Abstract
Combination therapy provides a useful therapeutic approach to overcome resistance until new antibiotics become available. In this study, the pharmacodynamics, including the morphological effects, of polymyxin B (PMB) and meropenem alone and in combination against KPC-producing Klebsiella pneumoniae clinical isolates was examined. Ten clinical isolates were obtained from patients undergoing treatment for mediastinitis. KPCs were identified and MICs were measured using microbroth dilution. Time–kill studies were conducted over 24 h with PMB (0.5–16 mg/L) and meropenem (20–120 mg/L) alone or in combination against an initial inoculum of ca. 106 CFU/mL. Scanning electron microscopy (SEM) was employed to analyse changes in bacterial morphology after treatment, and the log change method was used to quantify the pharmacodynamic effect. All isolates harboured the blaKPC-2 gene and were resistant to meropenem (MICs ≥8 mg/L). Clinically relevant PMB concentrations (0.5, 1.0 and 2.0 mg/L) in combination with meropenem were synergistic against all isolates except BRKP28 (polymyxin- and meropenem-resistant, both MICs >128 mg/L). All PMB and meropenem concentrations in combination were bactericidal against polymyxin-susceptible isolates with meropenem MICs ≤16 mg/L. SEM revealed extensive morphological changes following treatment with PMB in combination with meropenem compared with the changes observed with each individual agent. Additionally, morphological changes decreased with increasing resistance profiles of the isolate, i.e. increasing meropenem MIC. These antimicrobial effects may not only be a summation of the effects due to each antibiotic but also a result of differential action that likely inhibits protective mechanisms in bacteria.
Keywords: Polymyxin B, Meropenem, KPC carbapenemase, Klebsiella pneumoniae, Combination, Pharmacodynamics
1. Introduction
The incidence of carbapenem-resistant Enterobacteriaceae (CRE), particularly Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae, has increased over the past decade, and infections caused by these bacteria are associated with significantly high rates of treatment failure and mortality [1,2]. Carbapenemase-encoding genes are easily transferred by plasmid-mediated conjugation to other susceptible strains, thereby conferring carbapenem resistance to other organisms [3]. The US Centers for Disease Control and Prevention (CDC) estimates that CRE are responsible for ca. 9300 (ca. 6.6%) of the estimated 140,000 healthcare-associated Enterobacteriaceae infections in the USA each year [4].
The increasing burden of CRE infections, the high rate of treatment failure, the emergence of resistance during treatment [5,6] and the dearth of novel antimicrobial agents highlight the urgency to investigate effective approaches for treating these infections. Polymyxins [polymyxin B (PMB) and polymyxin E (colistin)] are rapidly bactericidal against KPC-producing K. pneumoniae [7]. It is believed that interactions between polymyxins and the lipopolysaccharide (LPS) present in the outer membrane of Gram-negative bacteria increase membrane permeability and the loss of intracellular contents, ultimately resulting in cell death [8]. However, concerns for the rapid emergence of resistance to polymyxins during treatment as well as dose-limiting nephrotoxicity favour the clinical use of combination therapy for treatment of CRE infections [9]. Furthermore, improved outcomes have been reported from select prospective clinical studies of polymyxin-based combination therapy for multidrug-resistant (MDR) Acinetobacter baumannii infections [10].
However, the role of polymyxin-based combinations in the treatment of infections caused by KPC-producing K. pneumoniae has not been well studied. The role of carbapenems as part of combination therapy against isolates with varying degrees of susceptibility needs to be explored in order to optimise dosage regimens. Meropenem, a broad-spectrum carbapenem, is active against Enterobacteriaceae owing to its high binding affinity for penicillin-binding protein 2 (PBP2), PBP3 and PBP4, which inhibits cell wall formation and facilitates bacterial cell lysis [11]. The objective of this study was to evaluate the pharmacodynamics of PMB in combination with meropenem against MDR KPC-producing K. pneumoniae with a range of susceptibility for both antibiotics and to assess the impact of this combination on bacterial morphology using scanning electron microscopy (SEM).
2. Materials and methods
2.1. Bacterial strains and antibiotics
Ten K. pneumoniae clinical isolates were obtained from ten different patients during outbreaks of KPC infections that occurred between June 2009 and June 2013 at Instituto Dante Pazzanese de Cardiologia (Sao Paulo, Brazil), a tertiary hospital specialising in cardiovascular surgery (Table 1) [12]. All isolates from this single cardiac treatment facility were stored at −80 °C and were subcultured onto Mueller–Hinton agar plates before each experiment.
Table 1.
Antibiotic minimum inhibitory concentrations (MICs) and genotyping results of ten multidrug-resistant KPC-2-producing Klebsiella pneumoniae strains isolated from mediastinitis patients.
| Isolate | MIC (mg/L)a
|
KPC-2 carbapenemase | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMB | CST | MEM | AMK | ATM | CAZ | CHL | CIP | FOF | GEN | MIN | RIFb | TGCc | TMP | ||
| BRKP30 | 0.5 S | <0.5 S | 8 R | 4 S | 64 R | >256 R | 128 R | 128 R | 16 S | 1 S | 4 S | 32 | 1 S | >256 R | + |
| BRKP20 | <0.5 S | 0.5 S | 16 R | 4 S | 64 R | >256 R | 32 R | 128 R | 16 S | <0.5 S | 8 I | 32 | 1 S | >256 R | + |
| BRKP22 | <0.5 S | <0.5 S | 16 R | 1 S | 64 R | 16 R | 64 R | 128 R | 16 S | <0.5 S | 32 R | 32 | 2 | >256 R | + |
| BRKP31 | <0.5 S | <0.5 S | 16 R | <0.5 S | 64 R | 64 R | 32 R | 64 R | 8 S | 32 R | 2 S | 32 | 1 S | >256 R | + |
| BRKP21 | 0.5 S | 0.5 S | 16 R | 2 S | 64 R | >256 R | 128 R | 128 R | 16 S | <0.5 S | 8 I | 32 | 2 | >256 R | + |
| BRKP27 | 1 S | 1 S | 16 R | 2 S | 64 R | 32 R | 32 R | 64 R | 8 S | 64 R | 4 S | 32 | 2 | >256 R | + |
| BRKP67 | 8 R | 16 R | 64 R | 4 S | 32 R | 128 R | 128 R | 128 R | 64 R | 128 R | 32 R | 32 | 8 R | >256 R | + |
| BRKP76 | <0.5 S | <0.5 S | 64 R | 2 S | >256 R | 32 R | >256 R | 64 R | 32 S | 128 R | 1 S | 64 | <0.5 S | 4 S | + |
| BRKP61 | <0.5 S | 0.5 S | 128 R | 8 S | 32 R | 32 R | 32 R | 128 R | 256 R | <0.5 S | 1 S | 32 | <0.5 S | >256 R | + |
| BRKP28 | >128 R | >128 R | 256 R | 2 S | 32 R | 128 R | 32 R | 64 R | 128 I | 64 R | 2 S | 16 | 1 S | >256 R | + |
PMB, polymyxin B; CST, colistin; MEM, meropenem; AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; FOF, fosfomycin; GEN, gentamicin; MIN, minocycline; RIF, rifampicin; TGC, tigecycline; TMP, trimethoprim; S, susceptible; I, intermediate; R, resistant; EUCAST, European Committee on Antimicrobial Susceptibility Testing; CLSI, Clinical and Laboratory Standards Institute.
The isolates were classified as susceptible, intermediate or resistant based on the 2015 EUCAST and CLSI breakpoints against Enterobacteriaceae.
Neither the CLSI nor EUCAST has defined rifampicin breakpoints for Gram-negative organisms.
EUCAST has defined breakpoint concentrations of tigecycline for susceptible and resistant, but no information has been provided for intermediate.
Fresh stock solutions of PMB and meropenem were prepared prior to each experiment by dissolving polymyxin B sulphate powder (Sigma-Aldrich, St Louis, MO; lot #WXBB4470V) and meropenem (AK Scientific, Union City, CA; lot #LC24337) in water and normal saline, respectively, and filter-sterilising through a 0.20 μm syringe filter (Corning Inc., Corning, NY).
2.2. K. pneumoniae genotyping
DNA was isolated from bacterial isolates using an E.Z.N.A.® Bacterial DNA Kit (Omega Bio-tek, Norcross, GA). Seven primer sets for β-lactamases of Ambler classes A (GES and KPC), B (NDM, VIM and IMP) and D (OXA-48 and OXA-40) [13] as well as one set of primers for the mgrB gene [14] were used in the PCR for characterisation of the isolates (Supplementary Table S1). Q5 Hi-Fidelity Taq DNA Polymerase (NEB, Ipswich, MA) was used in PCR reactions according to the manufacturer’s instructions. Reactions were carried out in an Eppendorf Mastercycler® (Eppendorf, Hamburg, Germany) and were analysed by agarose gel electrophoresis. PCR product sequencing was performed at the sequencing facility at the Roswell Park Cancer Institute (Buffalo, NY). The National Center for Biotechnology Information (NCBI) website was used for nucleotide and deduced protein sequence analysis (http://www.ncbi.nlm.nih.gov). Insertion sequences (IS) were analysed using the ISFinder website (http://www-is.biotoul.fr).
2.3. Bacterial isolates and antimicrobial susceptibility testing
The susceptibility of each isolate was determined for the antibiotics listed in Table 1 using the broth microdilution method in cation-adjusted (25.0 mg/L Ca2+ and 12.5 mg/L Mg2+) Mueller–Hinton broth (Becton Dickinson & Co., Franklin Lakes, NJ), according to Clinical and Laboratory Standards Institute (CLSI) guidelines [15]. The minimum inhibitory concentration (MIC) breakpoints for meropenem and colistin were defined according to the CLSI [15] and European Committee on Antimicrobial Susceptibility Testing (EUCAST) [16], respectively.
2.4. Time–kill studies
Static time–kill kinetics were examined to determine the rate and extent of bacterial killing in the absence (growth control) and presence of PMB and meropenem as monotherapy and in combination against the isolates. PMB concentrations of 0.5, 1, 2, 4, 8, 16, 64, 128 and 256 mg/L and meropenem concentrations of 10, 20, 40, 60 and 120 mg/L and a 5 × 4 concentration array of PMB (0.5, 1, 2, 4 and 16 mg/L) in combination with meropenem (20, 40, 60 and 120 mg/L) were evaluated against an initial inoculum of ca. 106 CFU/ mL. Antibiotic(s) was added to the bacterial suspension in log growth phase. Bacterial samples obtained at 0, 1, 2, 4, 6, 8 and 24 h were diluted with normal saline and the appropriate dilution of bacterial cell suspension (50 μL) was spirally plated on Mueller–Hinton agar using a Whitley automatic spiral plater (Don Whitley Scientific Ltd., Shipley, UK). Bacteria were quantified using a ProtoCOL HR automated bacterial colony counter (Synbiosis, Frederick, MD) following 24 h of incubation at 37 °C. The lower limit of quantification was 2.0 log10 CFU/mL.
The PMB concentrations selected were based on clinically achievable unbound plasma concentrations in critically ill patients following intravenous administration based on current dosing recommendations [17,18]. Meropenem plasma concentrations in critically ill patients following a 3-h extended infusion were 30.2 ± 10.7 μg/ mL with 1 g and 67.1 ± 31.0 μg/mL with 2 g [19]. Given that meropenem is only 2% bound to plasma [20], the concentrations employed in the current study (10, 20, 40, 60 and 120 mg/L) include the full range of clinically achievable unbound meropenem plasma concentrations.
The killing activity of the various individual and combination therapies was evaluated by characterising the pharmacodynamic effect (E) assessed according to the change in log10 CFU/mL at an early time point of 4 h and at later time points of 8 h and 24 h compared with the log10 CFU/mL at baseline (0 h) (point-based analysis [21]) according to the equation below:
Bactericidal activity was defined as a ≥3 log10 CFU/mL reduction from baseline. Synergy was defined as a ≥2 log10 CFU/mL reduction and additivity as ≥1 to <2 log10 CFU/mL reduction by the combination of PMB and meropenem compared with the most active single agent in the combination.
2.5. Scanning electron microscopy
A standard procedure was followed for SEM analysis [22] with minor modifications. Briefly, bacterial broth cultures in log phase (20 mL) at ca. 108 CFU/mL were treated with freshly prepared stock solutions of PMB (0.5 mg/L) or meropenem (20 mg/L) as monotherapy and in combination with 1.5 h of incubation at 37 °C in a shaking water-bath. All samples were centrifuged at 4000 × g for 10 min, were washed and were re-suspended in phosphate-buffered saline. Bacterial cells were fixed with 2.5% glutaraldehyde and were then incubated at 37 °C on a glass slide (22 × 22 mm) for 20 min. Samples were dehydrated by passing through graded ethanol (10%, 30%, 50%, 70%, 90% and 100%) for 5 min at each concentration. Slides were then dried at 37 °C for 20 min and a small amount of gold was spluttered onto the samples using a sputter coater system (SPI-Module™ Sputter Coater; Structure Probe Inc., West Chester, PA) to avoid charging in the microscope. For imaging, focused ion beam scanning electron microscopy (FIB-SEM) was performed using a Carl Zeiss AURIGA crossbeam microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY) at 2 kV.
3. Results
3.1. Characterisation of the isolates
PCR amplification predicted the blaKPC-2 gene (785-bp amplicon) in all isolates, confirmed by direct sequencing of the amplicon (Table 1). PMB-sensitive isolates generated an mgrB gene amplicon comparable in size with that in wild-type isolates. Polymyxin-resistant isolate BRKP67 generated a larger amplicon compared with wild-type isolates, suggestive of insertion of a DNA sequence in the mgrB gene. Sequencing of the amplicon revealed inactivation of mgrB by insertion of ISKpn13 (1148 bp), an IS5-like element, in the coding region between nucleotides 74 and 75. In another PMB-resistant isolate (BRKP28), sequencing analysis revealed a premature stop codon in mgrB resulting in a truncated 23-amino acid MgrB protein.
Antimicrobial susceptibility testing revealed a MDR profile for all of the isolates (Table 1). All of the isolates were resistant to meropenem (MICs ranging from 8 to 256 mg/L) and two isolates were resistant to PMB (MICs of 8 mg/L and >128 mg/L) (Table 1).
3.2. Polymyxin B and meropenem monotherapy
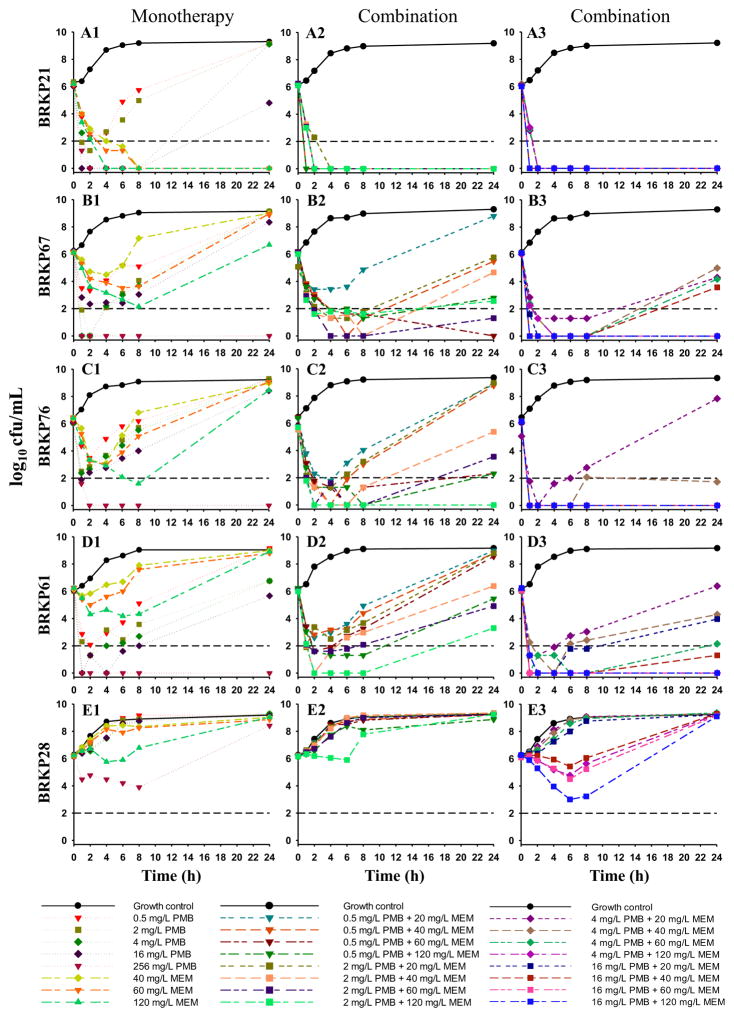
The lower PMB concentrations of 0.5, 1, 2 and 4 mg/L resulted in a ≥2 log10 reduction against all isolates by 4 h except for BRKP28 (resistant to both PMB and meropenem) (Fig. 1; Supplementary Fig. S1; Supplementary Table S2). This initial killing was not sustained, with bacterial re-growth observed by 24 h (Fig. 1; Supplementary Fig. S1). The higher PMB concentrations of 8 mg/L and 16 mg/L resulted in early bactericidal activity by 4 h against all isolates except BRKP28, followed by re-growth in seven (BRKP30, 31, 27, 21, 67, 76 and 61) of the nine isolates by 24 h. The highest PMB concentrations of 64, 128 and 256 mg/L resulted in a ≥4 log10 reduction against nine of the ten isolates with the exception of BRKP28 by 4 h. Furthermore, the bactericidal activity of PMB at 64 mg/L and 128 mg/L was sustained over 24 h against seven (BRKP30, 20, 22, 31, 21, 76 and 61) of the isolates with PMB MICs of ≤0.5 mg/L; the bactericidal activity of PMB at 256 mg/L was sustained against nine isolates except BRKP28 by 24 h (Fig. 1; Supplementary Fig. S1; Supplementary Table S2). The lower meropenem concentration of 10 mg/L did not show any significant effect on bacterial growth, but 20 mg/L was bactericidal against BRKP27 and BRKP31 by 24 h. Higher meropenem concentrations of 40, 60 and 120 mg/L resulted in sustained bactericidal activity against all isolates with meropenem MICs ≤16 mg/L (BRKP30, 20, 22, 27, 31 and 21), but activity was similar to the growth control by 24 h against isolates with meropenem MICs >16 mg/L (BRKP76, 67, 61 and 28) (Fig. 1; Supplementary Fig. S1; Supplementary Table S3).
Fig. 1.
Time–kill curves with polymyxin B (PMB) and meropenem (MEM) alone and in combination against an initial inoculum of ca. 106 CFU/mL of PMB-susceptible clinical isolates BRKP21, BRKP61 and BRKP76 and PMB-resistant isolates BRKP67 and BRKP28 over 24 h: PMB (0.5, 2, 4, 16 and 256 mg/L) and MEM (40, 60 and 120 mg/L) as monotherapy (A1–E1, monotherapy); PMB (0.5 mg/L and 2 mg/L) in combination with meropenem (20, 40, 60 and 120 mg/L) (A2–E2, combination); and PMB (4 mg/L and 16 mg/L) in combination with meropenem (20, 40, 60 and 120 mg/L) (A3–E3, combination). Black dashed line represents the limit of quantification for CFU/mL bacterial count.
3.3. Polymyxin B in combination with meropenem
PMB at all concentrations in combination with meropenem at 20, 40, 60 and 120 mg/L resulted in sustained bactericidal activity against the six PMB-susceptible isolates with meropenem MICs of ≤16 mg/L (BRKP20, 21, 22, 27, 30, and 31) (Fig. 1; Table 2; Supplementary Fig. S1). Against BRKP76 and BRKP61 (PMB-susceptible isolates with meropenem MICs ≥64 mg/L) and BRKP67 [PMB-resistant isolate (MIC = 8 mg/L), meropenem MIC = 64 mg/L], all PMB concentrations in combination with all meropenem concentrations resulted in early bactericidal activity (Fig. 1; Table 2). However, only PMB at 1, 2, 4 and 16 mg/L in combination with the highest meropenem concentration of 120 mg/L demonstrated sustained killing activity over 24 h. Combinations of PMB at 0.5, 1 and 2 mg/L with meropenem 120 mg/L were synergistic against BRKP67, BRKP76 and BRKP61. The higher PMB concentrations of 4 mg/L and 16 mg/L were synergistic when combined with lower meropenem concentrations of 40 mg/L and 60 mg/L and the highest meropenem concentration of 120 mg/L against BRKP67, BRKP76 and BRKP61. None of the PMB and meropenem combinations were effective against the highly PMB- and meropenem-resistant isolate BRKP28 by 24 h (Fig. 1; Table 2).
Table 2.
Summary of changes in bacterial density (log10 CFU/mL) at 4, 8 and 24 h compared with the initial inoculum (0 h) for polymyxin B (PMB) in combination with meropenem (MEM).
| Strain | Time (h) | Control | PMB 0.5 mg/L | PMB 1 mg/L | PMB 2 mg/L | PMB 4 mg/L | PMB 16 mg/L | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||
| MEM 20 mg/L | MEM 40 mg/L | MEM 60 mg/L | MEM 120 mg/L | MEM 20 mg/L | MEM 40 mg/L | MEM 60 mg/L | MEM 120 mg/L | MEM 20 mg/L | MEM 40 mg/L | MEM 60 mg/L | MEM 120 mg/L | MEM 20 mg/L | MEM 40 mg/L | MEM 60 mg/L | MEM 120 mg/L | MEM 20 mg/L | MEM 40 mg/L | MEM 60 mg/L | MEM 120 mg/L | |||
| BRKP30 | 4 | 2.28 | −4.60 | −4.37 | −6.18 | −4.93 | −3.81 | −4.07 | −4.84 | −6.03 | −5.03 | −6.18 | −6.21 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 |
| 8 | 2.80 | −6.20 | −4.85 | −6.18 | −6.23 | −6.22 | −6.27 | −6.14 | −6.03 | −6.33 | −6.18 | −6.21 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 | |
| 24 | 2.93 | −6.20 | −6.15 | −6.18 | −6.23 | −6.22 | −6.27 | −6.14 | −6.03 | −6.33 | −6.18 | −6.21 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 | |
| BRKP20 | 4 | 2.45 | −6.24 | −6.26 | −6.26 | −6.20 | −4.88 | −4.92 | −6.10 | −6.19 | −6.15 | −6.18 | −4.94 | −6.15 | −6.15 | −6.16 | −6.23 | −6.18 | −6.18 | −6.08 | −6.18 | −6.23 |
| 8 | 2.94 | −6.24 | −6.26 | −6.26 | −6.20 | −6.18 | −6.23 | −6.10 | −6.19 | −6.15 | −6.18 | −6.24 | −6.15 | −6.15 | −6.16 | −6.23 | −6.18 | −6.18 | −6.08 | −6.18 | −6.23 | |
| 24 | 3.10 | −6.24 | −6.26 | −6.26 | −6.20 | −6.18 | −6.23 | −6.10 | −6.19 | −6.15 | −6.18 | −6.24 | −6.15 | −6.15 | −6.16 | −6.23 | −6.18 | −6.18 | −6.08 | −6.18 | −6.23 | |
| BRKP22 | 4 | 2.42 | −6.24 | −6.22 | −6.23 | −6.41 | −6.06 | −6.01 | −6.01 | −6.13 | −6.03 | −6.02 | −6.01 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 |
| 8 | 2.89 | −6.24 | −6.22 | −6.23 | −6.41 | −6.06 | −6.01 | −6.01 | −6.13 | −6.03 | −6.02 | −6.01 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 | |
| 24 | 2.98 | −6.24 | −6.22 | −6.23 | −6.41 | −6.06 | −6.01 | −6.01 | −6.13 | −6.03 | −6.02 | −6.01 | −6.05 | −6.17 | −6.16 | −6.12 | −6.01 | −6.23 | −6.10 | −6.11 | −6.10 | |
| BRKP31 | 4 | 2.42 | −6.19 | −6.22 | −6.35 | −6.19 | −6.28 | −6.21 | −6.29 | −6.24 | −6.20 | −6.24 | −6.35 | −6.21 | −6.29 | −6.38 | −6.25 | −6.31 | −6.23 | −6.30 | −6.24 | −6.21 |
| 8 | 2.91 | −6.19 | −6.22 | −6.35 | −6.19 | −6.28 | −6.21 | −6.29 | −6.24 | −6.20 | −6.24 | −6.35 | −6.21 | −6.29 | −6.38 | −6.25 | −6.31 | −6.23 | −6.30 | −6.24 | −6.21 | |
| 24 | 3.17 | −6.19 | −4.92 | −6.35 | −6.19 | −6.28 | −6.21 | −6.29 | −6.24 | −6.20 | −6.24 | −6.35 | −6.21 | −6.29 | −6.38 | −6.25 | −6.31 | −6.23 | −6.30 | −6.24 | −6.21 | |
| BRKP27 | 4 | 1.30 | −5.04 | −5.11 | −6.31 | −6.34 | −6.09 | −6.11 | −4.73 | −6.20 | −4.78 | −6.04 | −6.09 | −6.21 | −6.11 | −6.19 | −6.20 | −6.09 | −6.09 | −6.15 | −6.25 | −6.11 |
| 8 | 2.51 | −6.34 | −6.41 | −6.31 | −6.34 | −6.09 | −6.11 | −6.03 | −6.20 | −6.08 | −6.04 | −6.09 | −6.21 | −6.11 | −6.19 | −6.20 | −6.09 | −6.09 | −6.15 | −6.25 | −6.11 | |
| 24 | 2.51 | −6.34 | −6.41 | −6.31 | −6.34 | −6.09 | −6.11 | −6.03 | −6.20 | −6.08 | −6.04 | −6.09 | −6.21 | −6.11 | −6.19 | −6.20 | −6.09 | −6.09 | −6.15 | −6.25 | −6.11 | |
| BRKP21 | 4 | 2.34 | −6.16 | −6.18 | −6.18 | −6.18 | −6.15 | −6.19 | −6.19 | −6.17 | −6.11 | −6.20 | −6.25 | −6.11 | −6.07 | −6.20 | −6.13 | −6.09 | −6.05 | −6.06 | −6.01 | −6.02 |
| 8 | 2.85 | −6.16 | −6.18 | −6.18 | −6.18 | −6.15 | −6.19 | −6.19 | −6.17 | −6.11 | −6.20 | −6.25 | −6.11 | −6.07 | −6.20 | −6.13 | −6.09 | −6.05 | −6.06 | −6.01 | −6.02 | |
| 24 | 3.07 | −6.16 | −6.18 | −6.18 | −6.18 | −6.15 | −6.19 | −6.19 | −6.17 | −6.11 | −6.20 | −6.25 | −6.11 | −6.07 | −6.20 | −6.13 | −6.09 | −6.05 | −6.06 | −6.01 | −6.02 | |
| BRKP67 | 4 | 2.50 | −2.77 | −4.31 | −4.31 | −4.24 | −3.30 | −4.52 | −4.88 | −4.87 | −3.77 | −4.79 | −6.14 | −4.22 | −4.70 | −6.00 | −6.01 | −6.00 | −6.09 | −6.10 | −6.11 | −6.11 |
| 8 | 2.83 | −1.33 | −4.91 | −4.61 | −4.85 | −1.86 | −6.12 | −6.18 | −6.17 | −3.29 | −6.09 | −6.14 | −4.40 | −4.70 | −6.00 | −6.01 | −6.00 | −6.09 | −6.10 | −6.11 | −6.11 | |
| 24 | 3.15 | 2.59 | −0.74 | −6.21 | −3.35 | 1.10 | −2.05 | −2.40 | −4.87 | 0.69 | −1.43 | −4.84 | −3.44 | −1.70 | −1.01 | −1.83 | −6.00 | −6.09 | −6.06 | −6.01 | −6.02 | |
| BRKP76 | 4 | 2.34 | −4.57 | −6.42 | −5.14 | −5.08 | −4.31 | −6.29 | −6.30 | −6.31 | −4.34 | −5.53 | −4.15 | −5.70 | −3.48 | −6.10 | −6.07 | −6.06 | −6.11 | −6.16 | −6.18 | −6.16 |
| 8 | 2.75 | −2.32 | −3.44 | −5.14 | −6.38 | −2.75 | −6.29 | −6.30 | −6.31 | −2.46 | −4.23 | −5.75 | −5.70 | −2.30 | −4.02 | −6.07 | −6.06 | −6.11 | −6.16 | −6.18 | −6.16 | |
| 24 | 2.89 | 2.51 | 2.34 | −4.14 | −2.05 | 2.93 | −3.54 | −1.77 | −4.54 | 3.32 | −0.16 | −2.20 | −5.70 | 2.76 | −4.37 | −6.07 | −6.06 | −6.11 | −6.16 | −6.18 | −6.16 | |
| BRKP61 | 4 | 2.37 | −3.26 | −3.05 | −4.28 | −4.93 | −3.22 | −3.88 | −4.86 | −6.23 | −3.55 | −4.45 | −4.45 | −6.00 | −4.11 | −6.03 | −4.70 | −6.10 | −6.03 | −6.03 | −6.05 | −6.24 |
| 8 | 2.94 | −1.28 | −1.84 | −2.93 | −4.93 | −1.68 | −2.53 | −3.61 | −4.45 | −2.38 | −3.08 | −3.98 | −6.00 | −2.98 | −3.62 | −6.00 | −6.10 | −4.26 | −6.03 | −6.05 | −6.24 | |
| 24 | 3.01 | 2.76 | 2.56 | 2.37 | −0.77 | 2.76 | 2.21 | 0.38 | −4.23 | 2.74 | 0.33 | −1.14 | −2.70 | 0.37 | −1.73 | −3.85 | −6.10 | −2.07 | −4.72 | −6.05 | −6.24 | |
| BRKP28 | 4 | 2.33 | 2.18 | 2.24 | 2.29 | 1.64 | 2.19 | 2.18 | 2.14 | .83 | 2.13 | 1.99 | 1.43 | −0.11 | 1.93 | 1.73 | 1.25 | −1.03 | 1.10 | −0.27 | −0.82 | −2.31 |
| 8 | 2.79 | 2.76 | 2.67 | 2.60 | 1.99 | 2.90 | 2.74 | 2.57 | 2.33 | 2.90 | 2.97 | 2.82 | 1.62 | 2.88 | 2.87 | 2.78 | −0.58 | 2.61 | −0.16 | −0.86 | −3.03 | |
| 24 | 2.98 | 2.96 | 2.99 | 3.03 | 2.76 | 3.20 | 2.93 | 2.98 | 3.07 | 3.18 | 3.12 | 3.04 | 3.08 | 3.11 | 3.15 | 3.20 | 3.03 | 3.07 | 3.09 | 3.06 | 2.84 | |
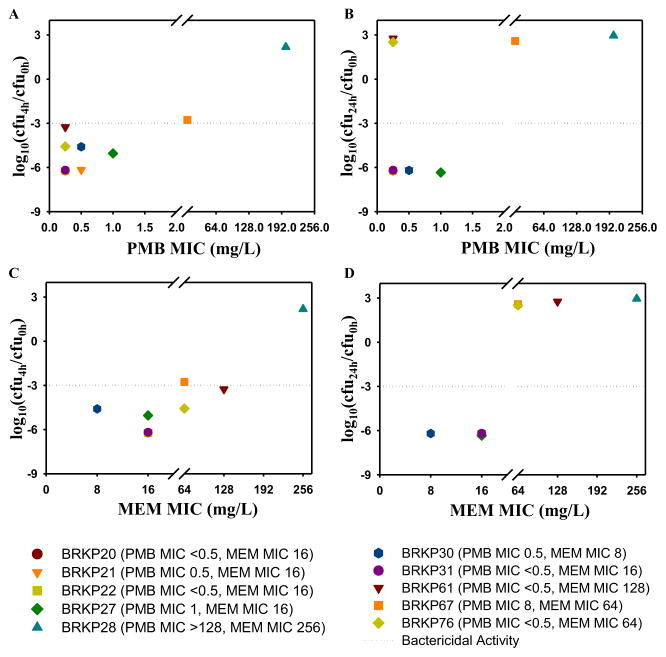
A summary of the point-based analysis for all of the isolates at clinically relevant concentrations (PMB 0.5 mg/L and meropenem 20 mg/L) is shown in Fig. 2. The pharmacodynamic effect at 4 h (Fig. 2A,C) and 24 h (Fig. 2B,D) for each of the isolates was plotted against the corresponding PMB (Fig. 2A,B) and meropenem (Fig. 2C,D) MICs of the isolates. Point-based analysis of these concentrations at the earlier time point of 4 h resulted in a >2.75 log10 reduction against all isolates except BRKP28, i.e. the isolate with the highest MICs both for PMB and meropenem (Fig. 2A). Isolates with increasing meropenem MICs revealed similar results at 4 h (Fig. 2C). Analysis at 24 h demonstrated that these concentrations did not result in sustained activity against isolates with meropenem MICs >16 mg/L (Fig. 2D) irrespective of their susceptibility to PMB (Fig. 2B).
Fig. 2.
Summary of point-based analysis of changes in bacterial density (log10 CFU/mL) at 4 h (A,C) and 24 h (B,D) compared with the bacterial density at baseline (0 h) for polymyxin B (PMB) 0.5 mg/L in combination with meropenem (MEM) 20 mg/L against an initial inoculum of ca. 106 CFU/mL of ten KPC-2-producing Klebsiella pneumoniae clinical isolates. The point-based analysis at 4 h (A,C) and 24 h (B,D) for ten clinical isolates was plotted based on their (A,B) PMB MIC and (C,D) MEM MIC.
3.4. Cell morphological changes assessed by scanning electron microscopy
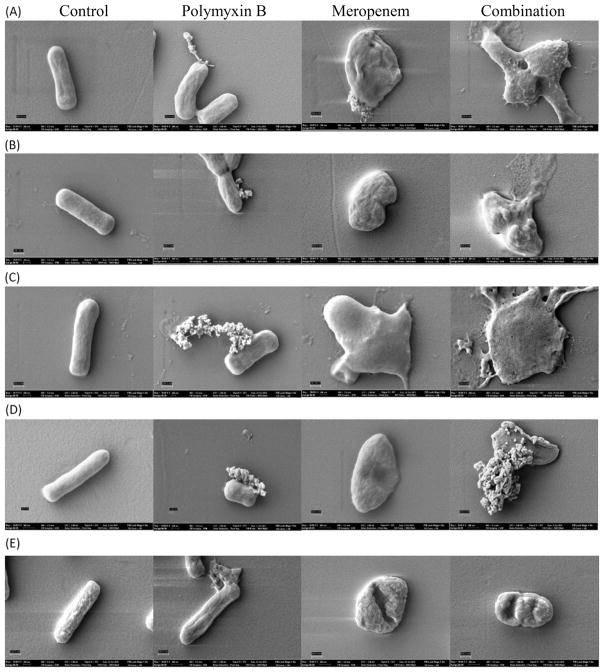
SEM analysis of the surface of the K. pneumoniae isolates revealed morphological changes of the bacterial cell surface treated with either drug as monotherapy or in combination (Fig. 3). Microscopy of five isolates (BRKP20, BRKP21, BRKP27, BRKP67 and BRKP28) with differing PMB and meropenem MICs was performed and images were captured at 50,000× magnification to determine the effect of the antibiotics on the individual bacteria (Fig. 3). The surfaces of cells in the untreated group (Fig. 3A–E, control) were relatively smooth, whilst isolates treated with PMB (Fig. 3A–E, polymyxin B) had numerous pits indicative of significant changes to the outer membrane. Increased permeabilisation of the outer membrane may explain the leakage of cytoplasmic material as seen in the SEM image. Bacteria exposed to meropenem monotherapy had a rounded appearance and cell wall disintegration (Fig. 3A–E, meropenem). The combination of PMB and meropenem resulted in more extensive damage, and the observed effect resembled a combination of changes observed with each individual agent (Fig. 3A–E, combination). PMB monotherapy appeared to have the same effect on bacterial cell populations with differing PMB MICs. In the case of meropenem monotherapy, the observed effect was reduced as the meropenem MIC of the isolate increased.
Fig. 3.
Scanning electron microscopy images of Klebsiella pneumoniae isolates in the absence of treatment (A–E, control) and in the presence of polymyxin B (PMB) alone at 0.5 mg/L (A–E, polymyxin B), meropenem alone at 20 mg/L (A–E, meropenem) and PMB in combination with meropenem (A–E, combination). Magnification at 50,000× with the scale bar set at 300 nm to image individual bacteria of (A) BRKP20 (PMB MIC <0.5 mg/L, meropenem MIC = 16 mg/L); (B) BRKP21 (PMB MIC = 0.5 mg/L, meropenem MIC = 16 mg/L); (C) BRKP27 (PMB MIC = 1 mg/L, meropenem MIC = 16 mg/L); (D) BRKP67 (PMB MIC = 8 mg/L, meropenem MIC = 64 mg/L); and (E) BRKP28 (PMB MIC >128 mg/ L, meropenem MIC = 256 mg/L).
4. Discussion
The global prevalence of carbapenemase-producing K. pneumoniae has increased significantly over the last decade [23]. These pathogens are resistant to almost all available antimicrobial agents, including carbapenems [24]. Infections due to these KPC-producing K. pneumoniae with increasing carbapenem MICs are associated with increased treatment failure and risk of mortality [1,2]. The continued unabated global dissemination of KPC enzymes in Gram-negative bacteria poses a serious threat to the treatment of these infections. In addition, current antimicrobials in the drug pipeline lack activity against the entire spectrum of carbapenemases and are not available in all countries. Lack of knowledge about effective treatments against these serious CRE infections highlights the pressing need for alternative strategies, such as optimising combinations of existing antibiotics.
This prompted us to systematically evaluate the in vitro activity of PMB and meropenem alone and in combination against ten clinical K. pneumoniae isolates expressing the blaKPC-2 gene. The bactericidal activity of polymyxin results in increased permeability of the outer membrane allowing for the enhanced entry of other agents [25]. Meropenem exerts its bactericidal effect via a different mode of action by binding to the high-molecular-weight PBPs, membrane-associated bacterial enzymes involved in the final steps of peptidoglycan biosynthesis [26]. Hence, the combination of PMB and meropenem will be able to maximise mechanistic synergy in addition to subpopulation synergy [27].
The time–kill results demonstrated that supra-MIC PMB concentrations of 64, 128 and 256 mg/L, much higher than clinically achievable concentrations, resulted in early bactericidal activity against all isolates except BRKP28, the isolate with a PMB MIC >128 mg/L. Although early bactericidal activity was observed with PMB monotherapy, the emergence of resistance observed at 24 h is concerning. Furthermore, the risk of dose-related nephrotoxicity associated with polymyxin treatment is high [28]; hence, polymyxin dose escalation to suppress the emergence of resistance is not a clinically viable approach. PMB monotherapy against these resistant bacterial populations, particularly in the case of severe infections with a high bacterial burden or infections in patients with an impaired immune system, would not be optimal.
Similarly, very high meropenem concentrations resulted in bactericidal activity against PMB-susceptible strains with meropenem MICs ≤16 mg/L. Increasing antimicrobial resistance has altered the usage of carbapenems, resulting in a proportional increase in both their infusion times and doses to ensure that a sufficiently high concentration above the MIC is maintained for 40% of the dosing interval [29]. As the MICs of these K. pneumoniae strains rise, monotherapy, even with maximal package insert-based dosing with carbapenems, may not be able to achieve this pharmacokinetic/pharmacodynamic (PK/PD) target and maintain therapeutic efficacy.
All PMB and meropenem combination regimens were bactericidal against PMB-susceptible isolates with meropenem MICs ≤16 mg/L. The findings presented here are consistent with previous studies that have suggested synergy between PMB and meropenem resulting in superior pharmacodynamic activity compared with monotherapy with either agent against these hard-to-treat isolates [30]. The combination of PMB and meropenem was synergistic against KPC-producing isolates with a range of meropenem MICs and this synergy was sustained at higher meropenem concentrations of 60 mg/L and 120 mg/L. In addition, pharmacodynamic analysis indicated that the pharmacodynamic effect of these combinations was attenuated as the meropenem MIC increased to >16 mg/L regardless of the PMB MIC.
These findings are consistent with clinical data investigating combination therapies including high-dose extended-infusion carbapenems for the treatment of KPC bloodstream infections. PK/PD studies have identified a range of meropenem MICs (8–16 mg/L) [31] with an increased probability of target attainment with high-dose prolonged infusions [32]. Tumbarello et al found a significant protective effect against mortality with the triple combination of colistin, meropenem (2 g every 8 h as a 3-h infusion) and tigecycline in patients with bloodstream infections due to KPC-producing K. pneumoniae [2]. Interestingly, when they analysed the impact of meropenem MIC on carbapenem-containing regimens, there was a stepwise increase in treatment failure as the carbapenem MIC increased [2]. Daikos et al showed similar results in an observational study that evaluated the clinical outcomes of 205 patients with bloodstream infections due to carbapenemase-producing K. pneumoniae [33]. In this retrospective analysis, the lowest mortality was observed in patients treated with carbapenem-containing combinations only when the carbapenem MIC was ≤ 8 mg/L [33].
The bacterial killing achieved by polymyxins against K. pneumoniae involves complex interactions with various surface components of the outer membrane of Gram-negative pathogens, which serve as a barrier to cationic polypeptides and, possibly, some intracellular components [8]. Following treatment with PMB alone, the formation of pits was indicative of increased cell permeability due to the pronounced disruption of the structural integrity of the bacterial outer membrane that could result in potential leakage of cytoplasmic material. These observations are consistent with the mechanism of action of PMB, which has a greater propensity to disrupt the outer membrane [34], thereby enabling meropenem to bind easily to high-molecular-weight PBPs [11] and maximise killing shortly after the start of the therapy to prevent bacteria from becoming tolerant or resistant to both antibiotics. Interestingly, the combination of PMB and meropenem caused even more pronounced changes, which morphologically appeared to be a summation of the antimicrobial effects of PMB and meropenem alone (Fig. 3). Similar outer membrane changes have been observed in Escherichia coli [35], Pseudomonas aeruginosa [35] and K. pneumoniae [36] following treatment with PMB. Treatment with meropenem alone resulted in only rounding of the bacterial cell, which has also been observed in P. aeruginosa [37]. Resistance to polymyxins in K. pneumoniae is hypothesised to be due to modification of LPS or a variation in capsule thickness occurring as a result of increased capsular polysaccharide production [38]. The mgrB gene is responsible for biochemical pathways involved in LPS modification. Sequencing of the mgrB amplicon from PMB-resistant isolate BRKP28 revealed a mutation in mgrB, which is predicted to result in a truncated and non-functional MgrB protein. Mutations resulting in modulation of mgrB gene expression have frequently been shown to mediate polymyxin resistance among KPC-producing K. pneumoniae in the clinical setting [39]. Surprisingly, in BRKP67, insertion of ISKpn13, an IS5-like sequence, was found to be the reason behind the disruption of mgrB and probably the reason for increased polymyxin resistance. A 5-bp target-site duplication (CTTAA), as the signature of the transposition event, was systematically observed at each extremity of the IS5-like element. Other studies have also observed similar results [14].
5. Conclusions
This study has demonstrated that combining a carbapenem with PMB is a viable treatment option against carbapenem-resistant KPC-2-producing K. pneumoniae clinical isolates with meropenem MICs ≤16 mg/L. In addition, the morphological changes of bacterial cells observed by SEM in response to antibiotic treatment decreased with increasing meropenem MIC of the bacterial isolate. Further in vitro studies to investigate humanised regimens using PMB in combination with meropenem over a longer duration are warranted to better characterise the time course of the pharmacodynamic effect and its impact on the emergence of resistance. This information will enable the design of antibiotic regimens better suited to treat infections due to such pathogens and prolong the efficacy of these antibiotics. PMB-based triple-drug combination regimens or regimens including agents with novel mechanisms of action are necessary to combat carbapenem-resistant K. pneumoniae strains with meropenem MICs >16 mg/L.
Supplementary Material
Acknowledgments
Funding: This study was partially supported by research grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [R01AI111695 and R01AI119446 to JL, KSK, JMP, JD and GGR]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Appendix. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ijantimicag.2016.10.025.
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
References
- 1.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–7. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 2.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis. 2012;55:943–50. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 3.Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, et al. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother. 2003;51:711–14. doi: 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- 4.US Centers for Disease Control and Prevention. Antibiotic/antimicrobial resistance: biggest threats. Atlanta (GA): CDC; 2013. [Google Scholar]
- 5.Zhu J, Ding B, Xu X, Zhu D, Yang F, Zhang H, et al. Klebsiella pneumoniae: development of carbapenem resistance due to acquisition of blaNDM-1 during antimicrobial therapy in twin infants with pneumonia. Front Microbiol. 2015;6:1399. doi: 10.3389/fmicb.2015.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott E, Brink AJ, van Greune J, Els Z, Woodford N, Turton J, et al. In vivo development of ertapenem resistance in a patient with pneumonia caused by Klebsiella pneumoniae with an extended-spectrum β-lactamase. Clin Infect Dis. 2006;42:e95–8. doi: 10.1086/503264. [DOI] [PubMed] [Google Scholar]
- 7.Lee GC, Burgess DS. Polymyxins and doripenem combination against KPC-producing Klebsiella pneumoniae. J Clin Med Res. 2013;5:97–100. doi: 10.4021/jocmr1220w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velkov T, Deris ZZ, Huang JX, Azad MAK, Butler M, Sivanesan S, et al. Surface changes and polymyxin interactions with a resistant strain of Klebsiella pneumoniae. Innate Immun. 2014;20:350–63. doi: 10.1177/1753425913493337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul M, Bishara J, Levcovich A, Chowers M, Goldberg E, Singer P, et al. Effectiveness and safety of colistin: prospective comparative cohort study. J Antimicrob Chemother. 2010;65:1019–27. doi: 10.1093/jac/dkq069. [DOI] [PubMed] [Google Scholar]
- 10.Teo J, Lim T-P, Hsu L-Y, Tan T-Y, Sasikala S, Hon P-Y, et al. Extensively drug-resistant Acinetobacter baumannii in a Thai hospital: a molecular epidemiologic analysis and identification of bactericidal polymyxin B-based combinations. Antimicrob Resist Infect Control. 2015;4:1–7. doi: 10.1186/s13756-015-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitzis MD, Acar JF, Gutmann L. Antibacterial activity of meropenem against Gram-negative bacteria with a permeability defect and against staphylococci. J Antimicrob Chemother. 1989;24(Suppl A):125–32. doi: 10.1093/jac/24.suppl_a.125. [DOI] [PubMed] [Google Scholar]
- 12.Abboud CS, Monteiro J, Franca JI, Pignatari AC, De Souza EE, Camargo EC, et al. A space–time model for carbapenemase-producing Klebsiella pneumoniae (KPC) cluster quantification in a high-complexity hospital. Epidemiol Infect. 2015;143:2648–52. doi: 10.1017/S0950268814003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67:906–9. doi: 10.1093/jac/dkr563. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Jayol A, Bontron S, Villegas M-V, Ozdamar M, Türkoglu S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Document M100-S23. Wayne (PA): CLSI; 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. [Google Scholar]
- 16.European Committee on Antimicrobial Susceptibility Testing. [Accessed 7 December 2016];Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0. 2016 Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf.
- 17.Zavascki AP, Goldani LZ, Cao G, Superti SV, Lutz L, Barth AL, et al. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis. 2008;47:1298–304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
- 18.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis. 2013;57:524–31. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 19.Jaruratanasirikul S, Sriwiriyajan S, Punyo J. Comparison of the pharmacodynamics of meropenem in patients with ventilator-associated pneumonia following administration by 3-hour infusion or bolus injection. Antimicrob Agents Chemother. 2005;49:1337–9. doi: 10.1128/AAC.49.4.1337-1339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drusano GL, Hutchison M. The pharmacokinetics of meropenem. Scand J Infect Dis Suppl. 1995;96:11–16. [PubMed] [Google Scholar]
- 21.Rao GG, Ly NS, Diep J, Forrest A, Bulitta JB, Holden PN, et al. Combinatorial pharmacodynamics of polymyxin B and tigecycline against heteroresistant Acinetobacter baumannii. Int J Antimicrob Agents. 2016;48:331–6. doi: 10.1016/j.ijantimicag.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Cheah S-E, Roberts KD, Nation RL, Thompson PE, Velkov T, et al. Transcriptomic analysis of the activity of a novel polymyxin against Staphylococcus aureus. mSphere. 2016;1 doi: 10.1128/mSphere.00119-16. pii: e00119-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–96. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J, Woodford N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents. 2011;37:415–19. doi: 10.1016/j.ijantimicag.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies TA, Shang W, Bush K, Flamm RK. Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:1510–12. doi: 10.1128/AAC.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landersdorfer CB, Ly NS, Xu H, Tsuji BT, Bulitta JB. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother. 2013;57:2343–51. doi: 10.1128/AAC.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubin CJ, Ellman TM, Phadke V, Haynes LJ, Calfee DP, Yin MT. Incidence and predictors of acute kidney injury associated with intravenous polymyxin B therapy. J Infect. 2012;65:80–7. doi: 10.1016/j.jinf.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Craig WA. The pharmacology of meropenem, a new carbapenem antibiotic. Clin Infect Dis. 1997;24(Suppl 2):S266–75. doi: 10.1093/clinids/24.supplement_2.s266. [DOI] [PubMed] [Google Scholar]
- 30.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, et al. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56:2108–13. doi: 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis. 2015;2:ofv050. doi: 10.1093/ofid/ofv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts JA, Kirkpatrick CM, Roberts MS, Robertson TA, Dalley AJ, Lipman J. Meropenem dosing in critically ill patients with sepsis and without renal dysfunction: intermittent bolus versus continuous administration? Monte Carlo dosing simulations and subcutaneous tissue distribution. J Antimicrob Chemother. 2009;64:142–50. doi: 10.1093/jac/dkp139. [DOI] [PubMed] [Google Scholar]
- 33.Daikos GL, Tsaousi S, Tzouvelekis LS, Anyfantis I, Psichogiou M, Argyropoulou A, et al. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother. 2014;58:2322–8. doi: 10.1128/AAC.02166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deris ZZ, Swarbrick JD, Roberts KD, Azad MAK, Akter J, Horne AS, et al. Probing the penetration of antimicrobial polymyxin lipopeptides into Gram-negative bacteria. Bioconjug Chem. 2014;25:750–60. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koike M, Iida K, Matsuo T. Electron microscopic studies on mode of action of polymyxin. J Bacteriol. 1969;97:448–52. doi: 10.1128/jb.97.1.448-452.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdul Rahim N, Cheah S-E, Johnson MD, Yu H, Sidjabat HE, Boyce J, et al. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’ antibiotics—polymyxin B and chloramphenicol. J Antimicrob Chemother. 2015;70:2589–97. doi: 10.1093/jac/dkv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siqueira VLD, Cardoso RF, Caleffi-Ferracioli KR, de Lima Scodro RB, Fernandez MA, Fiorini A, et al. Structural changes and differentially expressed genes in Pseudomonas aeruginosa exposed to meropenem–ciprofloxacin combination. Antimicrob Agents Chemother. 2014;58:3957–67. doi: 10.1128/AAC.02584-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004;72:7107–14. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arena F, De Angelis LH, Cannatelli A, Di Pilato V, Amorese M, D’Andrea MM, et al. Colistin resistance caused by inactivation of the MgrB regulator is not associated with decreased virulence of sequence type 258 KPC carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2016;60:2509–12. doi: 10.1128/AAC.02981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.