Abstract
Tankyrases (TNKSs) are enzymes specialized in catalyzing poly-ADP-ribosylation of target proteins. Several studies have validated TNKSs as anti-cancer drug targets due to their regulatory role in Wnt/β-catenin pathway. Recently a lot of effort has been put into developing more potent and selective TNKS inhibitors and optimizing them towards anti-cancer agents. We noticed that some 2-phenylquinazolinones (2-PQs) reported as CDK9 inhibitors were similar to previously published TNKS inhibitors. In this study, we profiled this series of 2-PQs against TNKS and selected kinases that are involved in the Wnt/β-catenin pathway. We found that they were much more potent TNKS inhibitors than they were CDK9/kinase inhibitors. We evaluated the compound selectivity to tankyrases over the ARTD enzyme family and solved co-crystal structures of the compounds with TNKS2. Comparative structure-based studies of the catalytic domain of TNKS2 with selected CDK9 inhibitors and docking studies of the inhibitors with two kinases (CDK9 and Akt) revealed important structural features, which could explain the selectivity of the compounds towards either tankyrases or kinases. We also discovered a compound, which was able to inhibit tankyrases, CDK9 and Akt kinases with equal µM potency.
Introduction
Tankyrases are enzymes (EC 2.4.2.30), which belong to a family of proteins known as human Diphtheria toxin-like ADP-ribosyltransferases. Two homologous tankyrases, TNKS1/ARTD5/PARP5a and TNKS2/ARTD6/PARP5b, catalyze a post-translational modification, poly-ADP-ribosylation, of target proteins. Tankyrases are very similar and both contain a catalytic ARTD domain, a sterile alpha motif (SAM) required for oligomerization and five ankyrin repeat clusters (ARCs) recognizing target proteins1. Tankyrases have overlapping functions in cellular signaling pathways and function e.g. in telomere homeostasis2–5, in elongation of centrioles and the formation of mitotic spindle during mitosis6, and in exocytosis of trans-Golgi vesicles containing IRAP and GLUT44,7,8. Recent studies have indicated tankyrases as anti-cancer drug targets as they are involved in regulating the Wnt/β-catenin pathway known to promote the survival of cancer cells9–11. In essence, the inhibition of TNKSs leads to a phosphorylation of β-catenin by a destruction complex formed by Axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3 β (GSK3β), and Casein kinase 1 (CK1)12,13. The subsequent proteasomal degradation of the phosphorylated β-catenin inhibits transcription of Wnt target genes. Tankyrases control β-catenin phosphorylation through regulation of the stability of destruction complex by ADP-ribosylating Axin and therefore inhibition of tankyrases offers an attractive strategy for treating cancers with increased Wnt-signaling14.
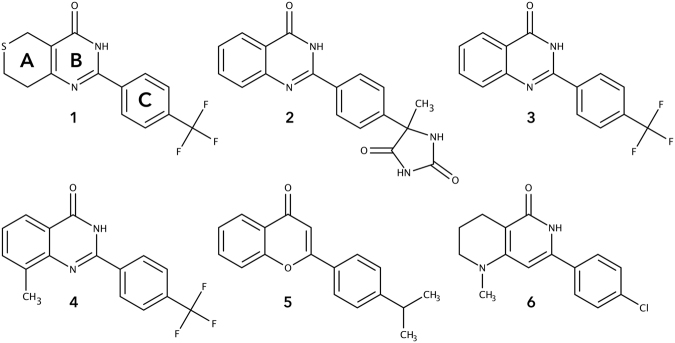
Recently, multiple studies have reported development of potent and in some cases selective small molecule tankyrase inhibitors targeting the catalytic domain15. Crystal structures of catalytic domain of both tankyrases are available16,17, which has facilitated structure-activity relationship (SAR) studies. Tankyrase inhibitors interact with the NAD+ binding groove of the catalytic domain and they bind either to the nicotinamide subsite, the adenosine subsite or span across both sites15. The first tankyrase inhibitor XAV939 (compound 1, Fig. 1) provided initial evidence of the involvement of tankyrase in cancerous Wnt signaling14. Multiple potent scaffolds resembling 1 in shape have been reported, including 2-phenylquinazolinones (2-PQS) (such as compounds 2–4)18,19, flavones (such as compound 5)20,21, arylnaphthyridinones (such as compound 6)22 (Fig. 1), and others such as 3-arylisoquinolin-1-ones23, tetrahydroquinazolin-4-ones24, pyrrolopyrimidinones25, and arylpyridopyrimidinones22. Here, we aimed at studying the 2-PQ class of TNKS inhibitors by synthesizing new analogues as well as assaying a large series of differently functionalized 2-PQS previously reported as Cyclin-dependent kinase 9 (CDK9) inhibitors26 and developed to interfere with HIV-1 Tat-mediated transcription27,28. We show that these compounds are in general more potent TNKS than CDK9 inhibitors and we rationalize the SAR towards tankyrases with a set of co-crystal structures.
Figure 1.
Examples of nicotinamide site binding tankyrase inhibitors.
As there was an evident potential for off targets within the kinase family for the 2-PQ compounds, we also evaluated this liability. It has been pointed out in the literature that simultaneous inhibition of certain kinases in combination with other enzymes could be used to potentiate the effect of kinase inhibitors offering a possibility for the development of double agents. For instance, it has been shown that the combination of telomerase inhibitors with tankyrase inhibitors enhances telomere shortening, leading to cell death29. Furthermore, synergistic effects were observed when inhibitors of telomere-regulating kinases, such as extracellular signal-related kinase 8 (ERK8), were used in combination with tankyrase inhibitors30. We profiled the best tankyrase-inhibiting 2-PQs against a panel of tankyrase and Wnt-related kinases and most of the analogs tested were significantly more potent against tankyrases than kinases, while one analog, compound 42, showed similar inhibition of TNKS2 and CDK9 as well as Akt kinase. These results identified potential routes for development of double agents, but also guide the design of analogs by revealing structure-based off-target liabilities.
Results
SAR and structural studies
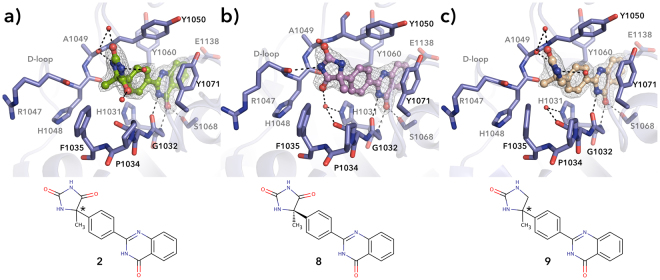
In a previous study we identified the hydantoin derivative 2 with nanomolar potency against tankyrases, which, however, was not potent in a cell-based assay (IC50 ≈ 1 µM)18. The compound was also not selective towards tankyrases, which we rationalized to be the result of polar substituent in the para-position of the C-ring. The low clogD of the compound (0.36) was intriguing taken into account that typically potent tankyrase inhibitors are rather hydrophobic and therefore the compound had a potential for further development. The original compound 2 was purchased commercially as a racemic mixture, and we decided to synthesize the pure (−)-enantiomer of compound 2 (hereafter named (−)-7) and (+)-enantiomer of compound 2 (hereafter named (+)-8) to evaluate the effect of the chirality (Supplementary Information).
In the TNKS2 complex structure with the commercial racemic mixture of 2 (Fig. 2a), we only observed the (−) enantiomer bound18. The other enantiomer, (+)-8 was also crystallized with TNKS2 (Fig. 2b) and similarly to (−)-enantiomer, the hydantoin moiety makes one hydrogen bond with the protein via an amide. However, instead of the main chain carbonyl of Ala1049, the (+)-enantiomer is hydrogen bonded to the main chain carbonyl of Arg1047. When evaluated for their ability to inhibit tankyrases (−)-7 and (+)-8 showed a similar potency against TNKS2 (IC50 = 2.4 and 6.7 nM, respectively) in agreement with that of the racemic mixture 2 (IC50 = 3.3 nM). It is likely that the crystal environment selects for the (−)-enantiomer. Because of modest cellular potency of compound 2 (68% inhibition at 2 µM), a small modification of the hydantoin substituent was attempted to improve the cellular potency while maintaining the inhibitory activity. Removal of one of the carbonyls led to 9 with a similar binding mode as compound 2 (Fig. 2c). Again only (−)-enantiomer was observed in the co-crystal of the racemic compound. Compound 9 showed lower potency against both TNKS1 and TNKS2 (IC50 = 10 nM) with respect to 2 (IC50 = 5.9 and 3.3 nM, respectively) (Table S1) but we did not observe improved potency in the Wnt-reporter assay (72% inhibition at 2 µM; Table S1). The modification however made the compound more selective towards the tankyrases (Table S1) likely due to the removal of interactions with the α-helical regulatory domains of ARTD1 and 2, which is not present in tankyrases (Figure S1)18.
Figure 2.
Crystal structures showing the binding mode of (a) 2 (PDB code 4BUY)18, (b) (+)-8 and (c) 9 to TNKS2 catalytic domain. Black dashed lines represent hydrogen bonds and red spheres represent water molecules. Sigma A weighted 2Fo – Fc electron density map around the ligands is contoured at 1.5 σ.
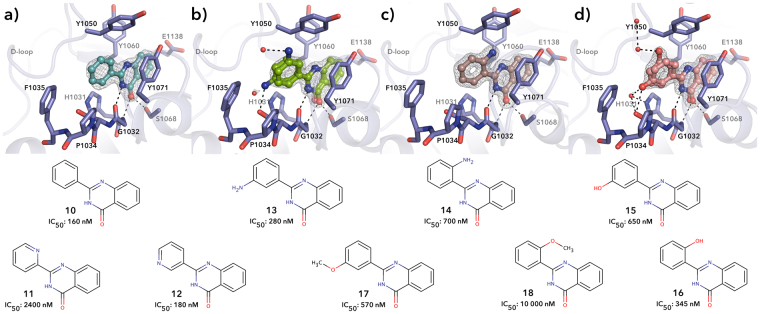
As previously reported, the basic 2-PQ scaffold 10 binds to the nicotinamide site of TNKS2 and makes typical hydrogen bonds to the active site glycine and serine residues as well as a stacking interaction with a tyrosine (Fig. 3a)18. In order to improve the compound we first studied the effect of the addition of hydrogen bond donors and acceptors to the ortho- and meta-positions of the phenyl ring (C-ring), which could form additional interactions with the protein (Fig. 3). Replacement of the phenyl group with a pyridine (found in 11 and 12) could provide a hydrogen bond acceptor and would enhance the solubility of the compound. The nitrogen in the ortho-position however caused a drop in the potency from 160 nM to 2.4 µM likely due to lost hydrophobic interactions, while nitrogen in the meta-position did not change the potency. We hypothesized that the nitrogen would not be close enough to form interactions and therefore we tested amine (13 and 14), hydroxyl (15 and 16) and methoxy groups (17 and 18) in both positions, which could act as hydrogen bond donor, donor as well as an acceptor, and an acceptor, respectively. All the substitutions were tolerated except methoxy in ortho-position, which likely causes a conformational change in the compound not compatible with the binding site. Crystal structures revealed that the amine in ortho-position (14) forms a dipole-dipole interaction with the side chain of Tyr1050, which however does not seem provide additional overall binding energy (Fig. 3c). In case of substituents in the meta-position (13 and 15) the compound forms additional water mediated interactions with the protein backbone, but the compounds are in two conformations in the crystal structures indicating that the binding poses would be of similar energy and the contribution of this hydrogen bond is not providing an affinity increase (Fig. 3b and d).
Figure 3.
Crystal structures showing the binding mode of (a) 10 (PDB code 4BU3)18, (b) 13, (c) 14, and (d) 15 to TNKS2 catalytic domain. The C-ring contains either the amino, hydroxyl or methoxy group in ortho- or meta-position. The compounds which have a meta-substitution are found in double conformations in the crystal structures. Black dashed lines represent hydrogen bonds and red spheres represent water molecules. Sigma A weighted 2Fo – Fc electron density map around the ligands is contoured at 1.5 σ.
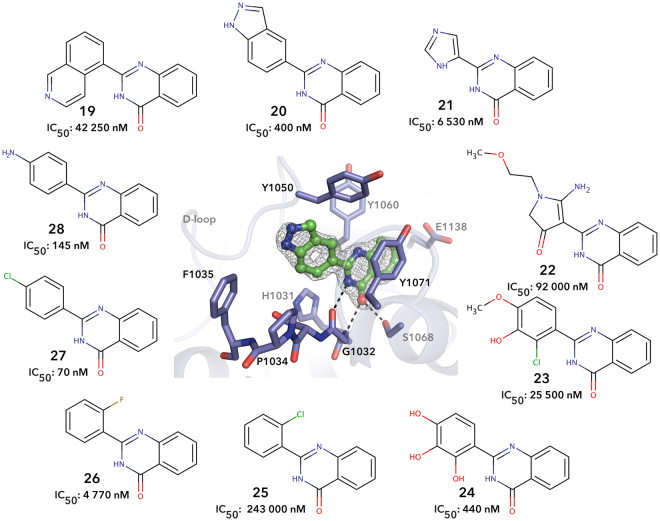
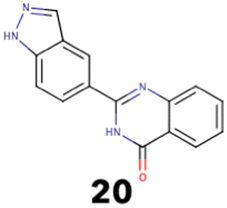
Some of the 2-PQs studied had a two ring-moiety instead of the phenyl group as a C-ring. The results demonstrated that the isoquinoline ring of 19 is too large to be accommodated in the tankyrase binding pocket, while the indazolyl of 20 is well tolerated. The crystal structure reveals that the compound extends out of the narrow binding pocket of the TNKS2 (Fig. 4).
Figure 4.
Schematic representation of 20 binding to the catalytic domain of TNKS2 as seen in the crystal structure. 20 showed higher potency against TNKS2 in comparison to ortho- and meta-substituted compounds. Black dashed lines represent hydrogen bonds and red spheres represent water molecules. Sigma A weighted 2Fo – Fc electron density map around the ligand is contoured at 1.5 σ.
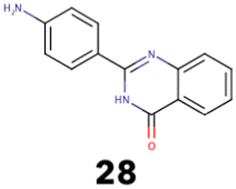
A replacement of the C-phenyl ring with a 5-membered ring in 21 was not tolerated and also highly substituted 22 showed poor inhibition (Fig. 4). Multiple substituents in the phenyl group gave contrasting results: a significant decrease in potency was observed for 23, while multiple hydroxyl groups in 24 are tolerated like in the flavone series20,21. The chlorine in ortho-position of 25 and 26 is likely to rotate the phenyl group out of the plane as we have reported earlier for similar methyl substitution in ortho-position of dihydroquinazoline and pyridopyrimidinone scaffold24. Also the fluorine in the ortho-position of 27 reduced the potency despite the small size of the substituent. On the other hand, when the chlorine was placed at the para-position of 28, a recovery in potency was observed with an IC50 of 70 nM.
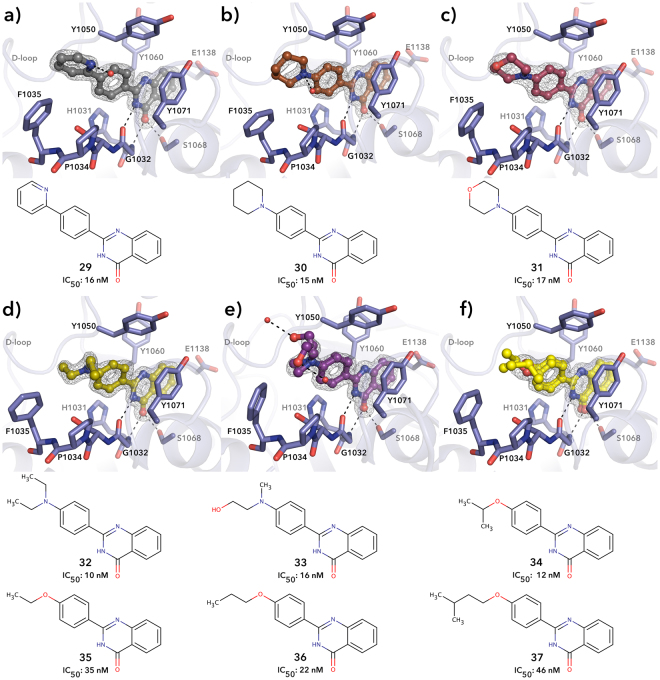
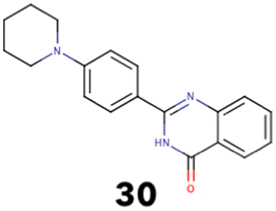
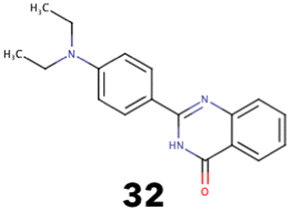
Substituents at the para-position of the phenyl group has been reported many times as a way to increase the potency of tankyrase inhibitors, and especially the addition of hydrophobic groups is known to improve specificity towards tankyrases18,19,21,31. The same trend was observed here. In particular, increasing the size of the para-substituent improved the potency in compounds 29, 30, and 31, all characterized by an additional ring, as well as in compounds 32, 33, and 34, having a branched side chain (Fig. 5). This increase in potency is caused by the hydrophobic interactions between the para-substituent and the side chain of Phe1035, Tyr1050 and Ile1075. A decrease in potency was however observed for compounds 35, 36, and 37 bearing a more linear side chain as para-substituent.
Figure 5.
Crystal structure showing the binding mode of (a) 29, (b) 30, (c) 31, (d) 32, (e) 33, (f) 34 to TNKS2 catalytic domain. Black dashed lines represent hydrogen bonds and red spheres represent water molecules. Sigma A weighted 2Fo – Fc electron density map around the ligands is contoured at 1.5 σ. Ile1075 was left out for clarity.
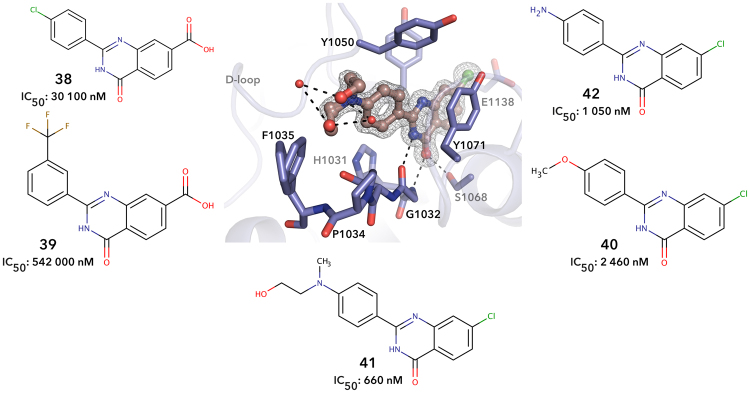
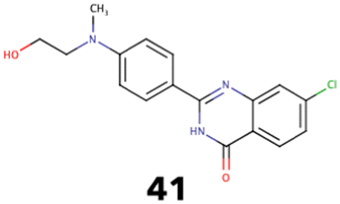
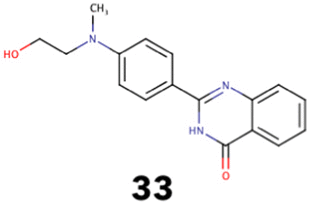
Chlorine and carboxyl substituents in the A-ring are not compatible with the TNKS2 inhibition (Fig. 6). However, while the carboxyl derivatives, 38 and 39 cause an unfavorable interaction with the active site glutamate, the chlorine substituent of compounds 40–42 fits to the binding pocket (Fig. 6). Notably, a chlorine atom in the A ring improved CDK9 inhibition26.
Figure 6.
Crystal structure showing 41. Black dashed lines represent hydrogen bonds and red spheres represent water molecules. Sigma A weighted 2Fo – Fc electron density map around the ligand is contoured at 1.5 σ.
Wnt signaling
In order to evaluate the potential of the best tankyrase inhibitors in cells, compounds 28–37 were tested in commercial TCF/LEF reporter cell line to explore their effect on Wnt/β-catening signaling. Out of 10 tested compounds, only 30 and 32 were active at 1 µM (>50% inhibition of Wnt/β-catening signaling, Table S2). In particular, 30 showed a sub-micromolar IC50 value of 570 nM, whereas 32 displayed a dual behavior: at 1 µM it showed 63% inhibition but at lower concentrations, it activated Wnt/β-catening signaling (Figure S2). 33 showed only a residual activity in the assay (20% inhibition).
Profiling of compounds against ARTDs
Compounds 30, 32 and 33, which were active in the reporter assay were profiled against a panel of human ARTDs to establish selectivity towards tankyrases (Table S2). Compounds 30 and 32 showed similar potencies against TNKS1/2 (30 nM/15 nM and 11 nM/10 nM, respectively), while 33 was four times more potent against TNKS2 than TNKS1 (16 nM/77 nM). Moreover, compounds 30 and 32 emerged as highly selective TNKS inhibitors, while 33 showed also inhibition of other enzymes of the family with potencies at low µM range (Table S2). Good selectivity profile coupled with the submicromolar activity in the reporter assay makes 30 the most promising tankyrase inhibitor among the studied 2-PQS.
Kinase profiling
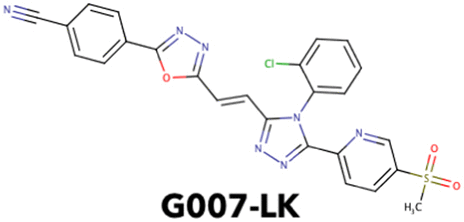
In order to elucidate the kinase vs. tankyrase inhibition of the 2-PQs, we profiled a selection of compounds against a panel of tankyrase and Wnt-signaling related kinases. We initially utilized compounds 30 and 32, because these compounds were more effective in cell-based assays, but we also included 20 and 42, which were previously reported to be active against CDK9, and the known tankyrase inhibitor XAV939 1. Compound G007-LK, which binds to a different location in the NAD+ binding groove, was also studied in parallel32.
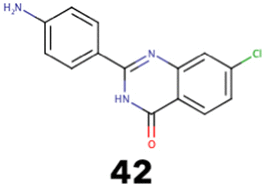
Results of the profiling (Tables 1, S3 and S4) showed that the best tankyrase inhibitor identified, compound 30, did not significantly inhibit the tested kinases although it showed modest inhibition at 10 µM concentration. This is similar to 20, which contains a bulkier two-ring substituent. Control tankyrase inhibitors 1 and G007-LK did not inhibit significantly the kinases either. Compound 32, which has a more flexibility at the para-substituent modestly inhibits most tested kinases at 10 µM. Compound 42 inhibited all the tested kinases except CK2 and the potencies against CDK9 and Akt kinases are at the same level as for TNKS2 (Table 1). We rationalized that this would be a result of the presence of the chlorine substituent in the quinazolinone core and therefore we tested three further compounds, its deschloro analog 28, and compounds 41 and 33, against CDK9 and Akt (Table 1). The results supported our hypothesis that the chlorine substituent transforms a specific tankyrase inhibitor into a double agent with micromolar potency.
Table 1.
Kinase profiling using selected TNKS inhibitors at 10 μM. Dose response was measured for selected kinase-inhibitor pairs and inhibition-% or an IC50 is shown.
| Compound | CDK9 | Akt | p42 | p38 | GSK3β | CK2 | TNKS2 |
|---|---|---|---|---|---|---|---|
|
34% | 1% | 14% | 3% | 57% | 12% | 400 nM |
|
31% | 2% | 3% | 3% | 11% | 1% | 15 nM |
|
16% | 48% | 18% | 20% | 4% | 2% | 10 nM |
|
6.2 µM | 14.7 µM | — | — | — | — | 660 nM |
|
16.1 µM* | 13.3 µM | — | — | — | — | 16 nM |
|
3.4 µM | 1.6 µM | 51% | 45% | 41% | 2% | 1,050 nM |
|
28.3 µM* | 13.8 µM | — | — | — | — | 145 nM |
|
12% | 7% | 2% | 3% | 7% | 1% | 5 nM |
|
9% | 1% | 3% | 3% | 3% | 1% | 25 nM |
TNKS2 IC50 values are shown for comparison.
*The compound does not completely inhibit the enzyme at concentrations used. IC50 value extrapolated by the analysis software might be not accurately estimated.
Binding modes of 41 and 42 into CDK9 and Akt
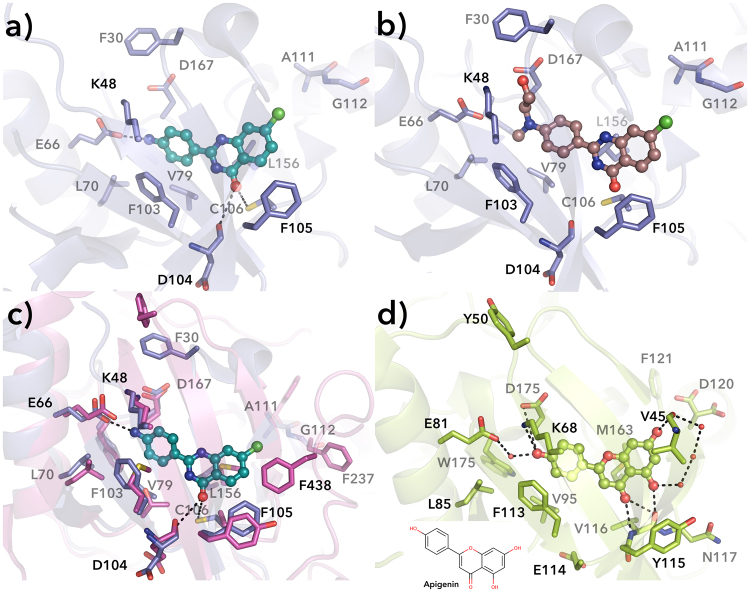
In order to determine structural features that make compounds 41 and 42 potent kinase/TNKS inhibitors, computational studies were performed. Docking of 42 into CDK9 structure (PDB code 3TNH) indicated potential binding modes of the compound to the kinase active site (Figure S3). In the best poses, compound 42 makes two hydrogen bonds with the side chain of Cys106 and the backbone amide of Asp104, while another hydrogen bond is formed between the para-amino group and the side chain of Glu66. In addition, the C-ring is well positioned for π-π stacking interaction with Phe103 (Fig. 7a). These interactions anchor 42 into the active site of CDK9 and may explain its high binding affinity in comparison to other compounds of the same series. The substitution of the para-amino group by a larger 2-(methylamino)ethanol ones in derivative 41 appears to abolish all of the interactions of 42 and the compound is moved out from the pocket towards Phe105 in poses similar to compound 42 (Fig. 7b, Figure S3). The loss of some interactions respect to amino derivative 42 is in line with the observed two-fold reduction in potency from 3.4 µM of 42 to 6.2 µM of 41 (Table 1).
Figure 7.
Structural representation showing the binding site of the inhibitors. (a) 42 and (b) 41 were docked into the active site of CDK9 (PDB code 3TNH). (c) Superposed structures of CDK9 (PDB code 3TNH, blue) and Akt (PDB code 4EKK, magenta)41,42 including compound 42. (d) Crystal structure of CK2 complexed with apigenin (PDB code 4DGM, green)43. Black dashed lines represent hydrogen bonds and red spheres represent water molecules.
The superposition of CDK9 (3TNH) and Akt (4EKK) structures revealed striking resemblance of the active sites with key residues being mostly conserved. Indeed, compounds 41 and 42 could maintain the same binding poses within the Akt active site as shown in Fig. 7c for compound 42. While compound 42 is a potent Akt inhibitor (1.6 µM), in the same order of CDK9 inhibition, compound 41 is a weaker inhibitor (14.7 µM). The difference could result from a smaller binding pocket in Akt lined by Phe438.
The binding mode of the compounds described here is similar to the one observed for apigenin. Apigenin has a similar shape as 2-PQs and a co-crystal structure with CK2 (PDB code 4DGM) (Fig. 7d) resembles the binding mode predicted for CDK9. However, we did not observe inhibition of CK2 by the 2-PQs (Table 1), which could be due to the unfavorable/missing interaction with Val45 (Fig. 7d). Compound potencies, co-crystal structures, and docking to the active sites of kinases indicate that the para-substitution is required for improving tankyrase potency and selectivity, whereas smaller substituents in the same position also fit into the binding pocket of CDK9, CK2 and Akt. In addition, a chlorine substituent in the A-ring is beneficial for the kinase inhibitory potency but leads to a potency decrease in TNKS inhibition.
Discussion
In this study we screened and profiled a family of CDK9 inhibitors against TNKS2. Even though most of these compounds were developed as CDK9 inhibitors, they had a quinazolinone-based scaffold, which is also a potent TNKS1/2 inhibitor scaffold. The compounds were able to inhibit TNKS2 with greater potency than CDK9. Furthermore, the SAR observed for these compounds were similar to other TNKS1/2 inhibitors that bind to the nicotinamide subsite such as flavones, arylquinazolinones, pyridopyrimidinones, dihydroquinazolinones and tetrahydroquinazolinones18–21,24. The most potent compounds were those characterized by a substitution in the para-position of the C-ring. Structural analyses revealed that the para-substituent improves the potency through hydrophobic interactions with Pro1034 and Phe1035 residues in the binding pocket of TNKS2. When larger para-substituents are present, new hydrogen bonds are created in addition to the described hydrophobic interactions, as exemplified by compounds 2 and 9.
Profiling of the most potent tankyrase inhibitors of the series against other ARTDs confirmed the selectivity of the scaffold in general towards tankyrases. Among the most potent TNKS inhibitors, compounds 30 and 32 were also able to interfere with the Wnt/β-catenin pathway in cells, when assayed at 1 μM in the TCF/LEF reporter assay.
Notable structural features emerged for selective TNKS inhibition and dual TNKS/kinases inhibition from the profiling a set of compounds against a panel of Wnt-signaling related kinases and CDK9. The presence of a chlorine atom on the A-ring imparted the same inhibitor potency towards TNKS and CDK9 as in compounds 41 and 42. The most potent CDK9 inhibitor of the series, compound 42 (IC50 = 3.4 µM), characterized by a small para-substituent, exhibited the same high potency also against Akt (IC50 = 1.6 µM). Interestingly, the anti-kinase activities are in the same order as the TNKS2 inhibition (IC50 = 1.1 µM) making compound 42 a promising dual agent. Thus, in order to achieve selectivity towards TNKS1/2 the para-substitution of the C-ring of the 2-PQs is crucial and A-ring substitution shifts the potency towards kinases. The study gives potential routes for the optimization of the 2-PQs scaffold to obtain selective TNKS inhibitors while avoiding kinase inhibition, and/or to generate more potent double agents in the future.
Even though the idea of develop double agent for intentional co-inhibition of kinases and TNKSs is fairly new, there are several examples where ARTD inhibitors were discovered to inhibit other therapeutic targets or potentiate the synergistic effect of other drugs. The typical and most studied example is that of Olaparib, which was discovered as an ARTD1 inhibitor and later found to inhibit other ARTDs including TNKSs31. Olaparib (AZD2281) is currently marketed under the trade name of Lynparza and it was approved by FDA for the treatment of homologous recombination (HR)-deficient cancers having BRCA1/2 mutations33. Co-inhibition of ARTDs and histone deacetylases (HDAC) with Olaparib and Suberoylanilide Hydroxamic Acid (SAHA) respectively is an example where a positive synergy was demonstrated using HR-proficient ovarian cancer cell lines against which Olaparib alone has minimal efficacy as a single agent34. The same combination was also proven to be effective in triple negative breast cancer cells and in a xenograft mouse model35. In another scenario, TNKS inhibitor XAV939 was shown to reduce ICP0- and ERK-dependent Herpes simplex viral replication36. Furthermore, TNKS inhibitors enhanced the inhibitory effect of Erlotinib, which is an EGFR inhibitor, by blocking the YAP signaling pathway37. This is a classical example where dual inhibition of two important pathways, YAP and Wnt/β-catenin, was achieved by the use of TNKS inhibitors. Considering that most TNKS inhibitors like 1 are widely used in cell-based assays and in different contexts, the possibility of an off-target effect should be taken into account by e.g. testing in parallel multiple compounds that are now available. On the other hand by careful crafting of the structure it could be possible to develop agents with dual activity on a single compound like in the case of 42.
Methods
Protein expression and purification
Human ARTD1–3 were expressed as full length proteins18,24 whereas ARTD4-7, ARTD10 and ARTD12 were expressed as protein fragments containing the catalytic domain and they were purified according to the reported protocols18,24,38,39.
Activity assay
The activity assay used to measure the potency of the compounds against TNKS2 is based on the quantification of NAD+38. The reactions were carried out in 96-well plate (Greiner bio-one, U-shaped) and the conditions were the same as reported earlier (Table S5)24.
Screening of compounds and potency measurements
The compounds were stored in 100% DMSO at −20 °C. All the compounds were diluted in the TNKS2 assay buffer (50 mM Bis-Tris Propane, 0.5 mM TCEP, 0.01% Triton-100x). Control reactions were used to exclude the effect of compound fluorescence and quenching from the calculations. One dose-response curve was measured using half-log dilutions and in quadruplicates for all the compounds and for the most potent compounds the measurement was repeated three times and on separate days. The pIC50 values were calculated and fitted with nonlinear regression using 4-parameters with GraphPad Prism (version 5.0 for windows).
Profiling of the compounds against ARTDs
The compounds showing high potency against TNKS2 were profiled against other human ARTDs (Table S2). The assay conditions used in the profiling were previously optimized for each enzyme (Table S5)24,40. For all compounds a dose-response curve was measured using half-log dilutions. The concentration of NAD+ was kept constant (500 nM) during the potency measurements.
Kinase profiling
The kinase functional assays were performed using a combination of kinase and 1.5 μM peptide substrate in the presence of ATP. The reaction mixture was analyzed on the Caliper LabChip 3,000 (Caliper, Hopkinton, MA) by the electrophoretic mobility shift of the fluorescent substrate and phosphorylated product. For the inhibition of human CK2α-1, the experiments were carried out with ADP-Glo kinase Assay Kit (Promega V9,101). Assays are described in detail in the Supplementary Methods and Table S6.
Reporter assay
The Wnt Signaling Pathway TCF/LEF Reporter HEK293 Cell Line (BPS Bioscience, catalog #60,501) was used to study the effect of the most potent tankyrase inhibitors on Wnt signaling. The Wnt assay was performed using a procedure modified from the BPS Bioscience protocol. Details are given in the Supplementary Methods.
Crystallization, data collection, refinement and docking
Crystallization of the catalytic domain of TNKS2 (residues 952–1,161) was done similarly as previously described20. Details are given in the Supplementary Methods and Table S7.
Chemistry
The majority of the 2-PQS herein reported were synthesized as previously described26, while compounds 10–12 and 22, 27, 38, 39 were purchased from vendors (10–11, Mir Biotech; 12, Life Chemicals; 22, BAS; 27, Vitas M Lab; 38 and 39, Enamine). Purity of the acquired compounds was measured by HPLC and were higher than 95%. Although compound 2 was commercially available as racemate its synthesis was never been reported before and is shown in (Scheme S1), together with the synthesis of its enantiomers (−)-7 and (+)-8 (Scheme S2), as well as compound 9 (Scheme S3).
Electronic supplementary material
Acknowledgements
We are grateful to Local Contacts at ESRF and at DLS for providing assistance in using the beamlines. This work was funded by Biocenter Oulu, the Sigrid Jusélius Foundation, the Jane and Aatos Erkko Foundation, Magnus Ehrnrooth Foundation and the Academy of Finland (grant no. 287063 and 294085 to LL, 266922 to TH, 294617 to JK and 284605 to TP). The research leading to these results has also received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement N°283570). The use of the facilities and expertise of the Biocenter Oulu core facility, a member of Biocenter Finland and Instruct-FI, is gratefully acknowledged. We also thank the COST action CA15135 (Multi-Target Paradigm for Innovative Ligand Identification in the Drug Discovery Process, MuTaLig) for support.
Author Contributions
Y.N. measured biochemical data and most of the structural data and compiled the manuscript. J.K. T.H., Su. M., E.O. and M.I.L. measured and analyzed biochemical data. J.D., L.S., Se.M. carried out chemical synthesis, and F.I. performed the e HPLC analyses. J.B., T.P., O.T. and L.L. conceived the study and supervised the work. All authors contributed to the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jenny Desantis, Jarkko Koivunen and Teemu Haikarainen contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19872-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Oriana Tabarrini, Email: oriana.tabarrini@unipg.it.
Lari Lehtiö, Email: lari.lehtio@oulu.fi.
References
- 1.Lehtiö L, Chi N-W, Krauss S. Tankyrases as drug targets. FEBS J. 2013;280:3576–3593. doi: 10.1111/febs.12320. [DOI] [PubMed] [Google Scholar]
- 2.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 3.Matsutani N, et al. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int. J. Oncol. 2001;19:507–512. [PubMed] [Google Scholar]
- 4.Sbodio JI, Lodish HF, Chi N-W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem. J. 2002;361:451–459. doi: 10.1042/bj3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seimiya H, Muramatsu Y, Smith S, Tsuruo T. Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol. Cell. Biol. 2004;24:1944–1955. doi: 10.1128/MCB.24.5.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MK, Dudognon C, Smith S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012;13:724–732. doi: 10.1038/embor.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 8.Yeh T-YJ, Sbodio JI, Tsun Z-Y, Luo B, Chi N-W. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem. J. 2007;402:279–290. doi: 10.1042/BJ20060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao R, et al. Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS ONE. 2012;7:e48670. doi: 10.1371/journal.pone.0048670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waaler J, et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012;72:2822–2832. doi: 10.1158/0008-5472.CAN-11-3336. [DOI] [PubMed] [Google Scholar]
- 11.Ma, L. et al. Tankyrase inhibitors attenuate WNT/β-catenin signaling and inhibit growth of hepatocellular carcinoma cells. Oncotarget (2015). [DOI] [PMC free article] [PubMed]
- 12.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 13.Cadigan, K. M. & Waterman, M. L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol4 (2012). [DOI] [PMC free article] [PubMed]
- 14.Huang S-MA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 15.Haikarainen, T., Krauss, S. & Lehtiö, L. Tankyrases: Structure, Function and Therapeutic Implications in Cancer. Curr. Pharm. Des. (2014). [DOI] [PMC free article] [PubMed]
- 16.Karlberg T, et al. Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J. Med. Chem. 2010;53:5352–5355. doi: 10.1021/jm100249w. [DOI] [PubMed] [Google Scholar]
- 17.Lehtiö L, et al. Zinc binding catalytic domain of human tankyrase 1. J. Mol. Biol. 2008;379:136–145. doi: 10.1016/j.jmb.2008.03.058. [DOI] [PubMed] [Google Scholar]
- 18.Haikarainen T, et al. para-Substituted 2-phenyl-3,4-dihydroquinazolin-4-ones as potent and selective tankyrase inhibitors. ChemMedChem. 2013;8:1978–1985. doi: 10.1002/cmdc.201300337. [DOI] [PubMed] [Google Scholar]
- 19.Nathubhai A, Wood PJ, Lloyd MD, Thompson AS, Threadgill MD. Design and Discovery of 2-Arylquinazolin-4-ones as Potent and Selective Inhibitors of Tankyrases. ACS Med. Chem. Lett. 2013;4:1173–1177. doi: 10.1021/ml400260b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narwal M, Haikarainen T, Fallarero A, Vuorela PM, Lehtiö L. Screening and structural analysis of flavones inhibiting tankyrases. J. Med. Chem. 2013;56:3507–3517. doi: 10.1021/jm3018783. [DOI] [PubMed] [Google Scholar]
- 21.Narwal M, et al. Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity. J. Med. Chem. 2013;56:7880–7889. doi: 10.1021/jm401463y. [DOI] [PubMed] [Google Scholar]
- 22.Kumpan K, et al. Structure-based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases. Bioorg. Med. Chem. 2015;23:3013–3032. doi: 10.1016/j.bmc.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Paine HA, et al. Exploration of the nicotinamide-binding site of the tankyrases, identifying 3-arylisoquinolin-1-ones as potent and selective inhibitors in vitro. Bioorg. Med. Chem. 2015;23:5891–5908. doi: 10.1016/j.bmc.2015.06.061. [DOI] [PubMed] [Google Scholar]
- 24.Nkizinkiko Y, et al. Discovery of potent and selective nonplanar tankyrase inhibiting nicotinamide mimics. Bioorg. Med. Chem. 2015;23:4139–4149. doi: 10.1016/j.bmc.2015.06.063. [DOI] [PubMed] [Google Scholar]
- 25.Johannes JW, et al. Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology. ACS Med Chem Lett. 2015;6:254–259. doi: 10.1021/ml5003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sancineto L, et al. Computer-aided design, synthesis and validation of 2-phenylquinazolinone fragments as CDK9 inhibitors with anti-HIV-1 Tat-mediated transcription activity. ChemMedChem. 2013;8:1941–1953. doi: 10.1002/cmdc.201300287. [DOI] [PubMed] [Google Scholar]
- 27.Massari S, Sabatini S, Tabarrini O. Blocking HIV-1 replication by targeting the Tat-hijacked transcriptional machinery. Curr. Pharm. Des. 2013;19:1860–1879. doi: 10.2174/1381612811319100010. [DOI] [PubMed] [Google Scholar]
- 28.Tabarrini O, Desantis J, Massari S. Recent advances in the identification of Tat-mediated transactivation inhibitors: progressing toward a functional cure of HIV. Future Med Chem. 2016;8:421–442. doi: 10.4155/fmc.16.3. [DOI] [PubMed] [Google Scholar]
- 29.Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell. 2005;7:25–37. doi: 10.1016/j.ccr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Cerone MA, Burgess DJ, Naceur-Lombardelli C, Lord CJ, Ashworth A. High-throughput RNAi screening reveals novel regulators of telomerase. Cancer Res. 2011;71:3328–3340. doi: 10.1158/0008-5472.CAN-10-2734. [DOI] [PubMed] [Google Scholar]
- 31.Narwal M, Venkannagari H, Lehtiö L. Structural basis of selective inhibition of human tankyrases. J. Med. Chem. 2012;55:1360–1367. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- 32.Voronkov A, et al. Structural basis and SAR for G007-LK, a lead stage 1,2,4-triazole based specific tankyrase 1/2 inhibitor. J. Med. Chem. 2013;56:3012–3023. doi: 10.1021/jm4000566. [DOI] [PubMed] [Google Scholar]
- 33.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 34.Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol. Oncol. 2014;133:599–606. doi: 10.1016/j.ygyno.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min A, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015;17:33. doi: 10.1186/s13058-015-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J. Virol. 2012;86:492–503. doi: 10.1128/JVI.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, et al. Tankyrase Inhibitor Sensitizes Lung Cancer Cells to Endothelial Growth Factor Receptor (EGFR) Inhibition via Stabilizing Angiomotins and Inhibiting YAP Signaling. J. Biol. Chem. 2016;291:15256–15266. doi: 10.1074/jbc.M116.722967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narwal M, Fallarero A, Vuorela P, Lehtiö L. Homogeneous screening assay for human tankyrase. J Biomol Screen. 2012;17:593–604. doi: 10.1177/1087057112436558. [DOI] [PubMed] [Google Scholar]
- 39.Venkannagari H, Fallarero A, Feijs KLH, Lüscher B, Lehtiö L. Activity-based assay for human mono-ADP-ribosyltransferases ARTD7/PARP15 and ARTD10/PARP10 aimed at screening and profiling inhibitors. Eur J Pharm Sci. 2013;49:148–156. doi: 10.1016/j.ejps.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Venkannagari, H. et al. Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem Biol10.1016/j.chembiol.2016.08.012 (2016). [DOI] [PubMed]
- 41.Baumli S, Hole AJ, Noble MEM, Endicott JA. The CDK9 C-helix exhibits conformational plasticity that may explain the selectivity of CAN508. ACS Chem. Biol. 2012;7:811–816. doi: 10.1021/cb2004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin K, et al. An ATP-site on-off switch that restricts phosphatase accessibility of Akt. Sci Signal. 2012;5:ra37. doi: 10.1126/scisignal.2002618. [DOI] [PubMed] [Google Scholar]
- 43.Lolli G, et al. Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight. Biochemistry. 2012;51:6097–6107. doi: 10.1021/bi300531c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.