Abstract
The genes that encode the α- and β-chain subunits of vertebrate hemoglobin have served as a model system for elucidating general principles of gene family evolution, but little is known about patterns of evolution in amniotes other than mammals and birds. Here, we report a comparative genomic analysis of the α- and β-globin gene clusters in sauropsids (archosaurs and nonavian reptiles). The objectives were to characterize changes in the size and membership composition of the α- and β-globin gene families within and among the major sauropsid lineages, to reconstruct the evolutionary history of the sauropsid α- and β-globin genes, to resolve orthologous relationships, and to reconstruct evolutionary changes in the developmental regulation of gene expression. Our comparisons revealed contrasting patterns of evolution in the unlinked α- and β-globin gene clusters. In the α-globin gene cluster, which has remained in the ancestral chromosomal location, evolutionary changes in gene content are attributable to the differential retention of paralogous gene copies that were present in the common ancestor of tetrapods. In the β-globin gene cluster, which was translocated to a new chromosomal location, evolutionary changes in gene content are attributable to differential gene gains (via lineage-specific duplication events) and gene losses (via lineage-specific deletions and inactivations). Consequently, all major groups of amniotes possess unique repertoires of embryonic and postnatally expressed β-type globin genes that diversified independently in each lineage. These independently derived β-type globins descend from a pair of tandemly linked paralogs in the most recent common ancestor of sauropsids.
Keywords: gene family evolution, genome evolution, gene expression, hemoglobin, gene duplication, amniotes
Introduction
Repeated rounds of gene duplication and divergence can lead to the functional and regulatory diversification of multigene families, where different members acquire distinct biochemical functions and/or patterns of expression. During the evolution of deuterostomes, the duplication and functional divergence of members of the globin gene superfamily has been an important source of physiological innovation (Hoffmann, Opazo, et al. 2010; Hoffmann, Storz, et al. 2010; Blank et al. 2011; Storz, Opazo, et al. 2011; Hoffmann, Opazo, and Storz 2012; Hoffmann, Opazo, Hoogewijs, et al. 2012; Hoogewijs et al. 2012; Schwarze and Burmester 2013; Storz et al. 2013; Burmester and Hankeln 2014; Schwarze et al. 2014, 2015; Opazo, Lee, et al. 2015). Within this diverse and ancient superfamily, vertebrate hemoglobin (Hb) genes comprise one of the most intensively studied gene families from a functional and evolutionary perspective (Hardison 2012; Storz 2016), providing an outstanding opportunity to assess the phenotypic consequences of changes in gene content.
The α- and β-globin genes of jawed vertebrates (gnathostomes) encode subunits of tetrameric (α2β2) Hb, the red blood cell protein that is responsible for the circulatory transport of oxygen and carbon dioxide. The ancestral α- and β-globin genes derive from the tandem duplication of a proto-Hb gene in the common ancestor of gnathostomes, and the resulting linked arrangement of α- and β-globin genes has been retained in cartilaginous fishes, ray-finned fishes, and amphibians (Jeffreys 1982; Hosbach et al. 1983; Gillemans et al. 2003; Fuchs et al. 2006; Opazo et al. 2013; Opazo, Hoffmann, et al. 2015; Opazo, Lee, et al. 2015). However, this ancestral arrangement has been lost in amniotes due to the translocation of the β-globin locus to a new chromosomal location, so that the α- and β-globin gene clusters are located on different chromosomes in this group (Hardison 2008; Patel et al. 2008; Hoffmann, Storz, et al. 2010; Hoffmann, Opazo, and Storz 2012).
For historical reasons, comparative studies of the α- and β-globin gene clusters of amniotes have mainly focused on mammals because of the greater availability of whole-genome assemblies. These studies have revealed very rapid rates of gene turnover, which result in high levels of variation in gene content among species (Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b, 2009; Storz et al. 2008; Runck et al. 2009; Gaudry et al. 2014). Our understanding of globin gene clusters in other amniotes is much more limited. The release of multiple avian genomes provided expanded opportunities for comparative studies of the α- and β-globin clusters among vertebrates and revealed far lower rates of gene turnover in birds than in mammals (Zhang et al. 2014; Opazo, Hoffmann, et al. 2015). Until recently, a comprehensive examination of variation in sauropsids, the sister groups of mammals, was not possible due to a lack of comparative data from crocodilians, turtles, squamates (lizards and snakes), and rhynchocephalians (represented by tuatara). This dearth of genomic data has limited our ability to decipher orthologous and paralogous relationships among the globin genes of different taxa. With the recent release of multiple nonavian sauropsid genomes (Alföldi et al. 2011; Castoe et al. 2013; Wan et al. 2013; Wang et al. 2013; Green et al. 2014; Liu et al. 2015), we can now extend these studies to include all major groups of amniotes.
Comparative studies suggest that most amniotes have retained copies of three tandemly linked α-type globin genes that were present in the last common ancestor of tetrapods: 5′-αE-αD-αA-3′ (Hoffmann and Storz 2007; Hoffmann, Storz, et al. 2010; Grispo et al. 2012). With the exception of mammals, where most species have multiple copies of αE- and αA-globin, most variation in gene family size among tetrapods is attributable to lineage-specific losses of one of the three ancestral paralogs. By contrast, the β-globin gene clusters have had a much more dynamic duplicative history, with multiple lineage-specific expansions of gene family size. As a result, the major lineages of amniotes share a common set of orthologous α-globin genes, but they have unique sets of β-globin genes that diversified via independent rounds of lineage-specific duplication and divergence (Opazo et al. 2008a; Hoffmann, Storz, et al. 2010; Storz, Hoffmann, et al. 2011; Storz 2016). In general, the linkage order of the globin genes reflects their temporal order of expression during development (Cirotto et al. 1987; Ikehara et al. 1997; Alev et al. 2008, 2009; Storz, Hoffmann, et al. 2011). This is the case in the mammalian and avian β-globin gene clusters, even though the lineage-specific gene repertoires diversified independently. In both cases, the β-type globin genes located at the 5′ end of the cluster are expressed during the earliest stages of embryogenesis whereas the genes at the 3′ end are expressed during adulthood.
In the current study, we take advantage of newly released whole genome sequences to investigate patterns of diversification of sauropsid α- and β-globin genes. Specifically, the objectives of the present study were 1) to characterize changes in the size and membership composition of the α- and β-globin gene families within and among the major sauropsid lineages, 2) to reconstruct the evolutionary history of the sauropsid α- and β-globin genes and to resolve orthologous relationships among them, and 3) to reconstruct evolutionary changes in the developmental regulation of gene expression. Our comparisons revealed contrasting patterns in the evolution of the α- and β-globin clusters. Evolutionary changes in the α-globin gene cluster are attributable to the differential retention of ancestral duplicates among lineages, whereas changes in the β-globin gene cluster are attributable to differential gene gains and losses. In addition, we identified differences in α- and β-globin expression between squamates and the group that includes testudines, crocodilians, and birds. In the case of β-globin, squamates and rhynchocephalians express adult Hb isoforms that incorporate the products of different β-type globin genes, whereas a single but independently derived gene encodes the β-type subunits of adult Hb in mammals, archosaurs, and turtles. Taken together, our results suggest that the regulatory architectures of the β-globin gene clusters evolved independently in mammals and in the group comprising archosaurs plus turtles.
Materials and Methods
Bioinformatic Searches
We implemented bioinformatic searches for α- and β-globin sequences in sauropsid genomes in two stages: we first focused on genomic data and we then incorporated auxiliary sequence data from transcriptomes. To characterize the sauropsid α- and β-globin gene clusters we searched for traces of α- and β-globin genes in the genomic sequence records from representative sauropsids in the Ensembl or NCBI (refseq_genomic, htgs, and wgs) databases using BLAST (Altschul et al. 1990). In all cases we used low stringency settings, searching with the BLASTn algorithm, with match/mismatch scores of 1 and −1, and gap existence and extension costs of 2 and 1, respectively. The full set of species is listed in supplementary table S1, Supplementary Material online, and includes ten squamates, four testudines, four crocodilians, and two birds. Even though there are more bird genome sequences available, variation in the α- and β-globin gene clusters of birds, which is the most studied group of sauropsids, has been covered recently (Opazo, Hoffmann, et al. 2015). Thus, we have included two species in the current study, zebra finch and chicken, which have high-quality genomes and provide good representation of avian diversity for comparative purposes. Once we located all genomic fragments containing traces of α- and β-like globin genes, we extracted the corresponding fragments and verified preliminary annotations by comparing identified sequences against the well-annotated α- and β-globin genes of human and chicken using BLAST2 (Tatusova and Madden 1999), as in Hoffmann, Storz, et al. (2010). In a second stage, we interrogated sauropsid transcriptomes available in NCBI and the Reptilian Transcriptomics database (www.reptilian-transcriptomes.org; last accessed June 2017; Tzika et al. 2015) for α- and β-type globin sequences in order to expand our taxonomic coverage. Information regarding the sources of all sequences in our analyses is provided in supplementary tables S1–S3, Supplementary Material online. For the purpose of our analyses, sequences were considered putatively functional if the predicted lengths of the exons matched expected lengths for amniote α- and β-globins, if there were no premature stop codons, and if start and stop codons were found at expected positions. Conversely, α- and β-like globin sequences were considered as pseudogenes if they include premature stop codons.
Data Curation
In the current assembly of the anole lizard genome, the αAglobin gene lies on a separate contig from the αD and GbY genes, whereas the king cobra assembly is missing the αA gene altogether. However, we suspect that these are issues related to the current assemblies of anole lizard and king cobra genomes because the gekko assembly places αA in the canonical genomic location, flanked by αD and GbY. In contrast to mammals, pseudogenes are rare in the α-globin clusters of sauropsids; the sole exception was a clearly recognizable αA pseudogene found in the python. In addition, our bioinformatic searches identified two different αA sequences in the common viper genome, two different αD sequences in the garter snake genome, and two identical αAparalogs in the same fragment of the Chinese softshell turtle genome. Given the nature of the current assemblies, we assume that this apparent sequence heterogeneity is attributable to assembly artifacts, although we cannot rule out the possibility that the sequences represent products of recent duplication events within each of the species.
The current assemblies of most of the sauropsid β-globin gene clusters are fragmented and appear to be missing genes in some cases. For example, the most current assembly of the anole lizard genome, AnoCar2.0, includes a single β-globin gene (Ensembl: ENSACAG00000012173, GeneID: 100552694), but analyses of transcriptomic and proteomic data indicate the presence of at least one additional β-type globin gene that is clearly distinct at the sequence level (Storz, Hoffmann, et al. 2011), and was present in AnoCar1.0 (ENSACAG00000010799). In the case of the Chinese alligator genome, the assembly includes an en bloc duplication involving the T1–T10 paralogs (supplementary fig. S1, Supplementary Material online). Because gene content is conserved between American alligator, saltwater crocodile and gharial, we use these three species as representatives of class Crocodilia for the comparative genomics portion of the study. Crocodilian β-globin gene clusters also stand out because of the relatively high number of pseudogenes, which are shared among the species.
Because some squamate genomes have incomplete coverage of the genes of interest, we completed the set of genomic sequences with additional transcriptome records whenever possible. For example, in the case of the corn snake and the python, the current assembly includes portions of two distinct genes, which correspond to the two β-globin gene sequences derived from their transcriptomes (available at the Reptilian Transcriptomics 2.0 database; Tzika et al. 2015). In the case of the two rattlesnake species for which genome sequence data were available, the current assemblies lack complete coverage of the α-globin gene cluster, so we also included the globin repertoire of the closely related Eastern diamond rattlesnake, derived from a venom-gland transcriptome (Rokyta et al. 2012). Similarly, in the case of the garter snake, the αA-globin gene is not present in the current assembly, so we included a copy of this gene from a transcriptome of the same species. In a second stage, we extended our analyses to include sequences derived from transcriptomes, with a view to increasing taxonomic coverage of squamates and testudines. Finally, in the case of the tuatara, because it is the sole representative of the order Rhynchocephalia, we included three transcriptome-derived sequences in the analyses of genome-derived sequences (two α-type globins and one β-type globin). We also compared amino acid sequences derived from direct peptide sequencing to conceptual translations of the coding sequences. The list of these additional amino acid sequences is provided in supplementary table S4, Supplementary Material online.
In many squamate transcriptomes we identified β-globin paralogs that appear to be assembly artifacts, as initial searches identified the presence of three or four putative paralogs, but careful examination revealed that these sequences represent different chimeric combinations of two closely related sequences. By default, we assumed that the paralogs more similar to genome-derived sequences were the “true” paralogs. Although chimeric fusion genes are relatively common among mammalian β-globins (Hoffmann et al. 2008b; Opazo et al. 2009; Gaudry et al. 2014), to err on the side of caution, we provisionally consider these transcripts as bioinformatic artifacts.
Phylogenetic Analyses
Phylogenetic relationships among the α- and β-type globin sequences were estimated using maximum likelihood (ML) and Bayesian analyses (BA). For these analyses, nucleotide and amino acid sequences of vertebrate globin genes were aligned using the program MAFFT version 7.304 (Katoh and Standley 2013), as implemented in the following server: http://mafft.cbrc.jp/alignment/server/, last accessed on January 2017. ML analyses were run using IQ-Tree ver 1.5.5 (Nguyen et al. 2015) in the implementation of IQ-Tree available from the IQ-Tree web server (Trifinopoulos et al. 2016) last accessed on June 2017, and support for the nodes was evaluated with 1, 000 pseudoreplicatesof the ultrafast bootstrap procedure (Minh et al. 2013). Bayesian Analyses were performed in MrBayes version 3.2 (Ronquist et al. 2012), running four simultaneous chains for 2×107 generations, sampling trees every 1,000 generations, and using default priors. We assessed convergence by measuring the SD of the split frequency among parallel chains. Chains were considered to have converged once the average split frequency was <0.01. We discarded trees collected before the chains reached convergence, and we summarized results with a majority-rule consensus of trees collected after convergence was reached. Alternative topologies were compared using the approximately unbiased test proposed by Shimodaira and Goldman (2002), as implemented in IQ-Tree.
Organismal Phylogeny and Divergence Dates
In all cases, we assumed that relationships among the different lineages studied: birds, crocodilians, rhyncocephalians, squamates, testudines, plus amphibians and mammals which were included as outgroups follow the arrangement reported by Crawford et al. (2012), based on the analyses of ultraconserved genetic elements. Estimates of divergence times among the lineages were obtained from the TimeTree server (Kumar, Stecher, et al. 2017).
Nomenclature
Several naming conventions have been proposed for the α- and β-globin genes of vertebrates, which in many cases result in inconsistent schemes (see Aguileta et al. 2006 for a review). As a result, in some schemes orthologous genes receive different names, such as the π- and ζ-globins in the α-globin gene clusters of birds and mammals, respectively, which are 1:1 orthologs. Conversely, in other schemes paralogous genes derived from independent duplications receive the same name, such as the ɛ-globin genes in the β-globin gene families of birds and mammals, which have the same name but are not 1:1 orthologs. In the case of the α-globin gene cluster, it is straightforward to reconcile nomenclature with the duplicative history of the genes. Accordingly, genes orthologous to the early expressed π-globin of chicken are labeled αE-globin, genes orthologous to the αD-globin of chicken are labeled αD-globin, and genes orthologous to αA-globin of chicken are labeled αA-globin. The case of the amniote β-globin genes is more complex. In the case of avian and mammalian β-globins, which derive from lineage-specific duplications, we will use the greek-letter labeling that is conventionally used for each, 5′-ρ-βH-βA-ɛ-3′ in birds, and 5′-ɛ-γ-η-δ-β-3′ in mammals, noting that the avian and mammalian ɛ-globin are not orthologous. Snake β-globins, which can be classified into two well defined groups are referred to as βI or βII, to faciliate comparisons with previous work (Gorr et al. 1998; Storz et al. 2015). Finally, in the case of crocodilian and testudine β-globins, we have labeled them with a T followed by a number indicating the presumptive position of the gene on the cluster in the forward direction, so that the β-globin gene on the 5′ end is labeled as Hbb-T1 and the genes downstream are labeled as T2, T3, and so forth. In this scheme, the bird ρ-βH-βA-ɛ globins would be labeled Hbb-T1 through T4.
Gene Expression Analyses
We collected publically available squamate, crocodilian, and turtle liver transcriptomes from McGaugh et al. (McGaugh et al. 2015). Sequences corresponding to the αA and αDgenes were identified using BLASTX and a database of previously annotated αA and αD amino acid sequences. Raw paired-end Illumina reads (SRA062458 and SRP071466) were cleaned and trimmed using Trimmomatic (Bolger et al. 2014) and were used to estimate expression of αA and αD-globin in each species with RSEM (Li and Dewey 2011). Transcripts per million (TPM) was used as the expression metric.
Results
We integrated synteny, phylogenetic, and expression analyses to reconstruct the evolution of the α- and β-globin gene clusters of sauropsids. Briefly, we surveyed genome databases to locate the sauropsid α- and β-globin gene clusters. We then annotated the α- and β-type globin genes in conjunction with the genes flanking the 5′ and 3′ ends of the globin clusters, and we used phylogeny reconstructions to resolve orthology and paralogy. We then compared the resulting trees with the organismal phylogeny to reconstruct ancestral repertoires and we incorporated gene and/or protein expression data to infer developmental patterns of expression. These surveys spanned all orders of sauropsids other than rhynchocephalians and included ten species of squamates (both lizards and snakes), four testudines, four crocodilians, plus zebra finch, and chicken as representative birds (see full list in supplementary table S1, Supplementary Material online).
Synteny
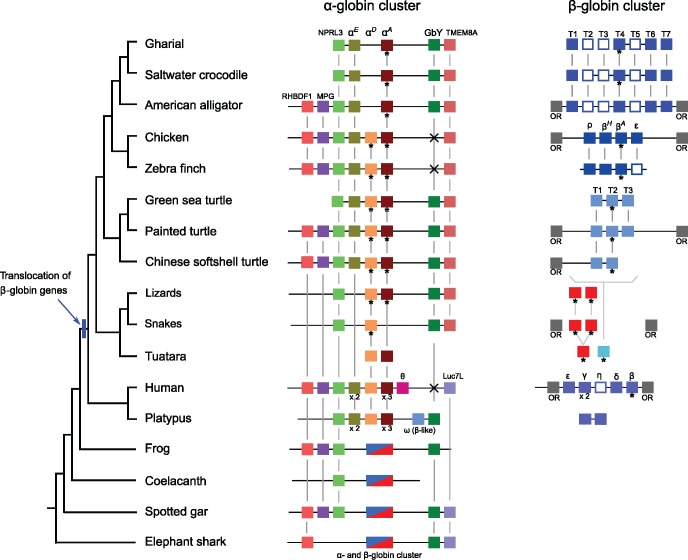
Comparative genomic analyses revealed that the α- and β-globin gene clusters of sauropsids both exhibit a high level of conserved synteny. The 5′ end of the α-globin cluster is flanked by orthologs of NPRL3, which spans multiple important cis-regulatory elements that govern the expression of α-type globins (fig. 1). Available data suggest that the conserved synteny of the genes flanking the sauropsid α-globin gene cluster extends to RBHDF1 and MPG (fig. 1). On the 3′ end of the α-globin gene cluster, sauropsids share a derived, small-scale inversion, so that the cluster is flanked by TMEM8A, rather than by Luc7L, as in mammals and other vertebrate groups (Opazo, Lee, et al. 2015). In addition, squamates, crocodilians, and testudines, but not birds, have retained a copy of Globin-Y (GbY) at the 3′ end of the α-globin gene cluster, an ancient vertebrate globin with an unknown functional role that also flanks the α-globin gene cluster of platypus, and the α- and β-globin gene clusters of frog (Xenopus), spotted gar, and elephant shark (fig. 1).
Fig. 1.
—Syntenty of the α- and β-globin gene clusters of representative vertebrates (arranged from left to right in 5′ to 3′ order) with a focus on saurospids. Genes inferred to be orthologs are linked by vertical lines, and adult-expressed genes are identified by an asterisk. White boxes correspond to pseudogenes. In the case of the to squamates, the lizard gene clusters combine data from Japanese gekko (α-globin) and the AnoCar1.0 release of the Green anole (β-globin), and the snake gene clusters combine data from king cobra (α-globin) and Burmese python (β-globin). Note that we lack a genome assempbly for the tuatara, the sole representive of the order Rhynchocephalia. The secondary loss of GbY in birds and humans is denoted by an “x.”
Patterns of conserved synteny in the β-globin gene cluster are less clear due to the fragmentary nature of this chromosomal region in the current genome assemblies of most sauropods. Among the species studied, flanking gene information for both sides of the β-globin gene clusters is available for chicken, the two alligators, and painted turtle, and in all these cases the β-globin gene cluster is flanked by olfactory receptors on both sides, as is the case with mammals (fig. 1). This also seems to be the case for king cobra and common viper as well, but the current assemblies are too fragmentary to draw definitive conclusions. Because of the high rate of gene gain and loss among the olfactory receptors of the different sauropsid lineages (Vandewege et al. 2016), establishing 1:1 orthologous relationships among olfactory receptors of these different lineages was not possible.
The α- and β-Globin Gene Clusters of Sauropsids
We found limited variation in the number of genes in the α- and β-globin gene clusters of most sauropsids relative to mammals (fig. 1). Birds and turtles have retained the ancestral complement of three α-type globin genes (αE, αD, and αA), whereas the other sauropsid taxa possessed alternative combinations of two genes: αD and αA in squamates, and αEand αA in crocodilians. We verified the absence of the αE-globin gene in squamates by comparing crocodilian and testudine αE-globin sequences against squamate sequences in GenBank and the Reptilian transcriptomics databases using low stringency searches. We were able to retrieve the αD and αA paralogs that were already present in our analyses, and the more distant β-globin paralogs as well, but we found no match that corresponded to an αE-like globin sequence. We also verified the absence of αD-globin in crocodiles by comparing avian and testudine αD-globin sequences against crocodilian sequences in GenBank and the Reptilian transcriptomics databases. In this case, we were able to retrieve the αE and αA sequences that were already present in our analyses, and we also recovered an intronless retroprocessed αD-globin pseudogene in the genomes of all crocodilians, located on a separate scaffold from the α-globin gene cluster, and flanked by the craniofacial development protein 1 (CFDP1) and the breast cancer antiestrogen resistance protein 1 (BCAR1) in each of the four crocodilian species surveyed. The fact that our protocols can identify β-type globin sequences when seeded with an α-like sequence, and can also recover divergent pseudogenes such as the αD-globin pseudogene of crocodilians, suggests that our searches were comprehensive. In addition, average pairwise distances among the αE, αD, and αA paralogs range from 38% to 40% at the nucleotide level, and from 48% to 50% at the amino acid level (supplementary table S5 and supplementary files S1 and S2, Supplementary Material online), indicating that these three α-globin paralogs are clearly distinct from each other. We therefore conclude that the apparent absence of αDin crocodilians and the absence of αE-globin in squamates are not artifacts of our search protocol.
The β-globin gene cluster of sauropsids is more variable than the α-globin cluster. In this case, we could not resolve orthology relationships using bioinformatic searches. Thus, we initially labeled the genes using numbers, which in the case of well-resolved clusters use the prefix “T,” and the numbers denote the order of the gene in the tandem array, so that the T1 is the first gene on the 5′ end, followed by T2 and so forth. We found two intact β-globin genes in most squamates, but could not determine their order in any of the species examined. On the other hand, we were able to resolve the tandem arrays of β-globin genes for birds, crocodiles, and testudines. There were two or three genes in turtles, labeled T1–T3, four in birds, labeled T1–T4 (which correspond to ρ-, βH-, βA-, and ɛ-globin), and four genes with three pseudogenes in crocodilians, labeled T1–T7 (fig. 1). The Chinese alligator was the only exception, as this species appears to have an en bloc duplication of the β-globin gene cluster, which includes five putatively funtional genes and seven pseudogenes labeled T1–T12, where the T7–T10 block of genes is a duplication of the T3–T7 paralogs (supplementary fig. S1, Supplementary Material online). Average pairwise distances for the different sets of β-globins paralogs were lower than the distances among α-globin paralogs, ranging from 8% to 33%, (supplementary table S6 and supplementary files S3 and S4, Supplementary Material online).
Phylogenies of the α- and β-Globin Genes of Sauropsids
We were able to resolve orthologous relationships of the sauropsid α- and β-globin genes using maximum likelihood and Bayesian phylogenetic analyses. In order to increase our taxonomic sampling, we combined the genome-annotated sequences with additional transcriptome records. In particular, the latter included two α- and one β-globin sequences from tuatara, so that representatives of all major groups of sauropsids were included in the analyses.
α-Globin Gene Cluster
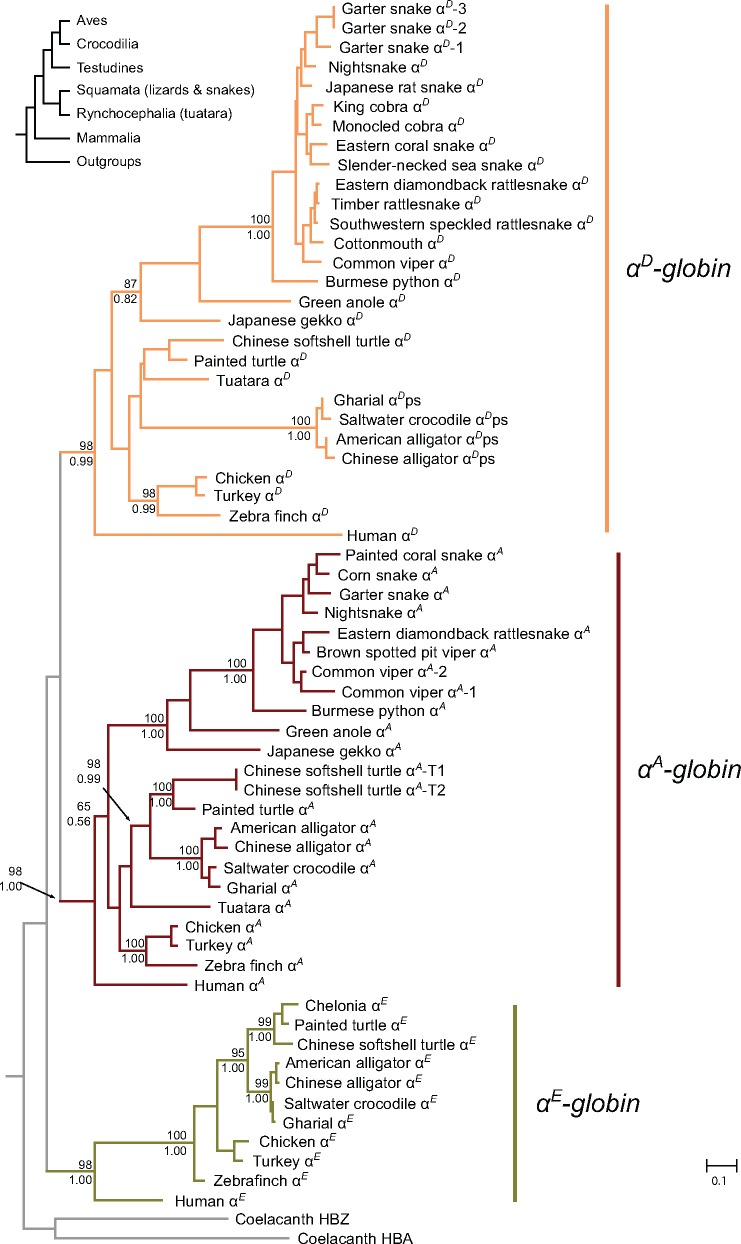
Our estimated phylogenies of the α-type globins confirmed our primary orthology assignments, as the sauropsid αE, αD, and αA genes group with their mammalian orthologs with high confidence, and the crocodilian αD-like pseudogene groups with the αD genes of birds, testudines, tuatara, and squamates (fig. 2). Within each of these α-globin clades, relationships among sequences did not deviate significantly from the expected organismal relationships. The tree indicates that variation in the number of α-globin genes is largely attributable to gene losses: the common ancestor of squamates appears to have lost the αE-globin gene whereas the common ancestor of crocodilians apparently lost αD-globin (fig. 2). We identified two α-like transcripts in the tuatara transcriptome available, which correspond to the αD and αA genes.
Fig. 2.
—Phylogeny of sauropsid α-type globin genes derived from genomes and transcriptomes. Support for the relevant nodes are indicated as bootstrap percentages from IQ-Tree above the nodes, and as Bayesian posterior probabilities from MrBayes below.
β-Globin Gene Cluster
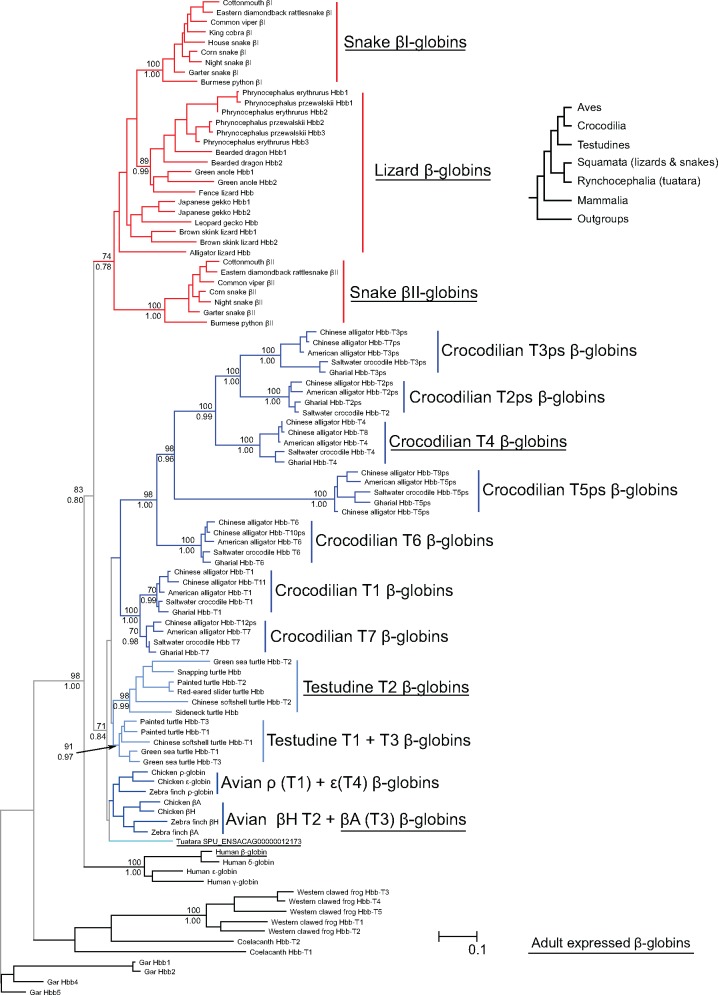
In the case of the β-globin gene family, our analyses based on nucleotide sequence suggest that birds, crocodilians, testudines, and squamates each posses an independently derived set of β-globin genes (fig. 3). Our analyses place sauropsid and mammalian β-globins as sister groups, and the former were divided into two clades, the first one containing the β-globin sequences from squamates, and the second one containing β-globin sequences of birds, crocodilians, and testudines. In general, the arrangements do not deviate significantly from the expected organismal relationships (Crawford et al. 2012). The one exception was the presence of one tuatara beta globin gene, SPU_ENSACAG00000012173, in the clade that includes birds, turtles, and crocodiles, which suggests that this gene lineage traces back to the ancestor of sauropsids and that it was secondarily lost in the ancestor of lizards and snakes. Forcing the β-globins of this second clade to follow the expected organismal arrangement, with tuatara as the deepest node, followed by testudines, with crocodilian β-globins as sister to avian β-globins did not result in a significant loss in likelihood score in an Approximately Unbiased topology test.
Fig. 3.
—Phylogeny of sauropsid β-type globin genes derived from genomes and transcriptomes. Genes or groups of genes that are expressed in adulthood are underlined. Support values for relevant nodes are indicated as bootstrap percentages (above) and as Bayesian posterior probabilities (below).
Within the squamate clade, there are two well-defined groups of snake β-globins, corresponding to the βI and βII clades defined by Storz et al. (2015). Lizard β-globins are paraphyletic relative to the snake βI-globin clade (fig. 3) and relationships among the lizard sequences do not deviate from known organismal relationships (Pyron et al. 2013). Orthology among lizard β-globins can only be resolved for a small subset of the genes because when multiple β-globin paralogs for a lizard species are present, they are usually very similar (fig. 3), a pattern that may reflect a history of lineage-specific gene duplications, interparalog gene conversion, or a combination of these two processes. For example, intraspecific distances among the β-globin paralogs of lizards range from 1% to 15%, lower than the comparisons between the snake βI- and βII-globins, which range from 22% to 27% (supplementary file S3, Supplementary Material online).
In our maximum-likelihood analyses, avian, crocodilian, and testudine β-globins form a monophyletic clade (fig. 3). In all cases, we found that genes at the 5′ and 3′ ends of the cluster grouped together in a clade, and the genes in center of the cluster are grouped in a second clade. Thus, the avian T1 (ρ) and T4 (ɛ) paralogs are sister to the clade that includes the avian T2 (βH) and T3 (βA) paralogs, the crocodilian T1 and T7 paralogs are sister to the clade that includes the T2–T6 paralogs, and the testudine T1 and T3 paralogs are sister to the clade of testudine T2s (fig. 3). It is noteworthy that crocodilian pseudogenes are shared across all species, which indicates they were already inactive in their last common ancestor, which existed ∼80 Ma.
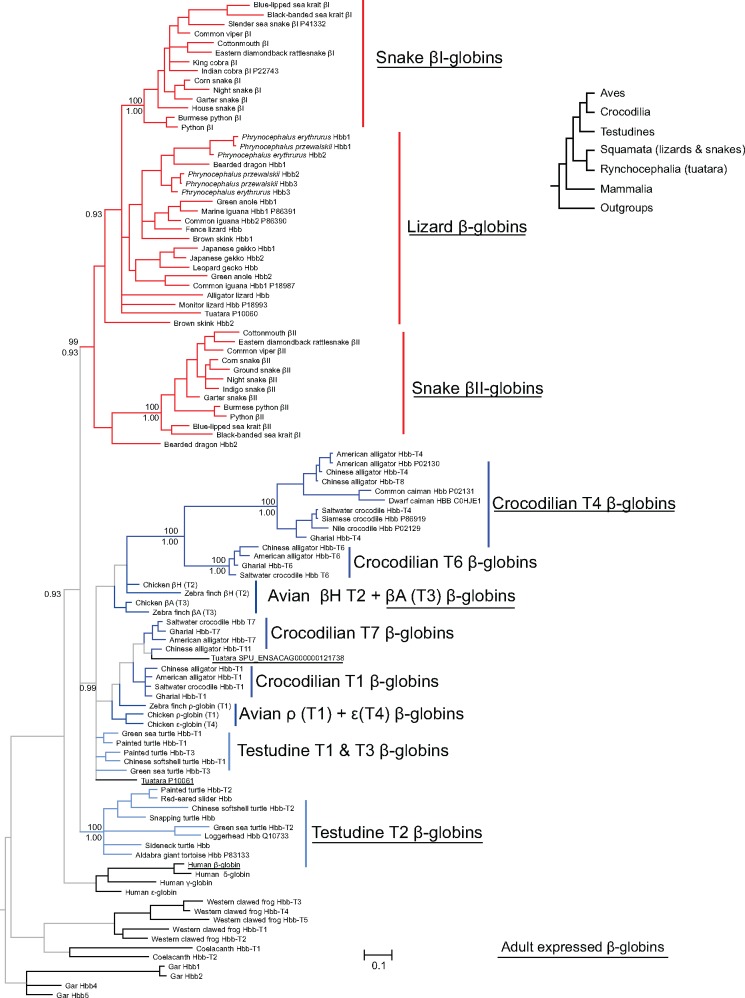
We then estimated phylogenetic affinities based on amino acid sequences in order to include two tuatara β-globin amino acid sequences, P10060 and P10061, as well protein records from testudines, squamates, and crocodilians that have no associated nucleotide sequence (fig. 4). The tree based on amino acid sequences was largely congruent with the one based on nucleotide sequences, but resolution was relatively poor due to the large number of sequences included relative to the number of characters (108 vs. 147). One of the tuatara β-globin amino acid sequences, P10060, grouped with the squamate β-globins, whereas the other two tuatara β-globins, protein record P10061 and the one derived from the trascriptome, fell in the clade that includes the β-globins of crocodilians, birds, and the T1 and T3 paralogs of testudines. Integrating the nucleotide and amino acid based phylogenies with protein and transcript sequences indicates that the β-chain subunits of adult Hb are products of single genes: Hbb-T3 (βA) in birds, Hbb-T4 in crocodilians, and Hbb-T2 in testudines.
Fig. 4.
—Phylogeny of β-type globin genes based on amino acid sequences, including additional protein records. Genes or groups of genes that are expressed in adulthood are underlined. Support values for relevant nodes are indicated as bootstrap percentages (above) and as Bayesian posterior probabilities (below).
Discussion
Our analyses capture the contrasting evolutionary pattern of the α- and β-globin gene families of sauropsids. In the case of the α-globin gene family, differences are restricted to the differential loss of one of the three paralogs present in the common ancestor. As a result, testudines and birds have retained copies of αE, αD, and αA, whereas squamates have retained αD and αA, and crocodilians have retained αE and αA (along with a vestige of αD in the form of processed pseudogene). By contrast, variation in gene copy number is more extensive in the β-globin gene cluster (fig. 1). Our previous study suggested that each of the major lineages of amniotes evolved distinct β-globin repertoires via repeated rounds of lineage-specific gene duplication (Hoffmann, Storz, et al. 2010). However, that study only included anole lizard as a representative of squamates and it did not include turtles or crocodilians. Interestingly, the phylogenies obtained with our increased taxonomic sampling suggest an even more extreme pattern of lineage-specific β-globin expansion (fig. 3), where the major groups of sauropsids (birds, crocodilians, testudines, and squamates) each possess repertoires that derive from lineage-specific duplications. In spite of the observed contrast in evolutionary dynamics between the α- and β-globin gene families, variation in gene content within the different sauropsid groups appears to be limited. This is in stark contrast to mammals, which exhibit high levels of variation in the size and membership composition of the α- and β-globin gene clusters (Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b; Gaudry et al. 2014).
Evolution of the Sauropsid α- and β-Globin Gene Families
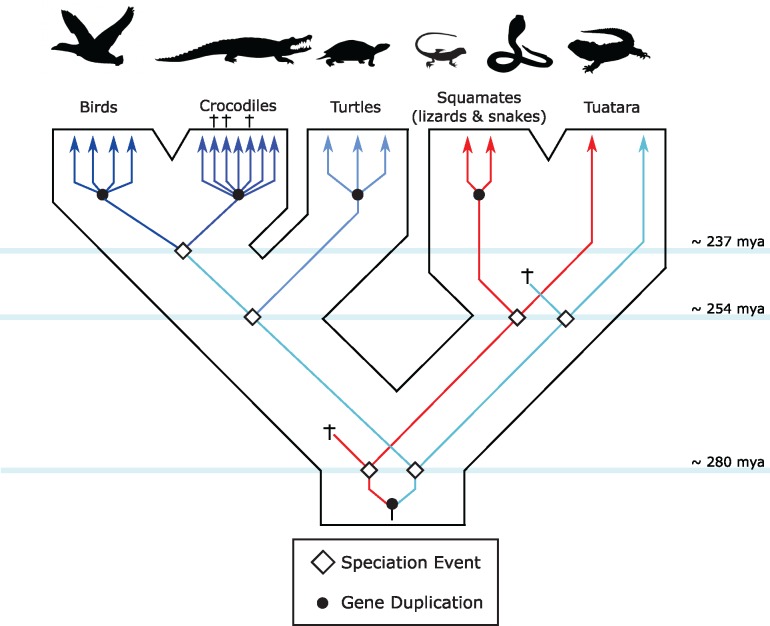
Reconciling individual gene trees with the organismal phylogeny is straightforward in the case of the α-globin gene family, as variation in gene content among groups is strictly attributable to lineage-specific losses of single members of the three-gene set that was present in the common ancestor of sauropsids. In the case of the β-type globins, we base our inferences on the tree based on nucleotide data, as it is better resolved and includes the crocodilian pseudogenes (fig. 3). The monophyly of amniote β-globins suggests that these genes can be traced back to a single gene in the last common ancestor of amniotes, and implies that a single β-globin was translocated to the novel chromosomal location. In turn, the affinity of the one tuatara β-globin transcript with the β-globins of birds, crocodilians, and turtles implies that the last common ancestor of sauropsids had at least two different β-globins: one retained by tuatara and squamates, and one retained by tuatara, archosaurs, and turtles. This inference is confirmed by the tree based on amino acid sequences, which includes two additional tuatara β-globin sequences, P10060 and P10061. The tuatara P10060 β-globin sequence groups with the β-globins of squamates, and the other two tuatara β-globins, P10061 and SPU_ENSACAG00000012173, group with the β-globins of birds, crocodiles, and turtles, indicating the presence of at least two β-globin genes in the last common ancestor of sauropsids (fig. 5).
Fig. 5.
—Reconstruction of the evolution of the sauropsid β-globin repertoires. Inferred duplication events are based on the phylogenies shown in figures 3 and 4. Crosses indicate gene losses due to either deletion or inactivations. Divergence times among the different lineages correspond to the estimated divergence times in the TimeTree server (Kumar, Stecher, et al. 2017).
Differences in the β-globin gene repertoires among sauropsid groups are the result of the differential retention of two ancestral genes, followed by lineage-specific duplications, which in the case of some squamates have been obscured by a history of interparalog gene conversion (Hoffmann, Storz, et al. 2010; Opazo, Hoffmann, et al. 2015). Among the β-type globin genes of squamates, the phylogenetic analyses indicate that the snake βII-globins represent the most early branching lineage (figs. 3 and 4), which suggests that the common ancestor of squamates had at least two β-globin genes, which have only been retained as a distinct pair in snakes. Most lizards analyzed have two β-globin genes that are most closely related to snake βI-globins, but robust orthology inferences are not possible due to an apparent history of interparalog gene conversion, gene turnover, or both.
The β-globin gene repertoires of birds, crocodilians, and testudines diversified independently via repeated rounds of lineage-specific duplication and descend from only one of the two ancestral sauropsid β-globin paralogs. Interestingly, even though bird, crocodilian, and testudine β-globins fall in reciprocally monophyletic groups, reflecting independent duplicative histories, in all cases the genes at the two ends of the cluster group together, and are sister to the clade of genes in the center of the cluster. Because there is evidence for gene conversion between the embryonically expressed T1 (ρ) and T4 (ɛ) globin genes of birds, we suspect a similar process is partly responsible for this arrangement in testudines and crocodilians. (Hoffmann, Storz, et al. 2010; Opazo, Hoffmann, et al. 2015). Interestingly, most gene turnover seems to have occurred during the early evolution of sauropsids, as the repertoires are different among the major lineages in this group, but highly conserved within them.
Evolution of Globin Expression
In all gnathostome vertebrates studied to date, the expression of the α- and β-type globin genes is ontogenetically regulated so that structurally and functionally distinct Hb isoforms are expressed during different stages of development (Storz 2016). Thus, changes in the size and membership composition of the α- and β-globin gene families may produce changes in the developmental regulation of Hb synthesis which can then constrain or potentiate the functional differentiation between Hb isoforms (Opazo et al. 2008a, 2013; Runck et al. 2009; Hoffmann, Storz, et al. 2010; Storz, Hoffmann, et al. 2011; Storz, Opazo, et al. 2011; Grispo et al. 2012; Damsgaard et al. 2013; Storz et al. 2013; Gaudry et al. 2014). In fact, the α- and β-globin gene clusters of teleost fish, lobe-finned fish, amphibians, squamates, birds, and mammals have all diversified in a lineage-specific manner such that each of those groups has distinct repertoires that are differentially expressed during embryonic development and postnatal life (Opazo et al. 2008a, 2013; Hoffmann, Storz, et al. 2010; Storz, Hoffmann, et al. 2011).
Adult birds typically coexpress two structurally and functionally distinct Hb isoforms in definitive red blood cells: the major HbA isoform, which incorporates products of the αA-globin gene, and the minor HbD isoform, which incorporates products of the αD-globin gene (Grispo et al. 2012; Opazo, Hoffmann, et al. 2015). Turtles also coexpress different adult Hb isoforms that incorporate the αA- and αD-globin genes. In both turtles and birds, the Hb isoform that incorporates the product of the αA-globin gene is the major isoform in adult red blood cells, and in both taxa the minor “D” type isoform exhibits an appreciably higher O2-binding affinity (Grispo et al. 2012; Damsgaard et al. 2013; Projecto-Garcia et al. 2013; Cheviron et al. 2014; Galen et al. 2015; Natarajan et al. 2015, 2016; Opazo, Hoffmann, et al. 2015; Kumar, Natarajan, et al. 2017). Squamates exhibit a different pattern of expression. In fact, in rattlesnakes and anole lizards, Hb isoforms that incorporate products of the αD-globin gene are actually expressed at higher levels than those that incorporate products of the αA-globin gene (Storz, Hoffmann, et al. 2011; Storz et al. 2015). Consistent with these measurements of protein abundance, the squamate transcriptome data revealed a higher abundance of αD transcripts relative to αA transcripts in adult red blood cells. An assessment of RNA libraries from adult liver provides further support for this observation, as the ratio of reads mapping to αD and αA is generally larger in squamates than in turtles (supplementary table S5, Supplementary Material online). In snakes, the major adult D-type Hb isoform actually exhibits a lower O2-affinity than the minor A-type isoform (Storz et al. 2015). Thus, in birds, turtles, and squamates, it appears that the major adult Hb isoform always has a lower O2-affinity than the minor isoform, but the subunit composition of the major and minor isoforms varies among lineages.
Analyses based on protein data suggest that Hb isoform composition is qualitatively different in tuatara and squamates relative to other amniotes (Abbasi et al. 1988; Gorr et al. 1998; Storz, Hoffmann, et al. 2011; Lu et al. 2015; Storz et al. 2015). Both squamates and tuatara express multiple, structurally distinct β-type globins during adulthood, whereas birds, crocodilians, turtles, and mammals generally possess a single adult β-type globin gene, or they possess two or more copies that are identical or nearly so (Abbasi et al. 1988; Gorr et al. 1998; Opazo et al. 2009; Runck et al. 2009, 2010; Storz, Hoffmann, et al. 2011; Storz et al. 2012, 2015; Weber et al. 2013; Gaudry et al. 2014; Lu et al. 2015; Schwarze et al. 2015). It is worth noting the long branch leading to the adult crocodilian β-type globin gene, Hbb-T4, reflects a high rate of amino acid substitution that may be related to the evolution of uniquely derived functional properties of crocodilian Hb (Bauer et al. 1981; Perutz et al. 1981; Komiyama et al. 1995; Weber et al. 2013). Interestingly, even though tuatara and squamates both possess two β-type globins, these genes clearly do not descend from the same ancestral gene pair (i.e., the tuatara and squamate β-globins are not 1:1 orthologs; figs. 3–5).
Incorporating an explicit evolutionary framework into the comparative genomic analysis of these differentially expressed globins can reveal important clues as to when and how the genetic control of their expression emerged. This is particularly interesting in the case of the amniote β-globin gene cluster, because it translocated to a novel position in the genome, altering its original genomic context (Hardison 2012). In mammals and birds, the switch from embryonic to fetal and then adult Hb during development is a complex process that involves interactions between distal cis regulatory sequences (located 40- to 60-kb upstream of the β-globin cluster) and more proximal elements (Alev et al. 2009; Wilber et al. 2011; Hardison 2012; Ulianov et al. 2012). Moreover, position effects appear to be important, since the 5′ to 3′ gene order generally matches the order of their expression during development. This also seems to be the case in crocodilians and turtles, where the genes in the 5′ end of the cluster apparently do not encode β-chain subunits of adult Hb.
Evolution of the α- and β-Globin Gene Clusters in Amniotes
Our evolutionary reconstructions show a clear contrast in the tempo and mode of evolution of the α- and β-globin gene clusters in amniotes. In the case of the α-globin gene cluster, which has remained in the ancestral genomic location, variation in gene content is largely limited to differential losses of α-globin paralogs that were present in the common ancestor of the group. However, expression changes have evolved: in squamates and tuarara, the major adult Hb isoform incorporates products of the αD-globin gene, whereas in archosaurs and turtles, the major isoform incorporates products of the αA-globin gene. In contrast to the high degree of conserved synteny in the α-globin gene cluster, the β-globin cluster, which translocated to a new chromosomal location in the common ancestor of amniotes, has experienced a much higher rate of gene gain and loss. Consequently, all major groups of amniotes (i.e., mammals, birds, crocodilians, turtles, rhynchocephalians, and squamates) possess unique repertoires of β-type globin genes that diversified independently in each lineage. These independently derived β-globin gene clusters trace back to two genes in the last common ancestor of sauropsids, which in turn trace back to a single copy gene in the last common ancestor of amniotes. Tuatara is the only extant taxon that retains descendant copies of both ancestral sauropsid β-globins. The two β-globin paralogs of squamates descend from one of these ancestral genes, whereas the β-globin paralogs of archosaurs and turtles descend from the other ancestral gene copy (fig. 5). In adult red blood cells, tuatara and squamates coexpress structurally distinct Hb isoforms that incorporate products of both β-type globin paralogs. By contrast, the adult Hbs of archosaurs and turtles incorporate the products of a single β-type globin gene. Interestingly, however, the adult β-type globin genes of the archosaurs and turtles are not 1:1 orthologs, and in each case the adult-expressed paralogs are located downstream from embryonically expressed paralogs. Thus, our results indicate that the stage-specific expression of early and late-expressed β-globins evolved at least twice in amniotes, once in mammals, and once in the lineage leading to birds, crocodilians and turtles.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Mary Clay Bailey and Amanda Black for editorial assitance. This work was supported by the National Institutes of Health/National Heart, Lung, and Blood Institute (HL087216 to J.F.S.), the National Science Foundation (MCB-1517636 and RII Track-2 FEC 1736249 to J.F.S; EPS-0903787, DBI-1262901, RII Track-2 FEC 1736026 and DEB-1354147 to F.G.H.), and the Fondo Nacional de Desarrollo Científico y Tecnológico from Chile (FONDECYT 1160627 to J.C.O.).
Literature Cited
- Abbasi A, Wells RM, Brittain T, Braunitzer G.. 1988. Primary structure of the hemoglobins from Sphenodon (Sphenodon punctatus, Tuatara, Rynchocephalia). Evidence for the expression of alpha D-gene. Biol Chem. 3698:755–764. [DOI] [PubMed] [Google Scholar]
- Aguileta G, Bielawski JP, Yang Z.. 2006. Proposed standard nomenclature for the alpha- and beta-globin gene families. Genes Genet Syst. 815:367–371.http://dx.doi.org/10.1266/ggs.81.367 [DOI] [PubMed] [Google Scholar]
- Alev C, et al. 2008. BetaA, the major beta globin in definitive red blood cells, is present from the onset of primitive erythropoiesis in chicken. Dev Dyn. 2374:1193–1197. [DOI] [PubMed] [Google Scholar]
- Alev C, Shinmyozu K, McIntyre BAS, Sheng G.. 2009. Genomic organization of zebra finch alpha and beta globin genes and their expression in primitive and definitive blood in comparison with globins in chicken. Dev Genes Evol. 2197:353–360. [DOI] [PubMed] [Google Scholar]
- Alföldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 4777366:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Bauer C, et al. 1981. Analysis of bicarbonate binding to crocodilian hemoglobin. J Biol Chem. 256:8429–8435. [PubMed] [Google Scholar]
- Blank M, et al. 2011. A membrane-bound vertebrate globin. PLoS One 69:e25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester T, Hankeln T.. 2014. Function and evolution of vertebrate globins. Acta Physiol. 2113:501–514. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 11051:20645–20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, et al. 2014. Integrating evolutionary and functional tests of adaptive hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis. Mol Biol Evol. 3111:2948–2962.http://dx.doi.org/10.1093/molbev/msu234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirotto C, Panara F, Arangi I.. 1987. The minor haemoglobins of primitive and definitive erythrocytes of the chicken embryo. Evidence for haemoglobin L. Development 1014:805–813. [DOI] [PubMed] [Google Scholar]
- Crawford NG, et al. 2012. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 85:783–786.http://dx.doi.org/10.1098/rsbl.2012.0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsgaard C, Storz JF, Hoffmann FG, Fago A.. 2013. Hemoglobin isoform differentiation and allosteric regulation of oxygen binding in the turtle, Trachemys scripta. AJP Regul Integr Comp Physiol. 3058:R961–R967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C, Burmester T, Hankeln T.. 2006. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 112(3–4):296–306. [DOI] [PubMed] [Google Scholar]
- Galen SC, et al. 2015. Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc Natl Acad Sci U S A. 11245:13958–13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudry MJ, Storz JF, Butts GT, Campbell KL, Hoffmann FG.. 2014. Repeated evolution of chimeric fusion genes in the β-globin gene family of laurasiatherian mammals. Genome Biol Evol. 65:1219–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillemans N, et al. 2003. Functional and comparative analysis of globin loci in pufferfish and humans. Blood 1017:2842–2849.http://dx.doi.org/10.1182/blood-2002-09-2850 [DOI] [PubMed] [Google Scholar]
- Gorr TA, Mable BK, Kleinschmidt T.. 1998. Phylogenetic analysis of reptilian hemoglobins: trees, rates, and divergences. J Mol Evol. 474:471–485.http://dx.doi.org/10.1007/PL00006404 [DOI] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 3466215:1254449.http://dx.doi.org/10.1126/science.1254449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grispo MT, et al. 2012. Gene duplication and the evolution of hemoglobin isoform differentiation in birds. J Biol Chem. 28745:37647–37658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. 2008. Globin genes on the move. J Biol. 79:35..http://dx.doi.org/10.1186/jbiol92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. 2012. Evolution of hemoglobin and its genes. Cold Spring Harb Perspect Med. 212:a011627.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Hoogewijs D. et al. 2012. Evolution of the globin gene family in deuterostomes: lineage-specific patterns of diversification and attrition. Mol Biol Evol. 297:1735–1745.http://dx.doi.org/10.1093/molbev/mss018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF.. 2008a. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 25:591–602. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF.. 2008b. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 25:2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF.. 2010. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc Natl Acad Sci U S A. 10732:14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF.. 2012. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Mol Biol Evol. 291:303–312.http://dx.doi.org/10.1093/molbev/msr207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Storz JF.. 2007. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol Biol Evol. 249:1982–1990. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Storz JF, Gorr TA, Opazo JC.. 2010. Lineage-specific patterns of functional diversification in the α- and β-globin gene families of tetrapod vertebrates. Mol Biol Evol. 275:1126–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, et al. 2012. Androglobin: a chimeric globin in metazoans that is preferentially expressed in mammalian testes. Mol Biol Evol. 294:1105–1114.http://dx.doi.org/10.1093/molbev/msr246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosbach HA, Wyler T, Weber R.. 1983. The Xenopus laevis globin gene family: chromosomal arrangement and gene structure. Cell 321:45–53.http://dx.doi.org/10.1016/0092-8674(83)90495-6 [DOI] [PubMed] [Google Scholar]
- Ikehara T, Eguchi Y, Kayo S, Takei H.. 1997. Isolation and sequencing of two α-globin genes α(A) and α(D) in pigeon and evidence for embryo-specific expression of the α(D)-globin gene. Biochem Biophys Res Commun. 2342:450–453. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ. 1982. Evolution of globin genes In: Dover GA, Flavell RB, editors. Genome evolution. New York: Academic Press; p. 157–176. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama NH, Miyazaki G, Tame J, Nagai K.. 1995. Transplanting a unique allosteric effect from crocodile into human haemoglobin. Nature 3736511:244–246.http://dx.doi.org/10.1038/373244a0 [DOI] [PubMed] [Google Scholar]
- Kumar A, Natarajan C. et al. 2017. Stability-mediated epistasis restricts accessible mutational pathways in the functional evolution of avian hemoglobin. Mol Biol Evol. 345:1240–1251.http://dx.doi.org/10.1093/molbev/msx085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 347:1812–1819.http://dx.doi.org/10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2015. Gekko japonicus genome reveals evolution of adhesive toe pads and tail regeneration. Nat Commun. 6:1–11.http://dx.doi.org/10.1038/ncomms10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, et al. 2015. Differences in hematological traits between high- and low-altitude lizards (Genus Phrynocephalus). PLoS One 105:e0125751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, et al. 2015. Rapid molecular evolution across amniotes of the IIS/TOR network. Proc Natl Acad Sci U S A. 11222:7055–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, Von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 305:1188–1195.http://dx.doi.org/10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, et al. 2015. Convergent evolution of hemoglobin function in high-altitude andean waterfowl involves limited parallelism at the molecular sequence level. PLOS Genet. 1112:e1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, et al. 2016. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 3546310:336–339.http://dx.doi.org/10.1126/science.aaf9070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 321:268–274.http://dx.doi.org/10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Butts GT, Nery MF, Storz JF, Hoffmann FG.. 2013. Whole-genome duplication and the functional diversification of teleost fish hemoglobins. Mol Biol Evol. 301:140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, et al. 2015. Gene turnover in the avian globin gene families and evolutionary changes in hemoglobin isoform expression. Mol Biol Evol. 324:871–887.http://dx.doi.org/10.1093/molbev/msu341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF.. 2008a. Genomic evidence for independent origins of beta-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci U S A. 1055:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF.. 2008b. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci U S A. 10535:12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Lee AP, et al. 2015. Ancient duplications and expression divergence in the globin gene superfamily of vertebrates: insights from the elephant shark genome and transcriptome. Mol Biol Evol. 327:1684–1694.http://dx.doi.org/10.1093/molbev/msv054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF.. 2009. Origin and ascendancy of a chimeric fusion gene: the β/δ-globin gene of paenungulate mammals. Mol Biol Evol. 267:1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VS, et al. 2008. Platypus globin genes and flanking loci suggest a new insertional model for beta-globin evolution in birds and mammals. BMC Biol. 6:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, et al. 1981. Allosteric regulation of crocodilian haemoglobin. Nature 2915817:682–684.http://dx.doi.org/10.1038/291682a0 [DOI] [PubMed] [Google Scholar]
- Projecto-Garcia J, et al. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 11051:20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ.. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K.. 2012. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 13:312.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 613:539–542.http://dx.doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck AM, Moriyama H, Storz JF.. 2009. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol Biol Evol. 2611:2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck AM, Weber RE, Fago A, Storz JF.. 2010. Evolutionary and functional properties of a two-locus β-globin polymorphism in Indian house mice. Genetics 1844:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze K, Burmester T.. 2013. Conservation of globin genes in the “living fossil” Latimeria chalumnae and reconstruction of the evolution of the vertebrate globin family. Biochim Biophys Acta Proteins Proteomics 18349:1801–1812. [DOI] [PubMed] [Google Scholar]
- Schwarze K, et al. 2014. The globin gene repertoire of lampreys: convergent evolution of hemoglobin and myoglobin in jawed and jawless vertebrates. Mol Biol Evol. 3110:2708–2721.http://dx.doi.org/10.1093/molbev/msu216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze K, Singh A, Burmester T.. 2015. The full globin repertoire of turtles provides insights into vertebrate globin evolution and functions. Genome Biol Evol. 77:1896–1913.http://dx.doi.org/10.1093/gbe/evv114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Goldman N.. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 513:492–508.http://dx.doi.org/10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- Storz JF. 2016. Gene duplication and evolutionary innovations in hemoglobin-oxygen transport. Physiology 313:223–232.http://dx.doi.org/10.1152/physiol.00060.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, et al. 2015. Oxygenation properties and isoform diversity of snake hemoglobins. Am J Physiol Regul Integr Comp Physiol. 3099:R1178–R1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Hoffmann FG, Opazo JC, Moriyama H.. 2008. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics 1783:1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Hoffmann FG, Opazo JC, Sanger TJ, Moriyama H.. 2011. Developmental regulation of hemoglobin synthesis in the green anole lizard Anolis carolinensis. J Exp Biol. 214(Pt 4):575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Opazo JC, Hoffmann FG.. 2011. Phylogenetic diversification of the globin gene superfamily in chordates. IUBMB Life 635:313–322.http://dx.doi.org/10.1002/iub.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Opazo JC, Hoffmann FG.. 2013. Gene duplication, genome duplication, and the functional diversification of vertebrate globins. Mol Phylogenet Evol. 662:469–478.http://dx.doi.org/10.1016/j.ympev.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Weber RE, Fago A.. 2012. Oxygenation properties and oxidation rates of mouse hemoglobins that differ in reactive cysteine content. Comp Biochem Physiol A Mol Integr Physiol. 1612:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL.. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1742:247–250. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ.. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzika AC, Ullate-Agote A, Grbic D, Milinkovitch MC.. 2015. Reptilian transcriptomes v2.0: an extensive resource for sauropsida genomics and transcriptomics. Genome Biol Evol. 76:1827–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulianov SV, Gavrilov AA, Razin SV.. 2012. Spatial organization of the chicken beta-globin gene domain in erythroid cells of embryonic and adult lineages. Epigenet Chromatin 51:16..http://dx.doi.org/10.1186/1756-8935-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewege MW, et al. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol Evol. 83:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q-H, et al. 2013. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 239:1091–1105.http://dx.doi.org/10.1038/cr.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 456:701–706.http://dx.doi.org/10.1038/ng.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RE, Fago A, Malte H, Storz JF, Gorr TA.. 2013. Lack of conventional oxygen-linked proton and anion binding sites does not impair allosteric regulation of oxygen binding in dwarf caiman hemoglobin. Am J Physiol Regul Integr Comp Physiol. 3053:R300–R312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber A, Nienhuis AW, Persons DA.. 2011. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood 11715:3945–3953.http://dx.doi.org/10.1182/blood-2010-11-316893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 3466215:1311–1320.http://dx.doi.org/10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.