Second, the pathways responsible for these phosphorylation events also have upstream components that are rhythmically expressed.47 However, in the mouse liver, the robust cycles of phosphorylation of the AKT-TOR pathway were not dramatically altered in a clock-deficient strain.47 Together these data suggest that rhythmic phosphorylation of basal translation factors may mediate the effect of feeding cycles on translation efficiency.
One output pathway for clock stimulated translation is represented by the clock-driven association of BMAL1 with the cytosolic translation initiation apparatus, which is stimulated when BMAL1 gets phosphorylated by S6 kinase67 in a diurnal fashion.41 This example of translational control, which was observed in mouse embryonic fibroblasts, may be global rather than mRNA-sequence specific.
In Arabidopsis, light-stimulated phosphorylation events are common in the translation apparatus.68,69 In contrast to the situation in the mouse liver, abundant rhythmic phosphorylation events have been documented in Arabidopsis seedlings even under free-running (constant light) conditions, including in ribosomal proteins.70 In addition, casein kinases are deeply involved in clock function in many organisms (reviewed by71), and phosphorylate translation initiation factors.72-74 Meanwhile, in Neurospora crassa, a bread mold, the clock controls translation via phosphorylation of eukaryotic elongation factor eEF-2. The clock causes the mitogen activated protein kinase (MAPK) OS-2 to be rhythmically phosphorylated. Both OS-2 and the core clock component FREQUENCY (FRQ) are necessary to create phosphorylation cycles of the eEF2 kinase, RCK-2, and in turn of translation elongation factor eEF2. This pathway leads to rhythmic, clock-dependent translation of glutathione-S-transferase as a target mRNA.75 These results open up the possibility that the clock might drive cycles of translation activity. However, at this time it is unclear in both mice and Arabidopsis how these events contribute to the bimodal, gene-specific translation cycles observed in the genome-wide studies.
Are there mRNA sequence elements that might explain the diurnal peaks of translation in mRNAs for ribosomal proteins, mitochondrial proteins or iron metabolism proteins? 5′-Terminal oligopyrimidine tracts (5′TOP motifs) are found preferentially in mRNAs with translational rhythms and flat transcript profiles such as the ribosomal protein mRNAs whose translation peaks at ZT17 in the mouse liver.47,49 TOP mRNA translation also varies with feeding pattern, and is not clock-dependent.49 The TORC1 kinase is regulated by nutrient availability and regulates 5′TOP mRNA translation through phosphorylation of eIF4E-binding protein (4E-BP) and S6K phosphorylation. Therefore, it appears plausible that TORC1 is responsible for the peak timing of translation of TOP mRNAs in the mouse liver,49 although this has not yet been shown directly. For comparison, in plants a sequence element known as the telobox is enriched in mRNAs whose translation is stimulated by light, although its specific role in this process remains to be identified.58
In Arabidopsis, a more recently proposed mechanism for diel translation involves the energy-sensing SnRK1 kinase. Under energy-replete conditions with high-level translational activity RPS6 is phosphorylated by the TOR kinase pathway,56 whereas under energy-limited conditions such as in an extended night, RPS6 is dephosphorylated dependent on SnRK1, in keeping with the idea that energy status impacts TOR pathway activity.76 Although plant ribosomal protein mRNAs do not contain canonical 5′ TOP motifs, it appears plausible that they are likewise translationally regulated by counteracting TOR and SnRK kinases.
The translation initiation sequence downstream of a short 5′UTR (TISU motif) is found preferentially in mRNAs for mitochondrial proteins, which are preferentially translated during the day. These mRNAs are known to be AMPK resistant, that is, these mRNAs remain translated when AMPK signaling depresses translation of other mRNAs. Therefore, the diurnal peak of translation of mitochondrial protein mRNAs during the day may be due to AMPK activity at this time, perhaps because AMPK relieves the competition for ribosomes by depressing translation of other mRNAs (discussed in49). Indeed, when mice were denied food during the day, this cohort of mRNAs experienced a translational phase advance by ∼3 hours.
For a small subset of genes in mice, iron response elements (IREs) in their mRNAs control translation cycles. In response to elevated iron levels, IRP1 forms a 4Fe–4S cluster, preventing the binding of IREs, allowing translation to occur. This can cause diurnal rhythms due to the fluctuating day/night levels of iron.50 None of these mRNA sequence motifs have been proven to be causal for clock-dependent translation; yet all merit further investigation.
Upstream open reading frames (uORFs) act as cis-regulatory elements controlling translation, and Density Regulated Protein (DENR) increases the translation of uORF-containing mRNAs by supporting translation reinitiation.77,78 Central clock mRNAs such as Arntl, Clock, Cry1, Nr1d1, and Nr1d2 all have ribosomes in their 5′ UTRs and contain at least one uORF.50 Deleting uORFs increased translation of the representative mRNA, Nr1d1, and disrupting the expression of DENR shortened the period of the circadian cycle by 1.5 hours.50 Meanwhile, Nr1d2 is also subject to uORF control; interestingly, a high degree of uORF occupancy on Nr1d2 predicts low main ORF occupancy for this mRNA, and this relationship may explain the different expression levels of Nr1d2 in liver and kidney.51 In summary, uORFs in clock mRNAs and elsewhere contribute to translational control of and by the clock.50 However, there is as yet no evidence for cyclical usage of uORFs independent of their main coding sequence.
Diurnal regulation of translation by P-bodies. Processing bodies (P-bodies) are cytoplasmic organelles that store and eventually degrade translationally silent mRNA. The percentage of cells with P-bodies cycles in a diurnal fashion, a cycle that requires clock activity, as was shown in human U2OS cells.53 Moreover, the mRNA for LSM1, a determinant of mRNA decapping and P-body formation, undergoes a clock-dependent diel cycle in ribosome loading. Thus, the clock may affect diel translation through P-body formation and potentially global mRNA decay.53
Integration of translation cycles with mRNA- and protein-level cycles
Cycles in translation efficiency must necessarily modulate any dynamic pattern at the level of transcript abundance, whether rhythmic or not. And assuming that translation efficiency influences the rate of protein production, the cycles will impact the dynamics of protein levels as well. What is the physiological relevance of the cycles in translation efficiency? In principle, there are two major scenarios. First, according to the ‘cyclical protein’ model, the cycle in translation efficiency should be multiplied with the cycle of transcript abundance to predict the diel cycle of the protein production rate, which in turn drives cycles of protein levels. Under this model, it should be noted that, unless the protein being produced is extremely unstable, the peak protein level will have a delayed phase as compared to the peak protein production rate. Assuming a constant and typical, slow rate of protein turnover79,80 the time of peak translation will be the time of the fastest increase in protein level and vice versa. Therefore, and further assuming a sinusoidal 24h-cycle of translation, the peak protein level will be reached 6 hours after the peak in protein production rate.81 Moreover, common phase shifts between peak mRNA level and peak translation will alter the waveform of the protein production rate.46 Second, according to the ‘invariant protein’ model, the translation cycle combines with a cycle of protein turnover and an optional cycle in the mRNA level to keep the protein level the same. Under this scenario, translational control is a potential mechanism to compensate for fluctuations in protein turnover that may be driven by cyclical environmental conditions. The following observations can be reasonably interpreted while considering these two models.
In Arabidopsis a large portion of the transcriptome cycles at the transcript level, while around 2000 genes have significant cycles of ribosome loading per mRNA.46 Analysis of the Arabidopsis proteome revealed that 30-40% of proteins with rhythmic protein levels do not have rhythmic transcripts,70 implicating translation and turnover in generating these rhythms. Meanwhile, many rhythmic mRNAs have no detectable protein rhythm,44,45 suggesting that Model 2 applies. Moreover, comparing between liver and kidney, ribosome occupancy is more similar than mRNA transcript levels for a large number of genes.51 These data are also consistent with model 2, especially its prediction that the cell should have mechanisms to compensate against fluctuations in gene expression in order to keep protein levels constant.
In the mammalian studies, translation cycles were often identified by focusing on mRNAs that did not cycle at the transcript level, which voids an analysis of phase shifts between the two. However, attempts were made to compare translation cycles with protein levels. For these analyses it should be understood that translation state is a proxy for the protein synthesis rate. Ribosomal protein mRNAs are subject to peak translation at ZT17.47,49 These proteins enter the nucleus for ribosome assembly. Indeed, in nuclear proteomics experiments, the 14 ribosomal proteins for which a cycle was detected peaked at ZT20.5 ± 1.3.30 A shorter-than-six-hour lag between translation peak and protein peak in this case is explained by the fact that the ribosomal proteins reside in the nucleus only transiently. The expected 4-6 h phase delay between peak translation rate (ribosome footprint density) and protein level was commonly detected using public protein level data from mouse liver.49 Interestingly, the levels of mitochondrial proteins did not cycle despite clear cycles in translation49; this may be a case of evidence for model 2, where the cycle in translation compensates for a phase-matched cycle of protein degradation. As an aside, the activity of many mitochondrial enzymes oscillates under clock control with a peak phase in the morning (ZT2-6),82 considerably later than the peak translation of mitochondrial protein mRNAs; there may be little overlap between the genes with cycling translation and cycling enzyme activity.
Janich et al. addressed in some detail how cycles in ribosome footprint density compare to cycles in protein levels. The phase shift between the two varied; for example, negative 5 h (equivalent to +19 h) for the transcription factor Deformed Epidermal Autoregulatory Factor 1 (DEAF1); +3 h and +5 h for ferritin heavy and light chain, respectively, and 0h for aminolevulinic acid synthase 2.50 In U2OS cells, SNRNP70 oscillates with no significant phase delay between ribosome footprint density and protein level, again a remarkably short lag time.53 These phase relationships imply that some of our simple assumptions are violated. For example, it is possible that in some cases a high ribosome footprint density does not equal high translation but non-canonical ribosome pausing.
In summary, there is now a solid foundation of quantitative transcriptome-wide data from multiple stages of gene expression, including translation. If our goal is to not just measure and understand, but also to predict and control83 diel cycles of gene expression, then we require corresponding quantitative mathematical models of gene expression that are founded on a rigorous theoretical framework from molecular biochemistry. Specifically, diurnal protein levels30,40,41,44,45,70,84 are determined by rates of mRNA synthesis and mRNA degradation, mRNA translation (protein synthesis), and protein turnover. Regarding these processes, the more widely available data are on mRNA transcript levels,85,86 which are the dynamic balance between transcription and turnover, in the same way that protein levels are the dynamic balance of protein synthesis and protein turnover. Protein turnover rates are being measured (e.g.79), but have yet to be measured in a diurnal context. Hence, data on mRNA translation states assume special significance, because they serve as a valuable albeit imperfect proxy for the rate of protein synthesis.
Future questions
The studies so far have pointed to the widespread existence of diel cycles of translation efficiency. Cycles in translation efficiency can enhance, modify, or even suppress cycles at the transcript level. Cycles of translation are driven by environmental changes and are influenced by the circadian clock to varying degrees. The clock can enhance, modify or suppress translational cycles. The significance of translational cycles at the level of protein abundance remains to be established in most cases; the extreme cases are that translation cycles can drive cycles of protein abundance, or that they compensate for cycles of protein turnover, keeping protein levels invariant. The mechanisms for the diel cycles are also barely beginning to come to light. They involve specific RNA sequence elements such as uORFs and 5′ TOP motifs, posttranscriptional modifications of the RNA, cellular trafficking of RNAs, for example into P-bodies, and regulatory signaling pathways such as TOR and MAP kinase pathways. The point of integration of light-dark signals, feeding signals, and output from the circadian clock also remains to be established.
Funding Statement
This work was supported by the National Science Foundation under Grants IOS-1456988 and MCB-1546402.
Glossary
• Translation efficiency. The degree to which a given RNA molecule is translated. Translation efficiency is most often estimated as follows. The ribosome footprint density or the polysome abundance (in reads per kilobase of sequence per million base pairs) of an mRNA is divided by its abundance in the total-RNA reference sample.
• Translation state. This term is sometimes defined as equal to translation efficiency. In other studies, the term simply means that the expression level of an mRNA was measured from a sample of ribosome-associated RNA rather than total RNA.
• Translation cycle. A cycle in translation efficiency with a circadian (∼24h) period. Translation cycles can be estimated in several ways. (i) Calculate the translation efficiency across the day and filter for statistically significant cycles. (ii) One can also preselect all mRNAs with bona fide invariable mRNA transcript levels in the total-RNA sample and examine this subset for a cycle in the polysome abundance or ribosome footprint density. (iii) Finally, it is possible to select mRNAs with a defined cycle at the transcript level and examine these mRNAs for deviating cycles at the level of the ribosome-associated RNA (polysome abundance or footprint density). If the ribosome-associated cycle differs from the transcript cycle, it is evidence for translational control.
Disclosure of interest
The authors report no conflict of interest.
Significance statement
The translation of mRNA into protein is modulated by cellular energy status and metabolic status. Because both factors cycle over the course of the day and are heavily influenced by the circadian clock, the question whether the circadian clock influences translation is not far fetched. Moreover, given that the circadian clock is referred to as a transcriptional-translational feedback loop, it is necessary to ask whether the clock is regulated at the level of translation. A thin veil has been lifted off this mystery by way of recent transcriptome-wide analyses in the mouse, humans, flies, and plants. A comparative analysis between these model systems reveals a common framework, apparent differences, as well as surprising similarities in the details of how the clock and the environment act together to regulate translation across different kingdoms.
Abbreviations
- KO
knockout
- RP
ribosomal protein
- RPF
ribosome-protected fragments, also known as ribosome footprints (RFPs)
- TE
translational efficiency
- TL
translation state
- 5′ TOP
5′ terminal oligopyrimidine motif
- TISU
translation initiator of short 5′UTR
- ZT
Zeitgeber time, time in hours after lights-on
References
- 1.Lee CD, Tu BP. Metabolic influences on RNA biology and translation. Crit Rev Biochem Mol Biol. 2017;52(2):176–84. doi: 10.1080/10409238.2017.1283294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413–6. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy B, von Arnim AG. Translational regulation of cytoplasmic mRNAs. The Arabidopsis book. 2013;11:e0165. doi: 10.1199/tab.0165g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sesma A, Castresana C, Castellano MM. Regulation of Translation by TOR, eIF4E and eIF2alpha in Plants: Current Knowledge, Challenges and Future Perspectives. Front Plant Sci. 2017;8:644. doi: 10.3389/fpls.2017.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishman B, Wurtman RJ, Munro HN. Daily rhythms in hepatic polysome profiles and tyrosine transaminase activity: role of dietary protein. Proc Natl Acad Sci U S A. 1969;64(2):677–82. doi: 10.1073/pnas.64.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MC, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al.. Diurnal changes of polysome loading track sucrose content in the rosette of wildtype Arabidopsis and the starchless pgm mutant. Plant Physiol. 2013;162:1246–65. doi: 10.1104/pp.112.212258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piques M, Schulze WX, Hohne M, Usadel B, Gibon Y, Rohwer J, Stitt M. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol. 2009;5:314. doi: 10.1038/msb.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchiyama Y, Asari A. A morphometric study of the variations in subcellular structures of rat hepatocytes during 24 hours. Cell Tissue Res. 1984;236(2):305–15. doi: 10.1007/BF00214231. [DOI] [PubMed] [Google Scholar]
- 9.Nohales MA, Kay SA. Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol. 2016;23(12):1061–69. doi: 10.1038/nsmb.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nature Rev Genet. 2017;18(3):164–79. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morse D, Milos PM, Roux E, Hastings JW. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc Natl Acad Sci U S A. 1989;86(1):172–6. doi: 10.1073/pnas.86.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–5. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 13.Jacklet JW. Neuronal circadian rhythm: phase shifting by a protein synthesis inhibitor. Science. 1977;198(4312):69–71. doi: 10.1126/science.897685. [DOI] [PubMed] [Google Scholar]
- 14.Taylor WR, Dunlap JC, Hastings JW. Inhibitors of protein synthesis on 80S ribosomes phase shift the Gonyaulax clock. J Exp Biol. 1982;97:121–36. [DOI] [PubMed] [Google Scholar]
- 15.Hartwig R, Schweiger M, Schweiger R, Schweiger HG. Identification of a high molecular weight polypeptide that may be part of the circadian clockwork in Acetabularia. Proc Natl Acad Sci U S A. 1985;82(20):6899–902. doi: 10.1073/pnas.82.20.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millar AJ. The Intracellular Dynamics of Circadian Clocks Reach for the Light of Ecology and Evolution. Annu Rev Plant Biol. 2016;67:595–618. doi: 10.1146/annurev-arplant-043014-115619. [DOI] [PubMed] [Google Scholar]
- 17.Hastings JW. The Gonyaulax clock at 50: translational control of circadian expression. Cold Spring Harb Symp Quant Biol. 2007;72:141–4. doi: 10.1101/sqb.2007.72.026. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, Beauchemin M, Dagenais-Bellefeuille S, Letourneau L, Cappadocia M, Morse D. The Lingulodinium circadian system lacks rhythmic changes in transcript abundance. BMC Biology. 2014;12:107. doi: 10.1186/s12915-014-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benegiamo G, Brown SA, Panda S. RNA Dynamics in the Control of Circadian Rhythm. Adv Exp Med Biol. 2016;907:107–22. doi: 10.1007/978-3-319-29073-7_5. [DOI] [PubMed] [Google Scholar]
- 20.Nolte C, Staiger D. RNA around the clock – regulation at the RNA level in biological timing. Front Plant Sci. 2015;6:311. doi: 10.3389/fpls.2015.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preussner M, Heyd F. Post-transcriptional control of the mammalian circadian clock: implications for health and disease. Pflugers Archiv: European J Physiol. 2016;468(6):983–91. doi: 10.1007/s00424-016-1820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romanowski A, Yanovsky MJ. Circadian rhythms and post-transcriptional regulation in higher plants. Front Plant Sci. 2015;6:437. doi: 10.3389/fpls.2015.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGlincy NJ, Valomon A, Chesham JE, Maywood ES, Hastings MH, Ule J. Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 2012;13(6):R54. doi: 10.1186/gb-2012-13-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filichkin S, Priest HD, Megraw M, Mockler TC. Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr Opin Plant Biol. 2015;24:125–35. doi: 10.1016/j.pbi.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Luck S, Thurley K, Thaben PF, Westermark PO. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–51. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 28.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–78. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang F, Sun X, Ma X, Wu R, Zhang D, Chen Y, Xu Q, Wu Y, Liu Y. Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nature Commun. 2016;7:12696. doi: 10.1038/ncomms12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, Dayon L, Sizzano F, Palini A, Kussmann M, Waridel P, et al.. Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab. 2017;25(1):102–17. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging. 2014;6(1):48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khapre RV, Patel SA, Kondratova AA, Chaudhary A, Velingkaar N, Antoch MP, Kondratov RV. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging. 2014;6(8):675–89. doi: 10.18632/aging.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakenfull RJ, Davis SJ. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017;40(11):2571–85. doi: 10.1111/pce.13033. [DOI] [PubMed] [Google Scholar]
- 34.Ronald J, Davis SJ. Making the clock tick: the transcriptional landscape of the plant circadian clock. F1000Res. 2017;6:951. doi: 10.12688/f1000research.11319.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu PY, Devisetty UK, Harmer SL. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife. 2013;2:e00473. doi: 10.7554/eLife.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JY, Song HR, Taylor BL, Carre IA. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. Embo J. 2003;22(4):935–44. doi: 10.1093/emboj/cdg075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502(7473):689–92. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan GB, Goncalves JM, et al.. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci U S A. 2011;108(12):5104–09. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shor E, Paik I, Kangisser S, Green R, Huq E. PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 2017;215:217–28. doi: 10.1111/nph.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choudhary MK, Nomura Y, Shi H, Nakagami H, Somers DE. Circadian Profiling of the Arabidopsis Proteome Using 2D-DIGE. Front Plant Sci. 2016;7:1007. doi: 10.3389/fpls.2016.01007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A. 2014;111(1):167–72. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al.. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 43.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Buhlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al.. Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol. 2012;8:606. doi: 10.1038/msb.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf A, Coman D, Uhrig RG, Walsh S, Flis A, Stitt M, Gruissem W. Parallel analysis of Arabidopsis circadian clock mutants reveals different scales of transcriptome and proteome regulation. Open Biol. 2017;7(3). doi: 10.1098/rsob.160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Missra A, Ernest B, Lohoff T, Jia Q, Satterlee J, Ke K, von Arnim AG. The Circadian Clock Modulates Global Daily Cycles of mRNA Ribosome Loading. The Plant Cell. 2015;27(9):2582–99. doi: 10.1105/tpc.15.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Y, Ainsley JA, Reijmers LG, Jackson FR. Translational profiling of clock cells reveals circadianly synchronized protein synthesis. PLoS Biol. 2013;11(11):e1001703. doi: 10.1371/journal.pbio.1001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A. 2015;112(47):E6579−6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015;25(12):1848–59. doi: 10.1101/gr.195404.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castelo-Szekely V, Arpat AB, Janich P, Gatfield D. Translational contributions to tissue specificity in rhythmic and constitutive gene expression. Genome Biol. 2017;18(1):116. doi: 10.1186/s13059-017-1222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mange F, Praz V, Migliavacca E, Willis IM, Schutz F, Hernandez N. Diurnal regulation of RNA polymerase III transcription is under the control of both the feeding-fasting response and the circadian clock. Genome Res. 2017;27(6):973–84. doi: 10.1101/gr.217521.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jang C, Lahens NF, Hogenesch JB, Sehgal A. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 2015;25(12):1836–47. doi: 10.1101/gr.191296.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Missra A, von Arnim AG. Analysis of mRNA translation States in Arabidopsis over the diurnal cycle by polysome microarray. Methods Mol Biol. 2014;1158:157–74. doi: 10.1007/978-1-4939-0700-7_10. [DOI] [PubMed] [Google Scholar]
- 55.Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11(1):e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. TOR Signaling and Nutrient Sensing. Annu Rev Plant Biol. 2016;67:261–85. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- 57.Juntawong P, Bailey-Serres J. Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci. 2012;3:66. doi: 10.3389/fpls.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu MJ, Wu SH, Chen HM. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, Schibler U. Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell. 2017;169(4):651–63 e614. doi: 10.1016/j.cell.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26(24):2724–36. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kojima S, Gendreau KL, Sher-Chen EL, Gao P, Green CB. Changes in poly(A) tail length dynamics from the loss of the circadian deadenylase Nocturnin. Sci Rep. 2015;5:17059. doi: 10.1038/srep17059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim DY, Kwak E, Kim SH, Lee KH, Woo KC, Kim KT. hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res. 2011;39(20):8901–14. doi: 10.1093/nar/gkr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol. 2012;32(3):717–28. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim I, Jung Y, Kim DY, Kim KT. HnRNP Q Has a Suppressive Role in the Translation of Mouse Cryptochrome1. PloS One. 2016;11(7):e0159018. doi: 10.1371/journal.pone.0159018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tataroglu O, Emery P. The molecular ticks of the Drosophila circadian clock. Curr Opin Insect Sci. 2015;7:51–57. doi: 10.1016/j.cois.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang Y, McNeil GP, Jackson FR. Translational Regulation of the DOUBLETIME/CKIdelta/epsilon Kinase by LARK Contributes to Circadian Period Modulation. PLoS Genet. 2014;10(9):e1004536. doi: 10.1371/journal.pgen.1004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161(5):1138–51. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boex-Fontvieille E, Daventure M, Jossier M, Zivy M, Hodges M, Tcherkez G. Photosynthetic control of Arabidopsis leaf cytoplasmic translation initiation by protein phosphorylation. PloS One. 2013;8(7):e70692. doi: 10.1371/journal.pone.0070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turkina MV, Klang Arstrand H, Vener AV. Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PloS one. 2011;6(12):e29307. doi: 10.1371/journal.pone.0029307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choudhary MK, Nomura Y, Wang L, Nakagami H, Somers DE. Quantitative Circadian Phosphoproteomic Analysis of Arabidopsis Reveals Extensive Clock Control of Key Components in Physiological, Metabolic, and Signaling Pathways. Mol Cell Proteomics. 2015;14(8):2243–60. doi: 10.1074/mcp.M114.047183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulekar JJ, Huq E. Expanding roles of protein kinase CK2 in regulating plant growth and development. J Exp Botany. 2014;65(11):2883–93. doi: 10.1093/jxb/ert401. [DOI] [PubMed] [Google Scholar]
- 72.Dennis MD, Browning KS. Differential phosphorylation of plant translation initiation factors by Arabidopsis thaliana CK2 holoenzymes. J Biol Chem. 2009;284(31):20602–14. doi: 10.1074/jbc.M109.006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin S, Wolgamott L, Roux PP, Yoon SO. Casein kinase 1epsilon promotes cell proliferation by regulating mRNA translation. Cancer Res. 2014;74(1):201–11. doi: 10.1158/0008-5472.CAN-13-1175. [DOI] [PubMed] [Google Scholar]
- 74.Tamaru T, Hattori M, Honda K, Nakahata Y, Sassone-Corsi P, van der Horst GT, Ozawa T, Takamatsu K. CRY Drives Cyclic CK2-Mediated BMAL1 Phosphorylation to Control the Mammalian Circadian Clock. PLoS Biol. 2015;13(11):e1002293. doi: 10.1371/journal.pbio.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caster SZ, Castillo K, Sachs MS, Bell-Pedersen D. Circadian clock regulation of mRNA translation through eukaryotic elongation factor eEF-2. Proc Natl Acad Sci U S A. 2016;(34):9605–10. doi: 10.1073/pnas.1525268113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nukarinen E, Nagele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Bornke F, Hanson J, Teige M, Baena-Gonzalez E, et al.. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep. 2016;6:31697. doi: 10.1038/srep31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuchler K, Stoecklin G, Duncan KE, et al.. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512(7513):208–12. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24(16):1787–801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, Nelson CJ, Trosch J, Castleden I, Huang S, Millar AH. Protein Degradation Rate in Arabidopsis thaliana Leaf Growth and Development. The Plant Cell. 2017;29(2):207–28. doi: 10.1105/tpc.16.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 81.Mermet J, Yeung J, Naef F. Systems Chronobiology: Global Analysis of Gene Regulation in a 24-Hour Periodic World. Cold Spring Harb Perspect Biol. 2017;9(3):a028720. doi: 10.1101/cshperspect.a028720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neufeld-Cohen A, Robles MS, Aviram R, Manella G, Adamovich Y, Ladeuix B, Nir D, Rousso-Noori L, Kuperman Y, Golik M, et al.. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc Natl Acad Sci U S A. 2016;113(12):E1673−1682. doi: 10.1073/pnas.1519650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157(1):151–61. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 84.Abraham PE, Yin H. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plants. 2016;2:16178. doi: 10.1038/nplants.2016.178. [DOI] [PubMed] [Google Scholar]
- 85.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–63. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 86.Sobel JA, Krier I, Andersin T, Raghav S, Canella D, Gilardi F, Kalantzi AS, Rey G, Weger B, Gachon F, et al.. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol. 2017;15(4):e2001069. doi: 10.1371/journal.pbio.2001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weger M, Diotel N, Dorsemans AC, Dickmeis T, Weger BD. Stem cells and the circadian clock. Dev Biol. 2017;431(2):111–23. doi: 10.1016/j.ydbio.2017.09.012. [DOI] [PubMed] [Google Scholar]


