Abstract
The baculovirus expression vector system (BEVS) is a widely used platform for the production of recombinant eukaryotic proteins. However, the BEVS has limitations in comparison to other higher eukaryotic expression systems. First, the insect cell lines used in the BEVS cannot produce glycoproteins with complex‐type N‐glycosylation patterns. Second, protein production is limited as cells die and lyse in response to baculovirus infection. To delay cell death and lysis, we transformed several insect cell lines with an expression plasmid harboring a vankyrin gene (P‐vank‐1), which encodes an anti‐apoptotic protein. Specifically, we transformed Sf9 cells, Trichoplusia ni High FiveTM cells, and SfSWT‐4 cells, which can produce glycoproteins with complex‐type N‐glycosylation patterns. The latter was included with the aim to increase production of glycoproteins with complex N‐glycans, thereby overcoming the two aforementioned limitations of the BEVS. To further increase vankyrin expression levels and further delay cell death, we also modified baculovirus vectors with the P‐vank‐1 gene. We found that cell lysis was delayed and recombinant glycoprotein yield increased when SfSWT‐4 cells were infected with a vankyrin‐encoding baculovirus. A synergistic effect in elevated levels of recombinant protein production was observed when vankyrin‐expressing cells were combined with a vankyrin‐encoding baculovirus. These effects were observed with various model proteins including medically relevant therapeutic proteins. In summary, we found that cell lysis could be delayed and recombinant protein yields could be increased by using cell lines constitutively expressing vankyrin or vankyrin‐encoding baculovirus vectors. © 2017 The Authors Biotechnology Progress published by Wiley Periodicals, Inc. on behalf of American Institute of Chemical Engineers Biotechnol. Prog., 33:1496–1507, 2017
Keywords: vankyrin, baculovirus, glycosylation, difficult to express proteins, SfSWT
Introduction
The baculovirus expression vector system (BEVS, aka Baculovirus insect cell system, BICS) is a recombinant protein production platform that combines insect cells with recombinant baculovirus vectors1, 2 and was recently reviewed by Refs. 3 and 4. In comparison to other higher eukaryotic recombinant protein production platforms, the BEVS quickly produces large amounts of properly folded proteins. Additional advantages of the BEVS include the ability to add eukaryotic post‐translational modifications, including O‐ and N‐linked glycosylation, at the correct sites. Moreover, multiple genes of interest can be encoded by the same recombinant baculovirus,5, 6, 7 and large DNA fragments can be cloned into the baculoviral vectors.3, 8 These advantages have led to widespread use of the BEVS for various applications, including the production of recombinant proteins for both basic and applied research, as well as the production of recombinant proteins for immunotherapy treatment (e.g., ProvengeTM), prescription medicine, and vaccine applications3, 4 such as the FDA‐licensed products Cervarix™ and FluBlok™.9, 10, 11
One major limitation of the BEVS is that baculovirus infection results in cell death and lysis, which limits baculoviral protein expression to the window of time between the onset of late viral gene expression and the time of cell death.12 Thus, protein expression is typically restricted to ∼3 days following infection. Furthermore, the insect cell secretory pathway is compromised during the later stages of baculovirus infection, limiting the extent to which secreted recombinant proteins can be folded and secreted into the extracellular medium. Secretory pathway impairment is caused, at least to some extent, by the accumulation of large amounts of virally encoded chitinase and cathepsin (a protease) in the secretory pathway.13, 14 Following lysis, viral cathepsin is released into the culture supernatant, and can degrade recombinant proteins after being activated by treatment with chaotropic reagents such as SDS or low pH. To address the negative impact of baculovirus chitinase and cathepsin on secretory pathway protein yield and integrity, baculovirus vectors lacking chitinase and cathepsin were developed.15, 16
Non‐lytic or delayed lytic baculovirus vectors have been used to delay cell death and lysis and improve production levels and integrity of recombinant proteins.17, 18 Gómez‐Sebastián et al. engineered a novel expression cassette containing various baculovirus genomic elements such as transactivators IE1 and IE0 and enhancer sequences.18 Insect cells infected with those viruses showed increased cell viability and integrity after infection, and an increase in recombinant protein yields. A similar effect was achieved when the baculovirus apoptotic inhibitor P35 was constitutively expressed from the insect cells.19 However, the overexpression of IAP‐1 and IAP‐2 did not consistently inhibit apoptosis in AcMNPV.20, 21
An alternative approach to delay lysis of baculovirus‐infected cells is the expression of viral ankyrins (vankyrins) derived from an insect polydnavirus, Campoletis sonorensis ichnovirus (CsIV).22 Baculovirus‐infected Sf9 cells constitutively expressing one of two vankyrin proteins (P‐vank‐1 or I2‐vank‐3) exhibit a delay in cell lysis due to inhibition of apoptosis, with some cells surviving several days longer than normal.22 The nature of the vankyrin proteins and studies of their activity suggest the antiapoptotic actions result from modulation of host cellular immune responses to virus infection.22 ‐ 24 Specifically, experimental evidence suggests vankyrin proteins are functional I‐κB homologs that act on the NF‐κB signaling pathway to alter cellular immunity at the transcriptional level to block apoptosis.25, 26
A second major limitation of the BEVS is the inability to produce N‐glycoproteins with human‐type N‐glycan structures. This is an important limitation, as glycosylation can affect protein half‐life, stability, function, structure, and/or immunogenicity,27, 28 and over 50% of human proteins are glycosylated.29 Because glycoproteins are involved in important physiological processes such as cell proliferation and differentiation, blood clotting and immunity, many glycoproteins are pharmaceutically relevant and used as therapeutics or in vaccines.8, 27, 30 Unfortunately, a large majority of glycoprotein therapeutics cannot be produced using conventional BEVS, because the N‐linked glycans on glycoproteins produced in the BEVS are different from those produced in mammalian cells, and do not provide efficient therapeutic effects. Specifically, the insect cell lines used in the BEVS do not produce activated sialic acid and do not express sufficient levels of several glycosyltransferases to produce complex, terminally sialylated glycoproteins. Instead, insect cells produce N‐glycoproteins with paucimannose glycans, where mammalian cells produce complex sugar groups with terminal sialic acids.31, 32, 33 Because the majority of medically relevant proteins are glycoproteins, this is an important limitation of the BEVS. Consequently, recombinant glycoprotein biologicals that require human‐type glycans for clinical efficacy have to be produced in mammalian expression platforms, although the BEVS is superior in many aspects.34, 35, 36
To address this limitation, both baculovirus vectors and insect cells have been engineered with the enzymes required to produce N‐glycoproteins with human‐type complex, terminally sialylated glycans.31, 32, 33, 37, 38 One such engineered cell line is SfSWT‐4, which is a Spodoptera frugiperda Sf9 cell line derivative that has been engineered to stably express glycosyltransferases necessary for N‐glycan elongation, as well as several enzymes required to produce and activate sialic acid.39
The present study was designed to expand the utility of the vankyrin technology and to address both of these major limitations of the BEVS. Our goal was to increase recombinant glycoprotein productivity and humanize N‐glycosylation in the BEVS by expressing vankyrin in glyco‐engineered insect cells. To achieve this goal, we stably transformed SfSWT‐4 cells with the P‐vank‐1 gene and demonstrated increased yields of secreted glycoproteins.
Furthermore, we demonstrated vankyrin expression improves protein yields in cell lines other than S. frugiperda cell lines. Several reports indicate Trichoplusia ni cells can produce significantly higher levels of secreted proteins than S. frugiperda cells.40, 41, 42 Here, we stably transformed High FiveTM insect cells, which are a T. ni cell line, to express P‐vank‐1. We found the resulting VE‐High Five cell line had enhanced cell viability and recombinant protein production as compared to the parental cell line.
Finally, we also describe new vankyrin‐enhanced (VE) baculovirus vectors. VE‐baculoviruses prolonged survival of infected insect cells compared to conventional baculoviruses, and accumulation of secreted proteins increased. In addition, a synergistic effect was seen when a VE‐baculovirus was used to infect VE‐insect cells.
In summary, we have addressed major limitations in the BEVS by demonstrating that vankyrin enhancement can significantly improve cell viability in several types of baculovirus‐infected cells, including a glycosylating cell line. As a result, secretion of recombinant proteins produced in the VE‐BEVS is prolonged with less protein degradation and thus, protein accumulation is considerably increased relative to conventional BEVS. Consequently, VE‐BEVS offer a novel, significant, adaptable, and proven improvement to the BEVS platform for various applications.
Materials and Methods
Cell lines and growth conditions
Spodoptera frugiperda Sf9 cells and Trichoplusia ni High Five™ cells were acquired from Thermo Fisher Scientific (Waltham, MA, USA). SfSWT‐4 cells39 were provided by Dr. Donald Jarvis from the University of Wyoming (Laramie, WY, USA) and VE‐Sf9 cells, which are referred to as VE‐CL02 cells,24 were developed at ParaTechs Corp. (Lexington, KY). These cells are also known as SuperSf9‐2 (Oxford Expression Technologies, Oxford, UK). Insect cells were maintained in suspension culture in 125 ml‐Erlenmeyer flasks at 27°C with shaking at 130–150 rpm. For each passage, insect cell cultures were diluted with insect cell culture medium to a seeding density of 1 × 106 cells mL−1 in a volume of 25–50 mL when the cell density reached 5 × 106 cells mL−1. Sf9 and VE‐CL02 cells were grown in Sf‐900™II serum‐free medium (Sf‐900TM II SFM; Thermo Fisher Scientific). High Five™ (Thermo Fisher Scientific) and VE‐High Five cells were grown in Express Five® serum‐free medium (Express Five® SFM; Thermo Fisher Scientific) supplemented with 18 mM L‐glutamine (Thermo Fisher Scientific) and 10 U of heparin per ml (Sigma–Aldrich, St. Louis, MO). SfSWT‐4 and VE‐SfSWT‐4 cells were routinely grown in TNM‐FH (Gemini Bio‐Products, West Sacramento, CA) supplemented with 10% heat‐inactivated fetal bovine serum (FBS) and 1% pluronic F‐68 (both Thermo Fisher Scientific).
VE‐High Five and VE‐SfSWT‐4 (“VE‐SWT”) cells were obtained by transforming cells with a Junonia coenia densovirus transformation vector encoding P‐vank‐1 from the Campolitis sonorensis ichnovirus (CsIV; accession no. AAX56953.1) as described for VE‐CL02 cells.24 The effect of vankyrin expression from several different insect and viral promoters on recombinant protein production were evaluated, and the VE‐High Five and VE‐SWT cell lines with P‐vank‐1 expression under the control of the constitutive CsIV AHv0.8 promoter were chosen for further evaluation. Stably transformed VE cells were selected with 400 µg mL−1 Geneticin G418 Sulfate (Thermo Fisher Scientific). Populations of antibiotic‐resistant cells were amplified to generate stable polyclonal VE‐High Five and VE‐SWT cell lines. The expression of P‐vank‐1 RNA in transformed cell lines was confirmed by RT‐PCR. Stable polyclonal cell lines were evaluated for recombinant protein production and performance relative to unmodified insect cells.
For monoclonal selection of VE‐High Five cells, limiting dilutions were prepared from individual polyclonal cell lines using 50% 48‐h‐conditioned Express Five® medium containing 400 µg mL−1 Geneticin G418 Sulfate. Each dilution containing a single cell was added to 96‐well flat bottom tissue culture plates. Plates were sealed and allowed to incubate at 27°C for 4 weeks, replacing the media once, before clonal populations of positive antibiotic‐resistant cells reached confluency and were reseeded into new wells in 96 well plates containing 200 µL of conditioned medium per well, and incubated for 1 week. Cells were seeded into a 48‐well plate for scale‐up and amplification, and grown to confluency in the presence of 400 µg mL−1 Geneticin G418 Sulfate prior to seeding into 24‐well, and finally six‐well plates. When cells in six‐well plates reached confluency, monoclonal cell lines were started in T25 flasks. RT‐PCR was performed to confirm expression of P‐vank‐1 in the monoclonal cell lines. YFP expression levels were then quantified in monoclonal isolates after infection with recombinant YFP‐BEVS (see below; Figure 1).
Figure 1.
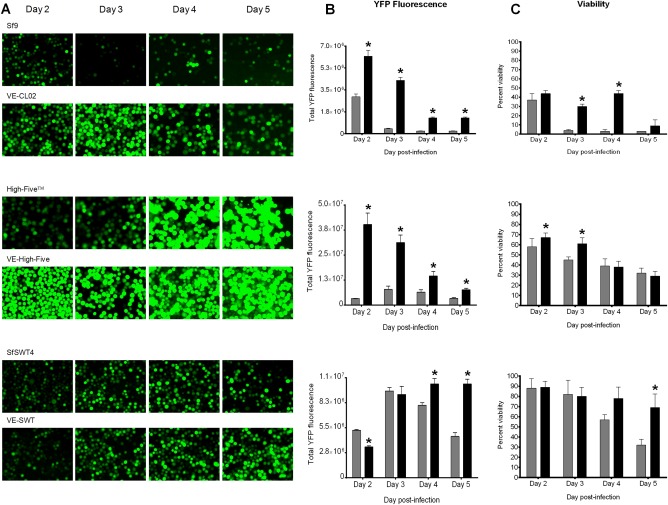
Vankyrin‐enhanced cells have increased YFP fluorescence and viability compared to their parental cell line.
Legend: (A) Fluorescent images (×200 magnifications), (B) measured YFP fluorescence, and (C) cell viability for VE‐CL02 and its parental cell line Sf9 (top panels; Sf‐900™II medium), VE‐High Five and its parental cell line High‐Five™ (middle panels; Express Five® SFM medium), and VE‐SWT and its parental cell line SfSWT4 (bottom panel; TNM‐FH medium with FBS) infected with a baculovirus encoding YFP (YFP‐AcMNPV) at a multiplicity of infection (MOI) of 5 is shown for days 2–5 post‐infection. All infections were in static cultures with a cell density of 5 × 105 cells at the time of infection. Total YFP fluorescence for each infection (B) was determined by flow cytometry using the Guava easyCyte Flow Cytometer as described in the Materials and Methods section. Percent viability (C) was determined by trypan blue staining as described in the Materials and Methods section. In (B) and (C), parental cell lines are indicated by gray bars, and Vankyrin‐enhanced (VE) cell lines are represented by black bars. The increase in cell viability in virus‐infected VE‐CL02 cells (C, top panel) from day 3 to day 4 can be explained by the difference in total cell number. The data presented are means and standard deviations for triplicate determinations for each cell line in a single experiment. The data presented here are representatives of multiple experiments performed from which equivalent results were obtained. Statistical significance (P ≤ 0.05) as determined by the Student two‐tailed t test for comparison of baculovirus‐infected parental cells vs. baculovirus‐infected VE‐cells is represented by an asterisk (*).
Monoclonal isolates of VE‐SWT cells were obtained as previously described39 by limiting dilution using conditioned TNM‐FH medium, yielding 34 monoclonal VE‐SWT cell lines stably expressing P‐vank‐1. Each monoclonal isolate was screened for the production of terminally sialylated glycoconjugates by cell surface staining with Texas Red conjugated Sambucus nigra agglutinin (SNA43). Cell surface fluorescence could be observed on all 34 VE‐SWT monoclonal isolates, as well on SfSWT‐4 positive control cells, whereas Sf9 control cells did not fluoresce, indicating VE‐SWT cells produced terminally sialylated glycoconjugates as expected. The 34 monoclonal isolates were further screened for growth characteristics and enhanced glycoprotein production. A clone designated VE‐SWT33 was selected for further experiments because it uniformly grew without clumping or floating in monolayer and consistently produced high levels of two mammalian model glycoproteins, erythropoietin (EPO) and secreted alkaline phosphatase (SEAP), when infected with recombinant baculoviruses encoding these proteins.
Baculovirus transfer vectors
pAcVE.02 and pAcVE.03 transfer vectors (Figure 3) are modified from pAcVE.01 (ParaTechs, Lexington, KY) with the addition of the honeybee melittin (HBM) signal peptide to increase protein secretion.44 pAcVE.01 was synthesized by GenScript (Piscataway, NJ, USA) and encodes, an 861 bp ampicillin resistance gene (derived from pUC57), a 131 bp SV40 polyadenylation signal, and genomic DNA fragments from AcMNPV (accession NC_001623) to target the polyhedrin locus for in vivo homologous recombination and that correspond to the polyhedrin promoter (nt 4425‐4521), the p10 promoter (nt 118728‐118839), ORF603 (nt 3759‐4364) and its promoter and ORF1629 (nt 5287‐6918) and its transcription termination signal. Downstream of the polyhedrin promoter is a 92 bp multiple cloning site (MCS) containing AvrII, BglII, BstZ17I, EagI, NcoI, NheI, SacII, SbfI, and XhoI restriction sites, followed by a 6x His‐tag to facilitate protein purification and a stop codon. On the complementary strand, downstream of the p10 promoter is the CsIV P‐vank‐1 gene flanked by AflII restriction sites. A 137 bp fragment comprising a 69 bp region encoding HBM signal peptide, a PmeI restriction site, an 8x His‐tag, and a MCS containing NotI, SbfI, and NheI restriction sites was synthesized by GenScript and cloned into pUC57 (HBM‐pUC57). This plasmid was digested with AvrII and NheI to excise the 137 bp insert, which was then ligated into the AvrII and NheI sites of pAcVE.01, thereby replacing its MCS with the 137 bp fragment, and chemically transformed into DH5α cells (Thermo Fisher Scientific). The resulting plasmid was designated as pAcVE.02 (Figure 3A). pAcVE.03 was constructed by inserting the HBM signal sequence upstream of the MCS of pAcVE.01 using Infusion cloning (Takara, Mountain View, CA). The HBM was PCR amplified from plasmid HBM‐pUC57 with primer set 335/343 (Table 1); each primer was designed to overlap both pAcVE.01 and the HBM sequences. The HBM PCR fragment was then ligated into the AvrII site of pAcVE.01, and the reaction product was chemically transformed into DH5α cells. Insertion of the HBM must be in frame with the restriction enzymes in the pAcVE.01 MCS and the C‐terminal 6x His‐tag, but insertion of the signal peptide resulted in three stop codons in this DNA region. The stop codons were removed using site‐directed mutagenesis (Agilent Technologies, Santa Clara, CA USA) to mutate G2732C (primer set 351/352), followed by deletions of A2684 (primer set 357/358) and A2768 (primer set 359/360). The resulting plasmid was designated pAcVE.03 (Figure 3A). To enable evaluation of the vankyrin‐harboring baculoviruses, control transfer vectors that are the non‐vankyrin encoding versions of pAcVE.02 and pAcVE.03 were constructed by deleting the P‐vank‐1 gene using the restriction enzyme AflII and were named pAc.02 and pAc.03, respectively. The sequences of all constructs were confirmed by dideoxy sequencing.
Figure 3.
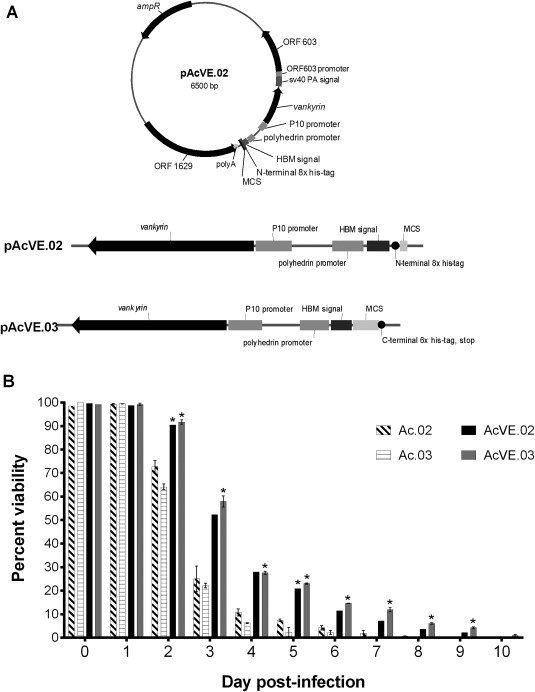
Vankyrin‐encoding baculoviruses increase insect cell viability compared to non‐vankyrin baculoviruses.
Legend: (A) New vankyrin‐encoding transfer vectors pAcVE.02 and pAcVE.03 are shown. Each vector contains an ampicillin resistance gene (ampR), the p10 promoter upstream of the vankyrin gene, a sv40 polyadenylation signal (sv40 PA signal), multi‐cloning site (MCS; pAcVE.02: NotI, SbfI, NheI; pAcVE.03: NcoI, SbfI, XhoI, BstZ17I, BglII, SacII, EagI), honey bee melittin signal peptide (HBM signal), his‐tag, and ORF1629 (including polyA signal) and ORF603 (including promoter)—the two open reading frames that flank the polyhedrin gene in the AcMNPV genome. (B) Viability of SfSWT4 cells infected with baculoviruses generated from transfer vectors pAc.02 and pAc.03, or the vankyrin‐encoding counterpart pAcVE.02 and pAcVE.03. Suspension cultures grown in Sf‐900™II were infected on day 0 with a MOI 5 of each baculovirus. The cell densities of each culture at the time of infections were ∼2 × 106 cells mL−1. Cell viability was determined at the on‐set of the experiment (0 dpi before viral infections) and 1–10 days post‐infection (dpi) by staining cells with trypan blue and counting living cells with the hemocytometer. The data presented are means and standard deviations for duplicate determinations for each infection in a single experiment. The data presented here are representative of multiple experiments performed from which equivalent results were obtained. Statistical significance (P ≤ 0.05) as determined by the Student two‐tailed t test for comparison of non‐vankyrin baculovirus infections vs. vankyrin‐encoding baculovirus infections using the same transfer vector backbone is represented by an asterisk (*).
Table 1.
Oligonucleotide Primers Used for PCR in this Study
| Designation | Footnote | Sequence | Type of PCR or Oligonucleotide |
|---|---|---|---|
| 335 | a | 5′‐ ATAAATATACCTAGGATGAAATTTCTAGTAAACGTTGCC‐3′ | Infusion |
| 343 | a | 5′‐ GGCCATGGACCTAGGCGGATCAGCATAGA‐3′ | Infusion |
| 351 | b | 5′‐ GTATACAAAGATCTCAAGTACCGCGGTCG‐3′ | Site‐directed mutagenesis |
| 352 | b | 5′‐ CGACCGCGGTACTTGAGATCTTTGTATAC‐3′ | Site‐directed mutagenesis |
| 357 | c | 5′‐ GCTGATCCGCCtgGTCCATGGCC‐3′ | Site‐directed mutagenesis |
| 358 | c | 5′‐ GGCCATGGacCAGGCGGATCAGC‐3′ | Site‐directed mutagenesis |
| 359 | c | 5′‐ CGCTCTATCTAGCtgCACATCACCATC‐3′ | Site‐directed mutagenesis |
| 360 | c | 5′‐ GATGGTGATGTGcaGCTAGATAGAGCG‐3′ | Site‐directed mutagenesis |
| 363 | d | 5′‐ CCATGGGCCCCCCCTAGATTAATT‐3′ | Amplifying |
| 364 | d | 5′‐ CTCGAGCCGATCGCCTGTACGGCA‐3′ | Amplifying |
Underlined nucleotides of infusion primers correspond to pAcVE.01 sequence; non‐underlined nucleotides correspond to HBM or signal peptide sequence.
Substituted nucleotides in the site‐directed mutagenesis primers are in bold and italicized.
Bases surrounding the deleted adenine are lowercase.
Primers used for routine PCR amplification of erythropoietin gene were synthesized with either a NcoI or XhoI restriction site (underlined) for ease of cloning.
Recombinant baculovirus generation
A baculovirus transfer vector (pVL‐YFP) encoding YFP was obtained by excising the YFP open reading frame from pEYFP‐C1 (Takara; discontinued) using BamHI and SmaI and cloning the fragment into the corresponding sites in the MCS downstream of the polyhedrin promoter of a pVL1392‐based transfer vector (ParaTechs, KY; in house vector). Genes encoding mature human secreted alkaline phosphatase (SEAP; accession no. NP_001623.3) and erythropoietin (EPO; accession no. NP_000790.2) were codon optimized using the OPTIMIZER program (http://genomes.urv.es/OPTIMIZER/) with the AcMNPV codon bias (http://www.kazusa.or.jp/), synthesized, and cloned into pUC57‐based vectors (GenScript), designated SEAP‐pUC57 and EPO‐pUC57 (Table 2). Neither gene included a start codon and native signal peptide, as these genes were designed to use the HBM start codon and signal peptide in pAcVE.02 and pAcVE.03 vectors. Further details regarding the construction of SEAP and EPO baculovirus transfer vectors are described in Table 2.
Table 2.
Bacterial Plasmids and Bacmids Used in this Study
| Plasmid | Description |
|---|---|
| pGEM®‐T Easy | ColE1‐based cloning vector; 3,015 bp, Apr (Promega #A1360) |
| pUC57 | ColE1‐based cloning vector; 2,710 bp, Apr (GenScript #SD1176) |
| pEYFP‐C1 | pBR322 origin vector containing Aequorea victoria GFP; 4,731 bp, Knr (Clontech, discontinued) |
| pFastbac™ Dual | pUC‐based vector containing MCSs after the polH and p10 promoters; 5,238 bp, Apr Gnr (Thermo Fisher Scientific #10712–024) |
| pAcVE.01 | Derivative of pUC57; described in materials and methods section |
| pAcVE.02 | Derivative of pAcVE.01; described in materials and methods and Figure 4 |
| pAcVE.03 | Derivative of pAcVE.01; described in materials and methods and Figure 4 |
| pAc.01 | Derivative of pAcVE.01 where the 516 bp P‐vank‐1 gene has been deleted |
| pAc.02 | Derivative of pAcVE.02 where the 516 bp P‐vank‐1 gene has been deleted |
| pAc.03 | Derivative of pAcVE.03 where the 516 bp P‐vank‐1 gene has been deleted |
| epo‐pUC57 | 607 bp codon optimized epo gene from human cells cloned into EcoRI site of pUC57 |
| pKH25 | 510 bp NcoI/XhoI DNA from epo‐pUC57 containing epo (PCR primers 363/364) cloned into pGEM®‐T Easy |
| epo‐pAc.03 | 504 bp NcoI/XhoI fragment from pKH25 containing epo with no stop codon cloned into pAc.03 |
| epo‐pAcVE.03 | 504 bp NcoI/XhoI fragment from pKH25 containing epo with no stop codon cloned into pAcVE.03 |
| seap‐pUC57 | 509 bp codon optimized seap gene from human cells cloned into EcoRI site of pUC57 |
| seap‐pAc.02 | Derivative of seap‐pAcVE.02 where the 516 bp P‐vank‐1 gene has been deleted |
| seap‐pAcVE.02 | 1,504 bp NotI/SbfI from seap‐pUC57 containing seap with stop codon cloned into pAcVE.02 |
| pVL‐YFP | BamHI/SmaI fragment from pEYFP‐C1 containing yfp cloned into pVL1392 |
Abbreviations: (Apr) ampicillin resistance, (Knr) kanamycin resistance, (Gnr) gentamicin resistance, (polH) polyhedrin, (seap) secreted embryonic alkaline phosphatase, (epo) erythropoietin.
Recombinant baculoviruses encoding YFP, SEAP, or EPO were generated through homologous recombination by transfecting Sf9 cells with the transfer vector and the flashBAC GOLD AcMNPV DNA backbone (which does not encode chitinase and cathepsin) using the manufacturer's instructions (Oxford Expression Technologies). The recombinant virus was amplified once or twice in 50‐mL Sf9 cultures; filter sterilized using a 0.22‐µm syringe filter, and titered using the plaque assay method.45
Virus infections
The 5 × 105 insect cells were seeded into 12‐well tissue culture plates in 1 mL of their corresponding growth medium. Once cells achieved confluency, cells were either left uninfected or infected with a specified multiplicity of infection (MOI) of recombinant baculovirus. Infections were incubated at 27°C for up to 10 days. On each day, baculovirus infected cells were monitored for cytopathic effects (nuclear and cellular hypertrophy, grainy appearance, and lysis), photomicrographs were taken with a Zeiss AX10 inverted microscope (Carl Zeiss Microscopy, Thornwood, NY) and acquired using an AxioCamMR3 digital camera (Carl Zeiss Microscopy). Samples were collected as specified for each experiment.
YFP quantification
Sf9, VE‐CL02, High Five, VE‐High Five, SfSWT‐4, and VE‐SWT cells were infected with YFP‐AcMNPV at an MOI of 5 in triplicate. On days 2–5 post‐infection, photomicrographs and YFP fluorescence (300 ms exposure time) for each infection well were captured using a Zeiss AX10 inverted fluorescence microscope with a 20× objective and the AxioVision Rel. 4.6 program (Carl Zeiss Microscopy). After gently collecting the cells, viability was determined by staining cells with trypan blue (Thermo Fisher Scientific) and counting viable and non‐viable cells using an improved Neubauer hemocytometer under magnification of a Zeiss AX10 inverted microscope (Carl Zeiss Microscopy). To quantify YFP fluorescence, insect cells were first counted in triplicate utilizing a Guava® easyCyte HT Sampling Flow Cytometer and the guava InCyte Assay software module (EMD Millipore, Hayward, CA). YFP fluorescence was then measured using a 405‐nm laser at a green spectral imaging band (525/30 nm) of the Guava® easyCyte™ HT Sampling Flow Cytometer. The total fluorescence of each infection was determined with the Guava® InCyte Assay software module.
Cell viability assay of vankyrin‐enhanced baculovirus infected cells
Sf9 and SfSWT‐4 insect cells were grown in 125 mL‐Erlenmeyer flasks at 106 cells mL−1 in their respective medium in a final volume of 25 mL (Sf9) or 40 mL (SfSWT‐4), followed by an overnight incubation at 27°C at 150 rpm. Next, cell density and viability were determined before infecting the cells with recombinant baculovirus at their optimal MOI (MOI of 5 for Ac.02, Ac.03, AcVE.02, and AcVE.03 baculoviruses and MOI of 1 for epo‐Ac.03 and epo‐AcVE.03 baculoviruses). Cultures were then incubated for another 10 days. On each day, 90 µL of culture from each flask was removed in duplicate, and the number of viable cells was determined by trypan blue staining as described above.
Western blotting analysis
To evaluate expression levels and processing of the five LDLa repeats of Manduca sexta pro‐hemolymph protease‐14 (proHP14), 1.6 × 106 Sf9 or VE‐CL02 cells mL−1 were seeded in duplicate six‐well plates in 2 mL Sf‐900TM II medium, and infected with a baculovirus encoding the five LDLa repeats of proHP14 at an MOI of 5. Cells were incubated at 27°C, and cell‐free medium samples were collected after 3, 5, and 7 days for SDS‐PAGE (12%) followed by Western blot analysis using 1:1,000 primary diluted anti‐His monoclonal antibody (GenScript) and 1:1,000 diluted goat anti‐mouse IgG‐AP conjugate as the secondary antibody (Bio‐Rad, Hercules, CA).
Erythropoietin (EPO; 34 kDa) protein levels were determined by Western blotting from the infected cultures used in the cell viability assay described earlier. On days 2–10 post‐infection, a small sample of each culture was collected, and cells were removed by centrifugation at 900 g for 10 min at 4°C. Supernatants were stored at 4°C until all of the samples were collected. To determine recombinant protein levels per mL of culture, 5 µL of supernatant were used for SDS‐PAGE (10%) followed by Western blot analysis using a 1:3,000 dilution of mouse monoclonal anti‐His IgG2 antibody (GE Healthcare, Wauwatosa, WI) and a 1:300 dilution of anti‐mouse IgG horseradish peroxidase secondary antibody (GE Healthcare). Membranes were exposed to CN/DAB substrate (Thermo Fisher Scientific) for 6 min, followed by rinsing the membrane with water to stop exposure. Membranes were scanned using a BioRad Universal Hood II Gel Doc UV transilluminator.
SEAP enzymatic assay
SfSWT‐4 and VE‐SWT cells were infected at an MOI of 1 with either seap‐Ac.02 or seap‐AcVE.02 baculoviruses in triplicate wells. On days 3–5 post‐infection, supernatants were collected, cells were removed by centrifugation (900 g, 10 min, 4°C), and the SEAP‐containing supernatant was stored at 4°C until all of the samples were collected. A previously described enzymatic assay was used to measure SEAP protein activity.46, 47 Triplicate samples were heated at 65°C for 5 min, then 1 µL of each sample was added to 200 µL SEAP buffer (1 M diethanolamine, 0.5 mM MgCl2, 10 mM homoarginine) in a 96‐well microtiter plate. Samples were incubated at 37°C for 10 min, pNPP working buffer [20 µL; 5 mg p‐nitrophenyl phosphate (Sigma) in pNPP stock buffer (1 M diethanolamine, 0.5 mM MgCl2, 3.1 mM NaN3, pH 9.8)] was added to each well, and the microtiter plate was incubated at RT for 5 min in the dark. SEAP enzymatic activity was read at an absorbance of 405 nm using the Epoch BioTek microplate spectrophotometer (Fisher Scientific).
Statistics
Data are reported as mean ± standard deviations. Statistical significance (P ≤ 0.05) between treatments was determined by the Student two‐tailed t test.
Results and Discussion
We previously reported that expression of the Campoletis sonorensis ichnovirus (CsIV) vankyrin gene P‐vank‐1 in the Sf9–derived VE‐CL02 cell line inhibits apoptosis and prolongs cell survival after baculovirus infection, UV irradiation, or treatment with an apoptosis‐inducing chemical.24 In the present study, we tested the hypothesis that expressing vankyrin increases heterologous protein yields following baculovirus infection of vankyrin‐expressing insect cell lines (VE‐insect cells), or through infection with a baculovirus vector encoding vankyrin (VE‐baculovirus), or both. We tested the previously established monoclonal VE‐CL02 cell line,24 and also tested Trichoplusia ni High FiveTM cells stably transformed with P‐vank‐1 expression constructs, as High FiveTM cells have been reported to provide higher recombinant protein yields than S. frugiperda cells.40, 41, 42 Finally, we also tested SfSWT‐4 cells stably transformed with a P‐vank‐1 expression construct, as SfSWT‐4 can produce recombinant proteins with human‐type N‐glycans.39 Thus, we aimed to increase yield of glycoproteins with authentic human‐type N‐glycans by combining vankyrin‐expression with humanized glycoprotein processing. Monoclonal vankyrin‐expressing cell lines are designated as VE‐CL02, VE‐High Five and VE‐SWT.
Different cell culture media were tested to establish optimal growth conditions for SfSWT‐4 and VE‐SWT cell. We found that the highest cell density (6 × 106 cells mL−1) can be reached when the VE‐SWT cells were subcultured in Sf900III medium. However, due to a faster doubling time of VE‐SWT cells grown in TNM‐FH with FBS (24 h) compared to cells grown in Sf900III medium (72 h) in the first 3 days of culturing, we decided to routinely use TNM‐FH medium with FBS. In contrary to Invitrogen's Mimic cells, which require FBS as a source of sialic acid, SfSWT‐4 and VE‐SWT cell lines are able to produce terminally sialylated proteins in the absence of FBS.39
To test if recombinant protein yields were increased in these three vankyrin‐enhanced cell lines compared to their respective parental cell lines, we infected cells with a YFP‐encoding baculovirus, and analyzed YFP fluorescence and cell viability (Figure 1). Fluorescence images show that YFP expression is considerably higher in VE‐CL02 and VE‐High Five cells for the duration of the experiment when compared to Sf9 and High FiveTM cells, respectively (Figure 1A). These results were confirmed when we quantified YFP fluorescence using flow cytometry (Figure 1B). YFP fluorescence in VE‐CL02 cells increases threefold on 3 days post‐infection (dpi) and YFP fluorescence in VE‐High Five cells increases fivefold on 2 dpi compared to their parental cell lines. Interestingly, average YFP fluorescence is higher in VE‐High Five cells compared to VE‐CL02 or VE‐SWT cells (Figure 1B), which was in line with earlier reports of higher protein expression in T. ni cell lines as compared to S. frugiperda cell lines. A significant increase in YFP fluorescence is also detected in VE‐SWT cells at 4 and 5 dpi (Figure 1B), which correlates with increased longevity (Figure 1C). VE‐CL02 cell viability is significantly higher than Sf9 on days 3 and 4 dpi, whereas the viability of YFP baculovirus‐infected VE‐High Five is significantly higher than High FiveTM on earlier days post‐infection (days 2 and 3; Figure 1C). Thus, early inhibition of apoptosis by vankyrin appears to be more important for improving protein yields than its effect on viability at later time points in VE‐CL02 and VE‐High FiveTM cells. Our observation of increased cell viability in cells expressing P‐vank‐1 as compared to their parental cell lines correlate with increased protein production in these cells (compare Figure 1B with 1C). Hence, our results indicate that constitutive expression of P‐vank‐1 in stably transformed insect cell lines leads to enhanced protein yields through an increase in cell viability following baculovirus infection. This study is especially relevant when considering the use of the BEVS for the production of recombinant proteins for use in vaccines,34, 48 as well as for use in applied and basic research.
Next, we investigated whether vankyrin expression can enhance yields of intracellularly processed proteins. Manduca sexta pro‐hemolymph protease 14 (proHP14) is an initiating protease found in the serine proteinase pathway that is involved in insect innate immunity.49, 50, 51 ProHP14 encodes a signal peptide, five LDLa repeats—the first one of which tends to be lost during intracellular processing50, 51—one Sushi domain, and one Wonton domain followed by a serine protease catalytic domain. We set out to compare expression levels of the five LDL repeats (LDLa1–5) of M. sexta proHP14 in Sf9 and VE‐CL02 cell lines. Immunoblotting showed that at 3, 5, and 7 dpi, VE‐CL02 cells have higher levels of the regulatory domain LDLa1–5 as compared to Sf9 cells (Figure 2). Furthermore, a majority of the recombinant protein had a molecular mass of 34 kDa when expressed from VE‐CL02 cells, whereas the processed product, with most likely the first LDLa domain removed by a Sf9 intracellular processing enzyme (e.g., furin, convertase),52 is detected at around 27 kDa when proHP14 is expressed in Sf9 cells (Figure 2). Even after 7 days post‐infection, VE‐CL02 cells contain mainly the full‐length protein, whereas only very low levels of protein of either size could be detected in Sf9 cells (Figure 2). Our observations support the notion that proteins expressed in VE‐Sf9 cells undergo less proteolysis, and that the integrity of the secretory pathway in those cells is preserved for an extended period of time after baculovirus infection.
Figure 2.
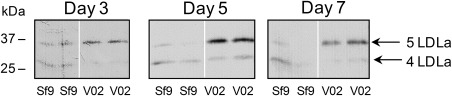
Vankyrin‐enhanced Sf9 cells, VE‐CL02, enhance protein yields of the five LDLa domains form of M. sexta pro‐hemolymph protease‐14.
Legend: Western blot analysis of cell free extracts determining protein levels of the five LDLa domain of M. sexta pro‐hemolymph protease 14 (proHP14) full‐length protein (5 LDLa; top band) and intracellularly processed protein (4 LDLa; bottom band). Sf9 or VE‐CL02 (designated V02) cells grown in static culture in Sf‐900™II medium with a seeding cell density of 1.6 × 106 cells mL−1 were infected with a baculovirus encoding the five LDLa domains in proHP14 at MOI 5. Samples were collected on days 3, 5, and 7 post‐infection, and protein extract from the same number of viable cells was analyzed. The experiment was carried out with duplicate samples.
Following baculovirus infection, host gene transcription is largely shut down and replaced with viral gene expression.53, 54, 55 Thus, vankyrin protein levels could potentially be increased further through the use of recombinant baculovirus vectors encoding P‐vank‐1. To produce and test such vectors, we first generated two new dual‐expression transfer vectors, pAcVE.02, and pAcVE.03 (Figure 3A). Each transfer vector has the P‐vank‐1 gene under transcriptional control of the late, very strong baculovirus p10 promoter.3, 56 These vectors also contain a multiple cloning site (MCS) downstream of the late, very strong polyhedrin promoter in the opposite orientation for insertion of a gene of interest.3, 56 Because the placement of purification tags are dependent on the type and function of the protein to be expressed, we designed pAcVE.02, which has an N‐terminal 8× His‐tag upstream and in frame with the MCS, and pAcVE.03, which has a C‐terminal 6× His‐tag in frame with the MCS. pAcVE.02 and pAcVE.03 both encode the honey bee melittin signal peptide (HBM) upstream of the MCS to enhance secretion.44, 57, 58
These vankyrin‐encoding transfer vectors and their counterparts lacking the P‐vank‐1 gene (as negative controls) were then used to generate recombinant baculoviruses. Each baculovirus was used to infect Sf9 insect cells, and cell viability was determined up to 10 days post‐infection. Cell viability is significantly increased in cells infected with the vankyrin‐encoding baculoviruses as early as day 2 post‐infection, and at 3 dpi cell viability is more than twice as high in cells infected with baculovirus harboring the vankyrin gene compared to cells infected with control viruses lacking the vankyrin gene (Figure 3B). A considerable number of cells are still viable 6 days after infection with a vankyrin‐enhanced baculovirus. These results indicate that the P‐vank‐1 gene also prolongs cell viability when expressed from the baculovirus vector.
To determine if baculovirus‐mediated vankyrin expression could also result in increased recombinant proteins yields, we inserted a gene encoding human erythropoietin (EPO) into pAcVE.03. Recombinant EPO is a glycoprotein hormone used to treat anemia, and its therapeutic efficacy requires human‐type N‐glycosylation.59, 60, 61, 62 SfSWT‐4 cells were infected with either a recombinant vankyrin‐enhanced baculovirus encoding EPO (EPO‐AcVE.03) or a recombinant baculovirus encoding EPO, but not vankyrin (EPO‐Ac.03). SfSWT‐4 cells remain viable until 8 dpi when infected with EPO‐AcVE.03, whereas cells infected with EPO‐Ac.03 are mostly nonviable by 5 dpi (Figure 4A). A concomitant increase in EPO yields was observed by immunoblotting (Figure 4B), indicating that baculovirus‐mediated vankyrin expression resulted in enhanced protein yields when the gene of interest was encoded by the same baculovirus.
Figure 4.
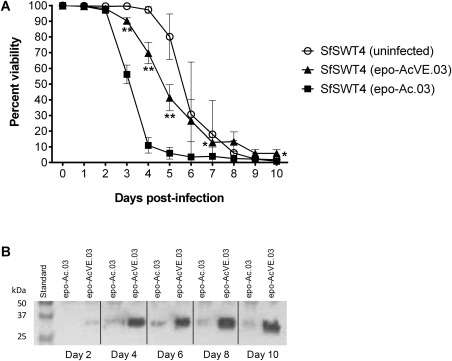
Enhanced mammalian glycoprotein yields when expressed from a vankyrin‐enhanced baculovirus.
Legend: (A) Cell viability was determined from suspension cultures grown in TNM‐FH medium with FBS of uninfected SfSWT4 cells, of SfSWT4 cells infected with a MOI 1 of the erythropoietin (epo) encoding baculovirus (epo‐Ac.03) or of the vankyrin‐enhanced epo‐baculovirus (epo‐AcVE.03) 0–10 days post‐infection (dpi). The cell densities of each culture at the time of infection (0 days) were ∼ 1 × 106 cells mL−1. The data presented here are representative of multiple (>3) experiments performed from which equivalent results were obtained. Statistical significance as determined by the Student two‐tailed t test for comparison of epo‐Ac.03 infected SfSWT4 cells vs. epo‐AcVE.03 infected SfSWT4 cells and is represented by either one asterisk (*; P ≤ 0.05) or two asterisks (**; P ≤ 0.01). (B) Western blot analysis determining protein levels of glycosylated erythropoietin (epo; 31 kDa compared to unglycosylated epo at 18.4 kDa69) in 5 μL culture supernatant for SfSWT4 cells infected with the epo‐encoding baculoviruse (epo‐Ac.03) or vankyrin enhanced baculovirus encoding epo (epo‐AcVE.03) at an MOI of 1. Samples were collected on days 2, 4, 6, 8, and 10 post‐infection.
Then, we explored if the prolonged cell viability and increased recombinant protein yields observed with baculovirus mediated‐vankyrin expression could be synergistically combined with cell lines engineered to stably express vankyrin. Thus, we evaluated the production of secreted alkaline phosphatase (SEAP) using a VE‐baculovirus in SfSWT‐4 and VE‐SWT cells. Clinical trials have shown promise for the use of SEAP in the treatment of acute renal failure, sepsis, and ulcerative colitis,63 and SEAP has been shown to improve outcomes in patients undergoing cardiac bypass surgery.64 Furthermore, SEAP can be accurately quantified using an enzymatic assay.46, 47
Higher alkaline phosphatase activity is detected in supernatants of infected VE‐SWT cells compared to those of SfSWT‐4 cells, irrespective of the type of baculovirus used (Figure 5). These observations further support the hypothesis that constitutive vankyrin expression increases recombinant protein yields. This result is consistent with results obtained by Lin et al. where Sf9 cells constitutively expressing AcMNPV P35 were infected with a recombinant SEAP baculovirus, and significantly higher protein levels were detected in that cell line compared to the parental cell line.19 Higher alkaline phosphatase activity is also observed when either SfSWT‐4 or VE‐SWT cells were infected with a vankyrin‐enhanced baculovirus encoding SEAP (SEAP‐AcVE.02) as compared to infection with a baculovirus encoding SEAP but not vankyrin (SEAP‐Ac.02; Figure 5). Similar data is seen when comparing SEAP‐AcVE.03 to SEAP‐Ac.03 virus infections (data not shown).
Figure 5.
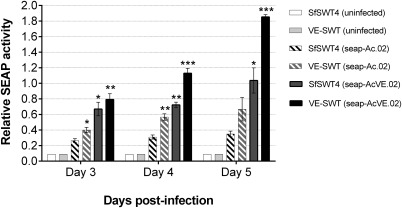
Synergistic effect on protein yield using a vankyrin‐encoding baculovirus to infect vankyrinenhanced insect cells.
Legend: Relative activity of secreted embryonic alkaline phosphatase (SEAP), measured at an absorbance of 405 nm, for uninfected SfSWT4 and VE‐SWT cells (solid light colored bars), SfSWT4 (dark striped bar) and VE‐SWT cells infected with SEAP encoded baculovirus (seap‐Ac.02; light striped bars), and SfSWT4 and VESWT cells infected with a vankyrin enhanced baculovirus encoding SEAP (seap‐AcVE.02; solid dark colored bars) at an MOI of 1 is shown. Static cultures were grown in TNM‐FH medium with FBS at a starting cell density of 5 × 105 cells mL−1. Samples were analyzed 3–5 days post‐infection. The protein activity data presented are means and standard deviations for triplicate determinations for each infection in a single experiment. The data presented here are representative of multiple (>3) experiments performed from which equivalent results were obtained. Statistical significance as determined by the Student two‐tailed t test for comparison of seap‐Ac.02 infected SfSWT4 cells (white bar with black stripes) vs. the other infected cells is represented by either one asterisk (*; P ≤ 0.05), two asterisks (P ≤ 0.01), or three asterisks (P ≤ 0.002).
The highest levels of alkaline phosphatase activity are detected when vankyrin‐enhanced insect cells (VE‐SWT) are infected with a vankyrin‐enhanced baculovirus encoding SEAP (SEAP‐AcVE.02). At 5 dpi, we observed a fivefold increase in SEAP activity in the VE‐SWT cells infected with SEAP‐AcVE.02 compared to SfSWT‐4 cells infected with SEAP‐Ac.02, a combination lacking any vankyrin expression. Taken together, these results suggest that the positive effects observed with cell‐mediated and baculovirus‐mediated vankyrin expression can be synergistically combined. Possibly, this combination provides vankyrin proteins early in infection from host cell expression, and during the late phase of infection from strong viral promoters, while vankyrin expression from stably integrated gene declines.65
In contrary to conventional BEVS where poor protein expression is often caused by loss of integrity of the secretory pathway during the late stages of baculovirus infection,66 we have previously shown that the secretory pathway is still functional in a vankyrin‐enhanced Sf9 cell line after infection with a baculovirus expressing a secreted protein.67 Here we report that the accumulation of two mammalian glycoproteins (Figures 4 and 5) in the medium continue to increase when expressed from a vankyrin‐enhanced baculovirus whereas protein accumulation ceased (Figure 5) or declined (Figure 4) over time when cells were infected with conventional baculovirus. Taken together, these results support the hypothesis that due to its anti‐apoptotic function, vankyrin has a positive effect on the integrity of the secretory pathway.
Several recombinant vaccines are produced in the BEVS,47 but this platform is not used to produce glycoprotein therapeutics, as most of these require complex, human‐type glycosylation patterns, which the insect cell lines used in the BEVS are unable to provide.32 In the present study, we showed that a cell line engineered to overcome this limitation of the BEVS can be combined with vankyrin‐enhancement technology to further increase production levels of humanized glycoproteins (Figures 4 and 5).
In summary, we document that vankyrin genes function to significantly improve cell viability and thus protein yields in baculovirus infected cells. Consequently, the VE‐BEVS offers a novel, significant, adaptable, and proven enhancement that substantially synergizes existing and improving BEVS technologies.
Conflict of Interest Disclosure
The transfer vectors pAcVE.02 and pAcVE.03 and the vankyrin enhanced VE‐CL02 cell line are currently being sold by ParaTechs Corporation. The VE‐BEVS transfer vectors are under patent protection United States Patent 7,629,160, and the VE‐CL02 cell line is protected under US patent 7,842,493.
Acknowledgments
The authors thank current and former members of the ParaTechs team for their guidance and input throughout this project, especially Esther Fleming for cell line passaging and Alisondra Maldanado for engineering the pVL‐YFP vector. They thank Algirdas Jesaitis for his evaluation of the VE‐CL02 cell lines and their superior performance in purifying the toxic components of flavocytochrome b558. Aspect of this research have been orally presented by KS at several scientific conferences: PEGS: the essential protein engineering summit (Boston MA, 2014), Peace Conference: protein expression in animal cells (San Diego CA, 2015), International society for bioprocess technology (Washington DC, 2016), and Cell line development & engineering conference (San Francisco CA, 2016). Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institute of Health under Award Numbers R42GM075628 and R44GM093411 to ParaTechs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. This technology was supported in part by an award from the Kentucky Cabinet for Economic Development, Office of Entrepreneurship, under the Grant Agreement KSTC‐184–512‐07–023 with the Kentucky Science and Technology Corporation to ParaTechs. This article is subject to the NIH Public Access Policy.
Literature Cited
- 1. Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3:2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pennock GD, Shoemaker C, Miller LK. Strong and regulated expression of Escherichia coli β‐galactosidase in insect cells with a baculovirus vector. Mol Cell Biol. 1984;4:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Contreras‐Gómez A, Sánchez‐Mirón A, García‐Camacho F, Molina‐Grima E, Chisti Y. Protein production using the baculovirus‐insect cell expression system. Biotechnol Prog. 2014;30:1–18. [DOI] [PubMed] [Google Scholar]
- 4. van Oers MM, Pijlman GP, Vlak JM. Thirty years of baculovirus‐insect cell protein expression: from dark horse to mainstream technology. J Gen Virol. 2015;96:6–23. [DOI] [PubMed] [Google Scholar]
- 5. Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22:1583–1587. [DOI] [PubMed] [Google Scholar]
- 6. Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. [DOI] [PubMed] [Google Scholar]
- 8. Vijayachandran LS, Viola C, Garzoni F, Trowitzsch S, Bieniossek C, Chaillet M, Schaffitzel C, Busso D, Romier C, Poterszman A, Richmond TJ, Berger I. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol. 2011;175:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. Molecular and structural characterization of the L1 virus‐like particles that are used as vaccine antigens in Cervarix™, the AS04‐adjuvanted HPV‐16 and −18 cervical cancer vaccine. Hum Vaccine. 2010;6:407–419. [DOI] [PubMed] [Google Scholar]
- 10. Cox MM, Izikson R, Post P, Dunkle L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv Vaccines. 2015;3:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lepenies B, Seeberger PH. The promise of glycomics, glycan arrays and carbohydrate‐based vaccines. Immunopharmacol Immunotoxicol. 2010;32:196–207. [DOI] [PubMed] [Google Scholar]
- 12. Blissard GW, Rohrmann GF. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35:127–155. [DOI] [PubMed] [Google Scholar]
- 13. Thomas CJ, Brown HL, Hawes CR, Lee BY, Min M‐K, King LA, Possee RD. Localization of a baculovirus‐induced chitinase in the insect cell endoplasmic reticulum. J Virol. 1998;72:10207–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodgson JJ, Arif BM, Krell PJ. Interaction of Autographa californica multiple nucleopolyhedrovirus cathepsin protease progenitor (proV‐CATH) with insect baculovirus chitinase as a mechanism for proV‐CATH cellular retention. J Virol. 2011;85:3918–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaba SA, Salcedo AM, Wafula PO, Vlak JM, van Oers MM. Development of a chitinase and v‐cathepsin negative bacmid for improved integrity of secreted recombinant proteins. J Virol Methods. 2004;122:113–118. [DOI] [PubMed] [Google Scholar]
- 16. Park EY, Abe T, Kato T. Improved expression of fusion protein using a cysteine‐ protease‐ and chitinase‐deficient Bombyx mori (silkworm) multiple nucleopolyhedrovirus bacmid in silkworm larvae. Biotechnol Appl Biochem. 2008;49:135–140. [DOI] [PubMed] [Google Scholar]
- 17. Ho Y, Lo HR, Lee TC, Wu CP, Chao YC. Enhancement of correct protein folding in vivo by a non‐lytic baculovirus. Biochem J. 2004;382:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gómez‐Sebastián S, López‐Vidal J, Escribano JM. Significant productivity improvement of the baculovirus expression vector system by engineering a novel expression cassette. PLoS One. 2014;9:e96562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin G, Li G, Granados RR, Blissard GW. Stable cell lines expressing baculovirus P35: resistance to apoptosis and nutrient stress, and increased glycoprotein secretion. In Vitro Cell Dev Biol Anim. 2001;37:293–302. [DOI] [PubMed] [Google Scholar]
- 20. Griffiths CM, Barnett AL, Ayres MD, Windass J, King LA, Possee RD. In vitro host range of Autographa californica nucleopolyhedrovirus recombinants lacking functional p35, iap1 or iap2. J Gen Virol. 1999;80:1055–1066. [DOI] [PubMed] [Google Scholar]
- 21. Ikeda M, Yamada H, Ito H, Kobayashi M. Baculovirus IAP1 induces caspase‐dependent apoptosis in insect cells. J Gen Virol. 2011;92:2654–2663. [DOI] [PubMed] [Google Scholar]
- 22. Kroemer JA, Webb BA. Divergences in protein activity and cellular localization within the Campoletis sonorensis Ichnovirus Vankyrin family. J Virol. 2006;80:12219–12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kroemer JA, Webb BA. Ikappabeta‐related vankyrin genes in the Campoletis sonorensis ichnovirus: temporal and tissue‐specific patterns of expression in parasitized Heliothis virescens lepidopteran hosts. J Virol. 2005;79:7617–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fath‐Goodin A, Kroemer JA, Webb BA. The Campoletis sonorensis ichnovirus vankyrin protein P‐vank‐1 inhibits apoptosis in insect Sf9 cells. Insect Mol Biol. 2009;18:497–506. [DOI] [PubMed] [Google Scholar]
- 25. Bitra K, Suderman RJ, Strand MR. Polydnavirus Ank proteins bind NF‐κB homodimers and inhibit processing of Relish. PLoS Pathog. 2012;8:e1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gueguen G, Kalamarz ME, Ramroop J, Uribe J, Govind S. Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF‐kappa B signaling in insect hosts. PLoS Pathog. 2013;9:e1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sinclair AM, Elliott S. Glycoengineering: the effect of glycosylation on the properties of therapeutic proteins. J Pharm Sci. 2005;94:1626–1635. [DOI] [PubMed] [Google Scholar]
- 28. Solá RJ, Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs Clin Immunotherap Biopharma Gene Ther. 2010;24:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS‐PROT database. Biochim Biophys Acta. 1999;1473:4–8. [DOI] [PubMed] [Google Scholar]
- 30. Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. [DOI] [PubMed] [Google Scholar]
- 31. Geisler C, Jarvis DL. Insect cell glycosylation patterns in the context of biopharmaceuticals In: Walsh G, editor. Post‐translational Modifications in the Context of Biopharmaceuticals, 1st ed Weinheim: Wiley‐VCH; 2009:165–191. [Google Scholar]
- 32. Geisler C, Mabashi‐Asazuma H, Jarvis DL. An overview and history of glyco‐engineering in insect expression systems. Methods Mol Biol. 2015;1321:131–152. [DOI] [PubMed] [Google Scholar]
- 33. Shi X, Jarvis DL. Protein N‐glycosylation in the baculovirus‐insect cell system. Curr Drug Targets. 2007;8:1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mena JA, Kamen AA. Insect cell technology is a versatile and robust vaccine manufacturing platform. Expert Rev Vaccines. 2011;10:1063–1081. [DOI] [PubMed] [Google Scholar]
- 35. Ikonomou L, Schneider Y‐J, Agathos SN. Insect cell culture for industrial production of recombinant proteins. Appl Microbiol Biotechnol. 2003;62:1–20. [DOI] [PubMed] [Google Scholar]
- 36. Vicente T, Roldão A, Peixoto C, Carrondo MJ, Alves PM. Large‐scale production and purification of VLP‐based vaccines. J Invertebr Pathol. 2011;107:S42–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmberger D, Klausberger M, Berger I, Grabherr R. MultiBac turns sweet. Bioengineered. 2013;4:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palmberger D, Wilson IB, Berger I, Grabherr R, Rendic D. SweetBac: a new approach for the production of mammalianised glycoproteins in insect cells. PLoS One. 2012;7:e34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mabashi‐Asazuma H, Shi X, Geisler C, Kuo CW, Khoo KH, Jarvis DL. Impact of a human CMP‐sialic acid transporter on recombinant glycoprotein sialylation in glycoengineered insect cells. Glycobiology. 2013;23:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krammer F, Schinko T, Palmberger D, Tauer C, Messner P, Grabherr R. Trichoplusia ni cells (High Five) are highly efficient for the production of influenza A virus‐like particles: a comparison of two insect cell lines as production platforms for influenza vaccines. Mol Biotechnol. 2010;45:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morais VA, Serpa J, Palma AS, Costa T, Maranga L, Costa J. Expression and characterization of recombinant human α‐3/4‐fucosyltransferase III from Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) cells using the baculovirus expression system. Biochem J. 2001;353:719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taticek RA, Choi C, Phan SE, Palomares LA, Shuler ML. Comparison of growth and recombinant protein expression in two different insect cell lines in attached and suspension culture. Biotechnol Prog. 2001;17:676–684. [DOI] [PubMed] [Google Scholar]
- 43. Shibuya N, Goldstein IJ, Broekaert WF, Nsimba‐Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(a2–6)Gal/GalNAc sequence. J Biol Chem. 1987;262:1596–1601. [PubMed] [Google Scholar]
- 44. Tessier DC, Thomas DY, Khouri HE, Laliberté F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. [DOI] [PubMed] [Google Scholar]
- 45. Jarvis DL. Recombinant protein expression in baculovirus‐infected insect cells. Methods Enzymol. 2014;536:149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schlaeger EJ, Kitas EA, Dorn A. SEAP expression in transiently transfected mammalian cells grown in serum‐free suspension culture. Cytotechnology. 2003;42:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang F, Murhammer DW, Linhardt RJ. Enzyme kinetics and glycan structural characterization of secreted alkaline phosphatase prepared using the baculovirus expression vector system. Appl Biochem Biotechnol. 2002;101:197–210. [DOI] [PubMed] [Google Scholar]
- 48. Felberbaum RS. The baculovirus expression vector system: a commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol J. 2015;10:702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ji C, Wang Y, Guo X, Hartson S, Jiang H. A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta . J Biol Chem. 2004;279:34101–34106. [DOI] [PubMed] [Google Scholar]
- 50. Takahashi D, Garcia BL, Kanost MR. Initiating protease with modular domains interacts with β‐glucan recognition protein to trigger innate immune response in insects. Proc Natl Acad Sci USA. 2015;112:13856–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Jiang H. Interaction of beta‐1,3‐glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta . J Biol Chem. 2006;281:9271–9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cieplik M, Klenk HD, Garten W. Identification and characterization of Spodoptera frugiperda furin: a thermostable subtilisin‐like endopeptidase. Biol Chem. 1998;379:1433–1440. [DOI] [PubMed] [Google Scholar]
- 53. Nguyen Q, Nielsen LK, Reid S. Genome scale transcriptomics of baculovirus‐insect interactions. Viruses. 2013;5:2721–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du X, Thiem SM. Responses of insect cells to baculovirus infection: protein synthesis shutdown and apoptosis. J Virol. 1997;71:7866–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mazzacano CA, Du X, Thiem SM. Global protein synthesis shutdown in Autographa californica nucleopolyhedrovirus‐infected Ld652Y cells is rescued by tRNA from uninfected cells. Virology. 1999;260:222–231. [DOI] [PubMed] [Google Scholar]
- 56. Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J Virol. 2013;87:6391–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sisk WP, Bradley JD, Leipold RJ, Stoltzfus AM, Ponce de Leon M, Hilf M, Peng C, Cohen GH, Eisenberg RJ. High‐level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus‐infected insect cells. J Virol. 1994;68:766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wicker‐Planquart C, Canaan S, Rivière M, Dupuis L, Verger R. Expression in insect cells and purification of a catalytically active recombinant human gastric lipase. Protein Eng. 1996;9:1225–1232. [DOI] [PubMed] [Google Scholar]
- 59. Browne JK, Cohen AM, Egrie JC, Lai PH, Lin FK, Strickland T, Watson E, Stebbing N. Erythropoietin: gene cloning, protein structure, and biological properties. Cold Spring Harb Symp Quant Biol. 1986;51(Part1):693–702. [DOI] [PubMed] [Google Scholar]
- 60. Fukuda MN, Sasaki H, Lopez L, Fukuda M. Survival of recombinant erythropoietin in the circulation: the role of carbohydrates. Blood. 1989;73:84–89. [PubMed] [Google Scholar]
- 61. Higuchi M, Oh‐eda M, Kuboniwa H, Tomonoh K, Shimonaka Y, Ochi N. Role of sugar chains in the expression of the biological activity of human erythropoietin. J Biol Chem. 1992;267:7703–7709. [PubMed] [Google Scholar]
- 62. Egrie JC, Browne JK. Development and characterization of novel erythropoiesis stimulating protein (NESP). Br J Cancer. 2001;84:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang Y, Millán JL, Mecsas J, Guillemin K. Intestinal alkaline phosphatase deficiency leads to lipopolysaccharide desensitization and faster weight gain. Infect Immun. 2015;83:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kats S, Brands R, Seinen W, de Jager W, Bekker MW, Hamad MA, Tan ME, Schönberger JP. Anti‐inflammatory effects of alkaline phosphatase in coronary artery bypass surgery with cardiopulmonary bypass. Recent Pat Inflamm Allergy Drug Discov. 2009;3:214–220. [DOI] [PubMed] [Google Scholar]
- 65. Jarvis DL. Effects of baculovirus infection on IE1‐mediated foreign gene expression in stably transformed insect cells. J Virol. 1993;67:2583–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jarvis DL, Oker‐Blom C, Summers MD. Role of glycosylation in the transport of recombinant glycoproteins through the secretory pathway of lepidopteran insect cells. J Cell Biochem. 1990;42:181–191. [DOI] [PubMed] [Google Scholar]
- 67. Fath‐Goodin A, Kroemer J, Martin S, Reeves K, Webb BA. Polydnavirus genes that enhance the baculovirus expression vector system. Adv Virus Res. 2006;68:75–90. [DOI] [PubMed] [Google Scholar]
- 68. Toth AM, Kuo CW, Khoo KH, Jarvis DL. A new insect cell glycoengineering approach provides baculovirus‐inducible glycogene expression and increases human‐type glycosylation efficiency. J Biotechnol. 2014;20:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]


