Abstract
A fraction of genetic variants segregating in any population are deleterious, which negatively impacts individual fitness. The domestication of animals and plants is associated with population bottlenecks and artificial selection, which are predicted to increase the proportion of deleterious variants. However, the extent to which this is a general feature of domestic species is unclear. Here, we examine the effects of domestication on the prevalence of deleterious variation using pooled whole-genome resequencing data from five domestic animal species (dog, pig, rabbit, chicken, and silkworm) and two domestic plant species (rice and soybean) compared with their wild ancestors. We find significantly reduced genetic variation and increased proportion of nonsynonymous amino acid changes in all but one of the domestic species. These differences are observable across a range of allele frequencies, both common and rare. We find proportionally more single nucleotide polymorphisms in highly conserved elements in domestic species and a tendency for domestic species to harbor a higher proportion of changes classified as damaging. Our findings most likely reflect an increased incidence of deleterious variants in domestic species, which is most likely attributable to population bottlenecks that lead to a reduction in the efficacy of selection. An exception to this pattern is displayed by European domestic pigs, which do not show traces of a strong population bottleneck and probably continued to exchange genes with wild boar populations after domestication. The results presented here indicate that an elevated proportion of deleterious variants is a common, but not ubiquitous, feature of domestic species.
Keywords: domestication, mutational load, artificial selection, effective population size, population bottleneck, natural selection
Introduction
Natural selection promotes the spread of beneficial mutations and removes harmful ones. However, it is not completely effective and all populations harbor genetic variants with deleterious effects (Charlesworth 2009). The number of deleterious variants in a population depends on the rate at which they enter by mutation and the efficacy of selection at removing them (the mutation–selection balance [Ohta 1992; Crow and Kimura 2009; Lynch 2010]). Most mutations in functional sites are expected to be deleterious, with a distribution of fitness effects ranging in severity from lethal to weakly deleterious (Eyre-Walker and Keightley 2007; Keightley and Eyre-Walker 2007; Boyko et al. 2008). The efficacy of selection at removing deleterious variants depends on their fitness effects, in addition to many other intrinsic factors such as dominance, epistatic and environmental interactions, and genetic linkage (Hill and Robertson 1966; Charlesworth B and Charlesworth D 1999; Henn et al. 2015). It also depends on demographic history, which can be summarized by the effective population size (Ne) (Charlesworth 2009). This parameter is reduced by population size fluctuations, such as bottlenecks and inbreeding. Despite being selected against, weakly deleterious mutations can persist in populations, resulting in a reduction of the overall fitness of the population, referred to as mutation load (Kimura et al. 1963; Ohta 1973).
Humans have experienced a bottleneck and expansion out of Africa resulting in reduction of Ne. Several studies have inferred that this has resulted in an increased proportion of deleterious mutations in non-African populations (Lohmueller et al. 2008; Fu et al. 2014; Henn et al. 2015). The deleterious mutation load has been reported to increase with distance from epicenter of human migrations (Henn et al. 2016). These observations could be explained by reduced efficacy of purifying selection caused by a bottleneck and population growth, or could reflect an effect of demography on alleles of different frequencies without differences in the efficacy of selection (Simons et al. 2014; Do et al. 2015). Deleterious mutations may also accumulate at the front wave of expansions, referred to as the expansion load (Peischl et al. 2013). There are therefore multiple ways in which demographic processes can alter the mutation load in a population. It has been suggested that an increased mutation load in human populations likely derives from rare deleterious variants that emerged recently, which can increase incidence of severe early-onset disease with simple genetic basis and cause elevated risk of common, complex diseases (Henn et al. 2015).
The process of domestication entails drastic changes in the nature and strength of selective forces acting on a population coupled with changes in its size and structure (Meyer and Purugganan 2013; Larson and Fuller 2014). During domestication, a number of individuals are isolated—either partially or completely—from a wild progenitor population and they are subjected to artificial selection for phenotypes desirable for humans. In many cases, these traits may be deleterious in the wild. For example, domesticated plants have often reduced or lost their ability for seed dispersal, have larger fruits and seeds, and altered breeding and seasonality—traits that increase yield but reduce survival in wild (Zohary et al. 2012). Domestic animals often exhibit increased tameness or toleration of human presence, shorter generation times, and extreme differences in size, morphology and behavior compared with their wild ancestors (Darwin 1876; Zeder 2012). There are also a number of ways in which the process of domestication, which entails population bottlenecks and expansions coupled to changes in the nature of selection, could result in an average increase in the prevalence of deleterious mutations.
First, factors such as inbreeding and population bottlenecks are predicted to reduce Ne, leading to increased genetic drift and a reduction in the efficacy of selection of removing deleterious variants (Charlesworth 2009). Second, selection can cause linked deleterious variants to rise in frequency by genetic hitchhiking, where positively selected variants are genetically linked to deleterious ones (Maynard Smith and Haigh 1974; Chun and Fay 2011; Hartfield and Otto 2011). Increased levels of artificial selection could therefore generate a general increase in the prevalence of weakly deleterious alleles in the genome if positive selection and genetic hitchhiking are common. Finally, domestication could also entail changes in the nature of selection. For instance, selection on some traits that are needed to survive and reproduce in the wild (e.g., speed, agility, sensory perception) could be relaxed due to management by humans.
Population genomic approaches have now been used in many domestic species to determine the effects of domestication on levels of genetic variation and the proportion of deleterious variants (Larson and Fuller 2014). Decreased levels of genetic variation, indicative of lower Ne, are commonly observed in domestic species (e.g., Lu et al. 2006; Xia et al. 2009; Lam et al. 2010; Vaysse et al. 2011; Groenen et al. 2012; Xu et al. 2011; Larson and Burger 2013). In addition, several studies have identified an elevated proportion of nonsynonymous compared with synonymous variants (pN/pS) in populations of domestic species compared with their wild relatives, which is interpreted as evidence of increased prevalence of deleterious mutations. It has been estimated that higher proportion of genetic variants in the mitochondrial (Björnerfeldt et al. 2006) and nuclear (Cruz et al. 2008; Marsden et al. 2016) genomes of dogs are nonsynonymous compared with wolves. It has also been reported that domestic horse genomes have an excess of mutations in highly conserved sites compared with ancient horse genomes (Schubert et al. 2014). An increased prevalence of deleterious mutations due to domestication could potentially explain why domestic dogs suffer from a high prevalence of heritable diseases (Ostrander and Kruglyak 2000; Karlsson and Lindblad-Toh 2008), which is also observed in other domestic animals (Mulvihill 1972).
A similar trend has been observed in the genomes of domestic plants. Coding alignments of single reference sequences from two subspecies of Asian rice compared with their wild ancestor revealed an elevated proportion of nonsynonymous changes in domestic lineages, indicating accumulation of deleterious mutations and a “cost” of domestication (Lu et al. 2006). This effect was larger for radical amino acid changes. An excess of nonsynonymous variants has also been reported in single samples of cultivated compared with wild tomato using transcriptome sequencing (Koenig et al. 2013). RNAseq of multiple accessions of domesticated sunflowers and related crops and their wild relatives also indicated increased proportions of nonsynonymous variants, observed in both rare and common variants (Renaut and Rieseberg 2015). This has also been reported in multiple accessions of barley (Kono et al. 2016), soybean (Lam et al. 2010; Kono et al. 2016), Asian rice (Liu et al. 2017), and African rice (Nabholz et al. 2014). An increased prevalence of deleterious alleles has also been inferred in populations of wild poplars with smaller Ne (Zhang et al. 2016).
There is therefore growing evidence of an increased proportion of deleterious variants in a number of domestic species, which is typically inferred from an observation of an excess proportion of variants with a predicted functional effect (e.g., nonsynonymous variants) compared with neutral variants. However, so far this has been examined in relatively few species, and particularly few animals. Furthermore, relatively few have surveyed variation within populations using multiple samples, which can give insight into the proportion of deleterious changes in rare and common genetic variants. Here, we investigate these questions using population-scale whole-genome resequencing data from pooled samples of domestic species compared with their wild ancestors from several animals and plants. The animal species comprise four vertebrates (dog, pig, rabbit, and chicken) and one insect (silkworm) and we also include two important crop plants (rice and soybean). We investigate evidence for reduced Ne and an elevated proportion of deleterious changes as a result of domestication of these species. In addition to examining the amino acid-changing variants, we also investigate whether the proportion of variants in functional noncoding elements increases as a result of domestication. Finally, we use a variety of bioinformatic predictors of the effect of amino acid changes to estimate how the proportion of genetic variants with small and large effects is affected by domestication.
Materials and Methods
Population Genomic Data Sets
We obtained data sets consisting of single nucleotide polymorphisms (SNPs) called from short-read next-generation sequencing data mapped to appropriate reference genomes. These data were derived from the following sources (see also supplementary table S4, Supplementary Material online):
Domestic rabbit (Oryctolagus cuniculus) compared with wild rabbit (Oryctolagus cuniculus) (Carneiro et al. 2014). These data consist of 6 pools, each consisting of multiple individuals from single domestic breeds, and 14 pools of wild rabbits, where each pool consisted of multiple individuals sampled from the same locality. These localities were spread across Spain and southern France. The pools contained a mean of 14.6 samples and were sequenced to a mean depth of 10.9× (Carneiro et al. 2014). A single sample of snowshoe hare (Lepus americanus) was sequenced to 10.2× coverage and used as an outgroup.
Dog (Canis familiaris) compared with gray wolf (Lupus lupus) (Axelsson et al. 2013). These data consisted of five pools, each consisting of domestic dog samples from one or more breeds, in addition to one pool of wolf samples. Each of the 6 pools contained 12 samples and were sequenced to a mean depth of 6.3×. As an outgroup, we utilized sequence from a single Andean fox (Lycalopex culpaeus) sequenced to 11× (Auton et al. 2013).
Pig (Sus scrofa domesticus) compared with wild boar (Sus scrofa) (Groenen et al. 2012). These data consisted of a total of 30 samples from 5 European pig breeds and 7 samples from 3 Asian domestic pig breeds, compared with samples of 6 wild boars from 4 localities in Europe and 5 wild boars from 3 localities in Asia. These samples were individually barcoded and sequenced to an average depth of 6.8×. Four Asian species of wild pig (Sus barbatus, Sus cebifrons, Sus celebensis, and Sus verrucosus) were included as outgroups (a single sample of each species, sequenced to an average coverage of 10.7×).
Chicken (Gallus gallus domesticus) compared with red jungle fowl (Gallus gallus) (Rubin et al. 2010). These data consisted of pools of eight domestic chicken strains or selection lines and one pool of red jungle fowl. Each pool contained an average of 9.7 samples from the same strain, sequenced to an average depth of 4.6×.
Domestic silkworm (Bombyx mori) compared with wild silkworm (Bombyx mandarina) (Xia et al. 2009). These data consisted of 29 individually barcoded samples of the domestic silkworm (average depth 2.9×) classified into 3 subgroups (supplementary table S1, Supplementary Material online) and 11 of its wild ancestor (average depth 3.1×). We classified 29 individually barcoded domestic silkworms into 3 subgroups based on phylogenetic relationship reported in Xia et al. 2009 (supplementary table S1, Supplementary Material online; subgroup 1: D1, D2, D3, D4, D6, D8, D9, D10, D11, D12, D13, D14, D25, D26, and D29; subgroup 2: D5, D7, D15, D16, and D24; subgroup 3: D30, D31, D32, D33, D34, D35, D36, D37, D38, D39, and D40).
Domestic soybean (Glycine max) compared with wild soybean (Glycine soja) (Lam et al. 2010). These data consisted of 10 individually barcoded samples of domestic soybean (average depth 5.4×) compared with 13 of its wild ancestor (average depth 5.7×).
Two subspecies of Asian domestic rice (Oryza sativa japonica and Oryza sativa indica) compared with two wild ancestors (Oryza rufipogon and Oryza nivara) (Xu et al. 2011). These data consisted of 24 samples of japonica domestic rice from 3 different varieties (sequenced to an average depth of 19.9×) and 12 samples of indica domestic rice (sequenced to an average depth of 18.3×). Five samples each of the two wild species were sequenced to an average depth of 18.7×.
Variant Calling
The raw sequence data used to call SNPs consisted of short reads produced on a number of different Illumina and SOLiD platforms specific to each study (supplementary table S4, Supplementary Material online). Sequencing library preparation methods and read lengths also varied between the original studies. The SNP data for rice, soybean, dog, pig, chicken, silkworm, and rabbit were defined relative to the genome assemblies IRGSP v4, Morex Assembly v3, Glycine max release v1.01, CanFam2, SusScr3, GalGal3, OryCun2, MelGal1, and Silkworm assembly v2, respectively. The methods used to map reads and call SNPs varied across original studies (see supplementary table S4, Supplementary Material online, for details). The alleles present in the reference genomes at each SNP were not included in the analysis. Genomic locations for SNPs on dog genome assembly CanFam2 and chicken genome assembly GalGal3 were converted to those based on CanFam3 (dog) and GalGal4 (chicken) by liftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver), respectively. We did not use admixed populations (F2 intercross pig; 12883_AUS, 8555_AUS, 43397_IND, and 60542_IV rice strains; C01, C12, C17, and C19 soybean strains) or those with low average depth (< 2.5×; W06, W09, W14, and W17 wild soybean strains) for our study.
High-quality SNPs for dogs and chickens were originally reported under conservative thresholds (Rubin et al. 2010; Axelsson et al. 2013), which decreases number of false positive SNPs while increasing the rate of false negatives. In particular, this leads to detection of a smaller number of rare SNPs, as power to detect them is lower than for common SNPs. In order to increase the sensitivity for detecting rare SNPs for the dog and chicken data sets, we repeated SNP calling on the original read mapping using less stringent thresholds. This was done by generating pileup files from pooled BAM files using SAMtools v0.1.18 (Li et al. 2009) and filtering away all bases with quality scores <30. No further filters were used to call SNPs from these data sets.
Inferring Ancestral State
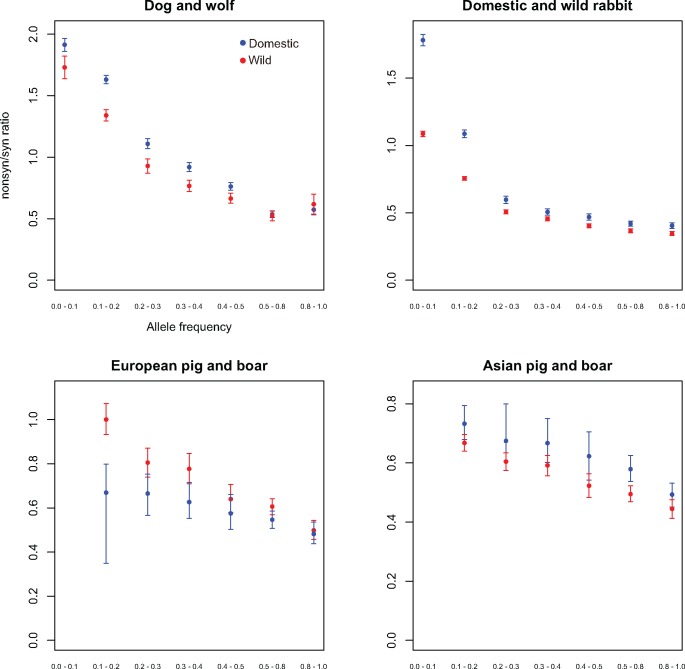
For inferring ancestral state of polymorphic sites, we used Snowshoe hare (L. americanus), Andean fox (Ly. culpaeus), and Asian pigs (S. barbatus, S. cebifrons, S. celebensis, S. verrucosus) as outgroups for rabbit, dog, and pig, respectively. We downloaded Illumina paired-end reads (SRX403460) for Andean fox from SRA (https://www.ncbi.nlm.nih.gov/sra). The short reads were mapped to the reference dog genome (build CanFam3) by Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009). SNPs for Andean fox were called by SAMtools v0.1.18 (Li et al. 2009). We obtained SNPs for pig (S. barbatus, S. cebifrons, S. celebensis, S. verrucosus) and rabbit (L. americanus) ancestors and from the studies mentioned earlier (Groenen et al. 2012; Carneiro et al. 2014). We estimated allele frequency for each polymorphic loci with fixed alleles in an ancestor where ancestral state was identifiable. The number of individually barcoded samples in pig was not the same as that in boar. Therefore, we resampled to use the same number of samples in pig and boar for the estimation of allele frequency. We compared those with randomly selected samples from Asian (or European) pigs of the same sample size (i.e., five Asian wild boars vs. randomly sampled five Asian pigs; six European wild boars vs. randomly sampled six European pigs). Note that we could not estimate low allele frequency (0.0–0.1) for pig and boar because of the small number of samples in wild boar pools (fig. 3).
Fig. 3.
—Relationship between the nonsyn/syn ratio and allele frequency classes. Relationship between derived allele frequency classes and nonsyn/syn ratio of SNPs in the dog/wolf, rabbit, and Asian and European pig/boar data. Ancestral alleles are determined by comparison with an outgroup. Blue and red circles indicate domestic and wild species, respectively. Error bars represent 95% confidence intervals.
Estimation of Watterson’s θ
We estimated Watterson's θ (Watterson 1975) for individually barcoded diploid samples (pig, rice, soybean, and silkworm) as an index of genetic diversity. We classified the samples into each pool (supplementary table S2, Supplementary Material online), and estimated θ for each pool based on the number of segregating sites (SNPs), genome size covered by reads for detecting SNPs (Xia et al. 2009; Lam et al. 2010; Xu et al. 2011), and sample size in a pool. Thus, we used whole-genome size as covered genome size to detect SNPs, resulting in slightly underestimation of θ for pigs and boars. Watterson's θ for rabbit, dog, and chicken, a pooled sequencing strategy was used, was estimated by popoolation (Kofler et al. 2011). We obtained bam files from previous studies (Rubin et al. 2010; Axelsson et al. 2013; Carneiro et al. 2014), and made pileup files from the bam files by SAMtools v0.1.18 (Li et al. 2009). We estimated θ for overall genome by sliding window analysis (window size: 40 kb, step size: 40 kb, minimum count: 2, minimum coverage 4, maximum coverage: 100, minimum quality: 20, minimum covered fraction: 0.3) implemented in popoolation. We estimated mean θ for all windows for each pool (supplementary table S2, Supplementary Material online).
Prediction of Deleterious Nonsynonymous Mutations
We obtained protein-coding sequences and their genomic locations (GTF files) from SilkDB (http://silkworm.swu.edu.cn/silkdb/) for silkworm, RIS (http://rice.genomics.org.cn/rice/index2.jsp) for rice, JGI (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Gmax/annotation/) for soybean and Ensembl release 73 for dog, chicken, and pig. We extracted only SNPs in protein-coding regions from SNP data sets and classified them into synonymous, nonsynonymous, missense (nonsense and read-through) changes.
We predicted deleterious effect of nonsynonymous SNPs by PROVEAN (threshold: <−2.5) in rice, soybean, silkworm, rabbit, dog, chicken, and pig. If SNPs were located in more than one alternative splicing forms, we estimated deleterious effect of the SNP for all alternative splicing forms and chose the most deleterious value as a representative one in the SNP locus. We evaluated an accumulation of deleterious mutations by estimating the proportion of PROVEAN damaging SNPs (number of damaging nonsynonymous SNPs/number of nonsynonymous SNPs).
Extraction of SNPs within UCNEs
We obtained 4,351 UCNEs having >200 bp length and 95% identity between human and chicken from UCNE database (Dimitrieva and Bucher 2013). Genomic locations for UCNEs on human genome assembly hg19 were converted to those based on CanFam3 (dog), OoryCun2 (rabbit), and SusScr3 (pig) by liftOver, resulting in 4,323, 4,278 and 3,647 UCNEs being mapped to the dog, rabbit, and pig genomes, respectively. Genomic locations for UCNEs on chicken genome assembly GalGal3 were converted to those based on GalGal4 (chicken) by liftOver, resulting in 4,338 UCNEs being mapped to the chicken genome. We counted the total number of SNPs within UCNEs for each domestic and wild population.
Statistical Analysis
We used a bootstrap approach to examine the statistical significance of differences between the number of total SNPs, the nonsyn/syn ratio, and the proportion of PROVEAN damaging SNPs between wild and domestic populations. We conducted a similar bootstrap procedure for the analysis of pooled samples and for individually barcoded samples. Individually barcoded samples were combined into pools for the pooled analysis and also analyzed individually. Most of the data sets consisted of multiple domestic and wild pools.
For analysis of pooled data, we estimated each of the measures above for each pool and calculated mean values for domestic and wild species by averaging across pools. The number of individually barcoded samples in pig was not the same as that in boar. Therefore, we resampled to use the same number of samples in pig and boar. We compared the measures for randomly selected samples from Asian (or European) pigs of the same sample size with those for pooled boars (i.e., five Asian wild boars vs. randomly sampled five Asian pigs; six European wild boars vs. randomly sampled six European pigs). We used a bootstrap procedure to estimate confidence intervals and determine significant differences in the measures above between domestic and wild species. We randomly sampled genes (or UCNEs) with replacement to produce an artificial data set with the same number of genes as the original one and repeated the above steps to calculate mean values. We repeated this procedure to produce 1,000 bootstrap replicates with which to compare the variables in domestic and wild populations. The sample sizes of individually sequenced data were resampled with each replicate so that domestic and wild pools of the same size were compared with each replicate.
The analysis of individual sequences proceeded in a similar way except we estimated the total number of heterozygous sites and used these sites to calculate the measures above. We then averaged the measures across all wild and all domestic sequences. As for pooled samples, confidence intervals and significance were estimated by bootstrapping by gene with 1,000 replicates.
In order to determine the significance of differences in the proportion of SNPs observed in UCNEs, we used a randomization approach to estimate the number of expected SNPs in UCNEs under neutrality. For each replicate, we randomly placed UCNEs of the same number and lengths as observed in the genome and counted the number of SNPs in these regions, which we refer to as the number of expected SNPs. We then calculated the observed/expected number of SNPs in UCNEs for each population and replicate. Significance was estimated by comparing this value in 1,000 replicates.
Results
SNP Data Sets from Domestic Species and Their Wild Ancestors
We constructed whole-genome SNP data sets of five domestic animal species (rabbit, dog, pig, chicken, and silkworm) and two domestic plant species (rice and soybean) compared with the present-day populations of the wild ancestors from which they are derived. These were produced by analyzing the next-generation data mapped to the appropriate reference genomes. For six of the species, the reference genome corresponded to the domestic species, whereas for chicken, the reference genome corresponded to red jungle fowl, the wild ancestor of chicken. Due to the low divergence and small proportion of fixed differences observed between domestic and wild populations studied here, the identity of the reference sequence is unlikely to lead to biases in SNP calling between domestic and wild samples. The allele at each SNP observed in the reference sequence was not included in our analyses. A previous evaluation of potential bias toward the reference sequence in a study of dogs and wolves has found it to be extremely low (Freedman et al. 2014).
For the three mammals: rabbit, dog, and pig, we also obtained sequences of species that are outgroups to both wild and domestic populations. For each species, multiple breeds and populations of wild and domestics were sampled. For rabbit, dog, and chicken, a pooled sequencing strategy was used, whereby a number of individuals from one or more breeds were sequenced in the same pool without individually barcoding samples, whereas for the other species, individually barcoded samples were available. A summary of all the populations sampled is presented in the supplementary table S1, Supplementary Material online, and in Materials and Methods.
Difference in Genetic Diversity between Domestic Species and Their Wild Relatives
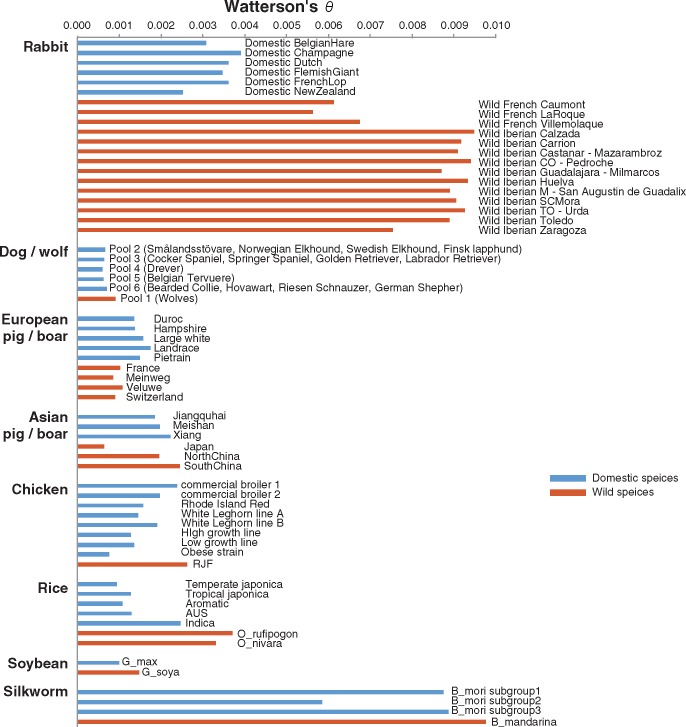
We first estimated the levels of genetic diversity in each of the wild and domestic populations sampled using Watterson’s θ (Watterson 1975). For the pooled samples, this was done using the popoolation software (Kofler et al. 2011), which specifically accounts for sampling of individuals among reads in pooled sequences (see Materials and Methods). There was a general trend for domestic species to have lower levels of genetic variation than extant populations of their wild ancestors. This observation was generally concordant across the wild and domestic populations we sampled (fig. 1; supplementary table S2, Supplementary Material online).
Fig. 1.
—Estimates of Watterson's θ from domestic and wild species. Watterson's θ values for rabbits, dog/wolf, pig/boar chicken, rice, soybean, and silkworm. Blue and red bars indicate Watterson's θ in pooled samples from domestic and wild species, respectively. Samples are grouped into pools based on the format of the original data sets.
The rabbit data included samples from 6 domestic breeds, 3 population samples from wild populations in France, and 11 from wild populations from the Iberian Peninsula. Within these three groups, levels of genetic variation were consistent. The mean values of θ for domestic, wild French, and wild Iberian rabbits were 0.0034, 0.0062, and 0.0090, respectively (fig. 1; supplementary table S2, Supplementary Material online). The dog data consisted of 5 pools of 12 dogs from one or more breeds and 1 pool of 12 wolves. Dog pools (mean θ = 0.00065) were all less diverse than wolf pool (mean θ = 0.00091). Pools with mixed breeds (pools 2, 3, and 6) tended to have higher variation than pools with a single breed, as expected due to their mixed composition. Pigs were believed to have been domestic independently in Asia and Europe, from two different subspecies of wild boars (Giuffra et al. 2000). On average, both wild and domestic Asian pig/boar samples showed higher levels of variation than European ones. Surprisingly, however, we found that domestic pigs from Europe (mean θ = 0.00151) and Asia (mean θ = 0.00202) were more genetically variable than corresponding wild populations (European wild boars, mean θ = 0.00096; Asian wild boars, mean θ = 0.00168), although wild boar samples from Japan had much lower levels of genetic variation (mean θ = 0.00064) than other Asian wild boars. For chicken, five pools from standard breeds (two broiler, two White Leghorn, and one Rhode Island Red) and three from lines maintained under strong selection (high growth line, low growth line, and obese strain) were investigated. The selection lines had consistently lower genetic variation (mean θ = 0.00113) compared with breed pools (mean θ = 0.00186) and both were consistently lower than the wild red jungle fowl. We observed that genetic variation of domestic silkworm pools (mean θ = 0.0078) was lower than wild silkworm (mean θ = 0.0098). For rice, θ decreased in the following order: Wild, indica, and japonica, as reported before (Xu et al. 2011). θ for wild soybeans (mean θ = 0.00164) which were collected from different geographical regions in China was higher than domestic soybean (mean θ = 0.00105). We therefore find the reduced levels of genetic variation in all the domestic populations we have studied with the exception of pigs.
Increased Proportion of Nonsynonymous Changes in Domestic Species
We extracted SNPs in protein-coding regions for each pool of domestic and wild species, and classified them into synonymous, nonsynonymous, missense (nonsense or read-through) changes. We compared the numbers of each category of SNPs across wild and domestic pools using a multistage bootstrap procedure (see Materials and Methods). In this procedure, for each gene, a randomly chosen wild and domestic pool are compared. This allows the total number of SNPs in each category for a randomized wild and domestic samples to be estimated and compared. Table 1 shows the average numbers of SNPs in each category derived from the bootstrap procedure that resamples genes from domestic and wild populations. In concordance with the estimates of θ, the total number of SNPs within protein-coding regions for domestic species was significantly lower than that for their wild relatives except for European pigs. The ratio of nonsynonymous to synonymous polymorphisms (nonsyn/syn ratio) was also significantly higher in domestic species than in wild species for all species analyzed except for European pigs (table 1). These results mirror levels of genetic variation, whereby domestic species with lower levels of variation than their extant ancestors tended also to have a greater proportion of nonsynonymous variants.
Table 1.
Average Number of SNPs in Coding Sequences
| Species | Average Number of SNPs in Coding Sequences |
Average nonsyn/syn Ratio |
Average Proportion of PROVEAN Damaging SNPs (#damaging SNPs/#nonsyn SNPs) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Domestic | Wild | P-Value | Domestic | Wild | P-Value | Domestic | Wild | P-Value | |
| Rabbit | 45,477.8 | 123,534.9 | <0.001 | 0.83 | 0.70 | <0.001 | 0.36 | 0.35 | 0.026 |
| Dog | 31,932.6 | 38,934.0 | <0.001 | 1.19 | 1.04 | <0.001 | 0.48 | 0.46 | <0.001 |
| Pig (Europe) | 60,405.6 | 34,084.3 | NS | 0.62 | 0.80 | 1.00 | 0.24 | 0.28 | NS |
| Pig (Asia) | 48,365.5 | 61,963.5 | <0.001 | 0.68 | 0.62 | <0.001 | 0.24 | 0.25 | NS |
| Chicken (domestic breeds) | 15,057.5 | 30,942.3 | <0.001 | 0.46 | 0.42 | <0.001 | 0.27 | 0.24 | <0.001 |
| Chicken (selection lines) | 9,947.0 | 30,970.0 | <0.001 | 0.57 | 0.42 | <0.001 | 0.32 | 0.24 | <0.001 |
| Rice (japonica) | 35,863.3 | 85,096.0 | <0.001 | 1.83 | 1.20 | <0.001 | 0.42 | 0.30 | <0.001 |
| Rice (indica) | 39,401.0 | 67,852.0 | <0.001 | 1.40 | 1.20 | <0.001 | 0.34 | 0.32 | <0.001 |
| Soybean | 65,130.0 | 103,270.0 | <0.001 | 1.39 | 1.36 | <0.001 | 0.21 | 0.23 | NS |
| Silkworm | 131,370.0 | 179,755.0 | <0.001 | 0.32 | 0.30 | <0.001 | 0.15 | 0.15 | NS |
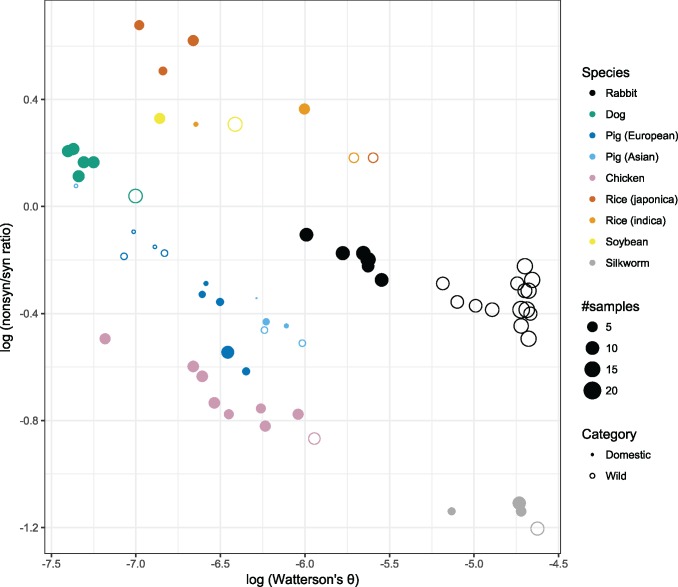
There is a general negative correlation between nonsyn/syn ratio and θ for each pool across all species investigated (fig. 2 andsupplementary table S2, Supplementary Material online). This trend is evident in all species and demonstrates that domestic populations have both lower genetic variation and higher nonsyn/syn ratio compared with their wild relatives. For each wild–domestic pair, there is no indication that nonsyn/syn ratio is further elevated in domestic pools relative to the expectation from θ. This is consistent with the hypothesis that Ne is the major determinant of average nonsyn/syn ratio.
Fig. 2.
—Relationship between the nonsyn/syn ratio and Watterson's θ for each pool. Relationship between Watterson's θ and the nonsyn/syn ratio in the pools of sequence data analyzed in this study.
Prediction of Effect Size of Nonsynonymous SNPs
We predicted the effect size of nonsynonymous SNPs for domestic and wild species using PROVEAN (Choi and Chan 2015). There was a general trend for the proportion of amino acid changes estimated as potentially damaging by PROVEAN to be higher in domestic species compared with wild species, although this trend was not observed in European and Asian pigs, or soybean, which all exhibited the opposite pattern (table 1). This result indicates a tendency for segregating amino acid variants with large phenotypic effects to be particularly enriched in domestic species, although in general the differences are not striking. The largest difference in the proportion of PROVEAN damaging SNPs is observed between Oryza rufipogon (wild rice) 0.30 versus Oryza japonica (domestic rice) 0.42. These results suggest that, in addition to an excess proportion of nonsynonymous SNPs, domestic populations also tend to harbor deleterious variants of larger effect size.
Relationship between Nonsyn/Syn Ratio and Allele Frequency
Inclusion of an outgroup sequence allowed us to determine the ancestral and derived allele for each SNP for dog, rabbit, and Asian and European pig populations (see Materials and Methods). We then estimated the nonsyn/syn ratio in each derived allele frequency bin in each species. We found a negative correlation between the nonsyn/syn ratio and allele frequency, with a higher proportion of nonsynonymous changes in the low frequency category (fig. 3). This is consistent with the higher prevalence of rare deleterious variants compared with common ones due to the influence of purifying selection. We also observed a strong tendency for domestic populations to have a higher nonsyn/syn ratio across allele frequency classes, with the exception of European pigs, which do not show this pattern overall (table 1). These differences between wild and domestic were most pronounced in the low frequency category (fig. 3). The trend was also observed in individual pools of dog (supplementary fig. S1, Supplementary Material online) and rabbit (supplementary fig. S2, Supplementary Material online). However, for variants at high frequencies close to fixation (0.8–1), we find that these differences are not significant (dog, rabbit, and European pig).
SNPs in Ultraconserved Noncoding Elements
Conserved noncoding sequences are known to harbor functionally important gene regulatory elements. Ultraconserved noncoding elements (UCNEs) are defined as genomic regions that show a high degree of conservation over extended evolutionary distances. Mutations within UCNEs are therefore more likely to have deleterious effects compared with those in neutrally evolving regions of the genome. UCNEs are common in vertebrate genomes (Dimitrieva and Bucher 2013), but are rarer in plant genomes (Inada et al. 2003). We analyzed the distribution of SNPs in UCNEs in our vertebrate data sets, defined as regions of >200 bp length and 95% identity between human and chicken (Dimitrieva and Bucher 2013). We found that the ratio of the number of observed and expected SNPs in UCNEs was higher in domestic species compared with wild ones, except for Asian pigs and European pigs, where there were no significant differences (table 2). These results are therefore consistent with those from coding sequences indicating an increased proportion of functional genetic variation segregating in populations of domestic species.
Table 2.
SNPs in UCNEs
| Species | Average Ratio (#observed UCNE SNPs/#expected UCNE SNPs) |
||
|---|---|---|---|
| Domestic | Wild | P-Value | |
| Rabbit | 0.20 | 0.16 | <0.001 |
| Dog | 0.65 | 0.51 | <0.001 |
| Pig (Europe) | 0.30 | 0.30 | NS |
| Pig (Asia) | 0.28 | 0.28 | NS |
| Chicken (domestic breeds) | 0.21 | 0.16 | <0.001 |
| Chicken (selection lines) | 0.24 | 0.16 | <0.001 |
Analysis of Heterozygous Sites in Individual Sequences
In the pig, rice, soybean, and silkworm data sets, samples were individually barcoded rather than sequenced in pools. Therefore, in addition to analyzing the nonsyn/syn ratios in segregating SNPs, we were able to analyze heterozygous sites in each sample. We examined the number nonsyn/syn heterozygous single nucleotide variations (SNVs) for each individual. We observed a significant enrichment of nonsynonymous SNVs in heterozygous sites in domestic rice, soybean, and silkworm, but not Asian or European pigs, compared with their wild relatives, consistent with the results from pooled sequence data (table 3 andsupplementary table S3, Supplementary Material online). Also consistent with the results from pooled data, we found a negative relationship between the number of heterozygous sites and the nonsyn/syn ratio among these sites (supplementary table S3 and fig. S3, Supplementary Material online). However, although domestic samples had significantly fewer heterozygous sites and higher nonsyn/syn ratios on average (apart from pigs), several wild individuals of rice, soybean, and silkworm had a low number of heterozygous sites and nonsyn/syn ratios similar to domestic samples. We also observed that heterozygous nonsynonymous sites in domestic samples had significantly more damaging effects measured by PROVEAN in rice, soybean, and silkworm (table 3), consistent with results from pooled samples.
Table 3.
Average Number of Heterozygous Sites in Individual Sequences
| Species | Average Number of Heterozygous Sites (SNVs) in Coding Sequences |
Average nonsyn/syn Ratio |
Average Proportion of PROVEAN Damaging SNVs (#damaging SNVs/#nonsyn SNVs) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Domestic | Wild | P-Value | Domestic | Wild | P-Value | Domestic | Wild | P-Value | |
| Pig (Europe) | 14,715.4 | 9,324.0 | NS | 0.73 | 0.90 | NS | 0.25 | 0.28 | NS |
| Pig (Asia) | 16,470.9 | 15,353.2 | NS | 0.68 | 0.72 | NS | 0.25 | 0.28 | NS |
| Rice (japonica) | 4,454.9 | 19,132.2 | <0.001 | 2.70 | 1.38 | <0.001 | 0.51 | 0.30 | <0.001 |
| Rice (indica) | 5,461.1 | 13,590.0 | <0.001 | 2.04 | 1.69 | <0.001 | 0.42 | 0.34 | <0.001 |
| Soybean | 3,045.1 | 6,095.9 | <0.001 | 1.84 | 1.73 | <0.001 | 0.24 | 0.24 | NS |
| Silkworm | 13,383.8 | 40,321.7 | <0.001 | 0.38 | 0.26 | <0.001 | 0.16 | 0.14 | <0.001 |
Note.—syn SNVs, synonymous SNVs; nonsyn SNVs, nonsynonymous SNVs.
Discussion
Increased Proportion of Deleterious Variation in Domestic Species
Here, we have analyzed the effects of domestication on genetic diversity in five animals and two crop plants using comparisons of whole-genome sequencing data. The main trends that emerge are that domestic populations have lower levels of genetic variation, a higher nonsyn/syn SNP ratio, and a higher proportion of SNPs in UCNEs. We find that the proportion of nonsynonymous variants is higher in domestic populations across a range of allele frequencies. In addition, there is a slight tendency for nonsynonymous variants in domestic populations to have more severe effects, as predicted by PROVEAN. These trends are not present across all the species we investigated. In European domestic pigs, genetic variation is not lower than their wild relatives, and there is no evidence for an increased proportion of nonsynonymous changes. These results are likely indicative of an increased proportion of deleterious mutations segregating in domestic populations as a consequence of historical population bottlenecks associated with the domestication process.
We find that the nonsyn/syn ratio is negatively correlated with genetic variation between sequenced pools for all species and that there are no apparent deviations from this trend dependent on whether a pool is from a wild or domestic population (fig. 2). In addition, neutral genetic variation is typically reduced in domestic pools and the proportion of nonsynonymous SNPs is elevated, consistent with this general pattern. This pattern likely reflects that differences in population size and demography, which affect Ne, result in differences in the average proportion of deleterious genetic variation segregating in a population. In particular, population bottlenecks are a common feature of domestication events, which result in a reduction of genetic variation. A reduction of Ne is predicted to lead to a reduction in the efficacy of purifying selection, which could result in the increased proportion of nonsynonymous variants and those in conserved noncoding elements observed here. It is however important to note that demographic changes such as bottlenecks may have different effects on the evolutionary trajectories of nonsynonymous and synonymous variants because they segregate at different frequencies on average, which implies that the proportion of deleterious variants can rise in a population without a relaxation of selection if populations are not at demographic equilibrium (Do et al. 2015; Brandvain and Wright 2016).
Genetic hitchhiking is another process that can lead to an increase in the prevalence of deleterious variants. Here, deleterious variants rise in frequency because they are in genetic linkage with nearby variants under positive selection (Maynard Smith and Haigh 1974). This possibility has been investigated in dogs, where it has been shown that the number of nonsynonymous SNPs is elevated around regions identified as selective sweeps (Marsden et al. 2016). This suggests that sweep regions contribute proportionally more to excess deleterious genetic variation in dogs. However, because selective sweep regions only comprise a small fraction of the genome they only make minor contribution to overall patterns of variation and cannot explain the overall excess of deleterious genetic variation. It is therefore likely that genetic hitchhiking makes a contribution toward generating the excess proportion of deleterious variation in domestic populations investigated here although we have not quantified this effect.
The majority of nonsynonymous variants segregating in a population is expected to be either neutral or weakly deleterious, as new advantageous mutations occur relatively infrequently and mutations with large fitness effects contribute little to polymorphism (Eyre-Walker and Keightley 2007). It is therefore unlikely that the excess proportion of nonsynonymous variants we observe in domestic populations is comprised positively selected variants. The largest excess of nonsynonymous variants occurs at low-frequency alleles, consistent with the view that they tend to be weakly deleterious. We infer a substantial proportion of nonsynonymous variants (15–50%) to be damaging using PROVEAN in both wild and domestic populations (table 1), and this proportion is significantly higher in domestic populations for certain comparisons (see also below). A small proportion of these variants may in fact have positive consequences for fitness. However, this is unlikely to lead to the significant genome-wide differences between genetic variation in domestic and wild populations that we observe here.
Simulations of the demographic history associated with domestication, based on the evolution of dogs, indicate that an early population bottleneck associated with an initial domestication event is likely to be the most important factor leading to an increased proportion of deleterious variants (Marsden et al. 2016). Relative to this population bottleneck, the effects of inbreeding associated with breed creation and maintenance are inferred to be minor. As the time since humans began domesticating wild species is short in evolutionary terms, it is likely that most deleterious variants are already present in ancestral populations, but genetic bottlenecks allow deleterious variants to drift to higher frequency (Eyre-Walker et al. 1998). It is also interesting to note that European pigs, which have not experienced strong population bottlenecks but presumably similar levels of artificial selection compared with other domestic animals (Groenen et al. 2012; Rubin et al. 2012; Frantz et al. 2015), do not show evidence of an increased proportion of deleterious variants. Our results are therefore consistent with the hypothesis that early population bottlenecks that likely occurred in the domestication of most animals and plants have resulted in an increased proportion of deleterious variation in these species. However, it is clear that every domesticated species has a unique demographic history and the process of domestication differs for each species.
The extent to which the spectrum of effect sizes of segregating nonsynonymous genetic variation is expected to be influenced by domestication is not clear. One previous study found that radical amino acid changes occurred in cultivated rice much more frequently than those in wild rice (Lu et al. 2006). A reduction in the efficacy of selection would be expected to allow genetic variants with negative fitness coefficients of slightly higher magnitude to evade the effects of purifying selection. In general, amino acid changes with highly deleterious effects would be removed by purifying selection even in populations with reduced Ne. An exception to this could occur if a deleterious variant was linked to a variant under strong artificial selection. We detect an excess of damaging amino acid-changing variants in domestic populations in some of the species under investigation here, although this is not the case with all populations and the effects are minor in some cases.
It has been suggested that an increase in the proportion of weakly deleterious variants could represent a cost of domestication (Björnerfeldt et al. 2006; Cruz et al. 2008; Schubert et al. 2014; Renaut and Rieseberg 2015; Kono et al. 2016; Marsden et al. 2016). Our results do not directly quantify the mutational load per genome. Such an estimation is problematic using pooled samples. An increased proportion of deleterious variation in a population does not necessarily imply an increase in the total number of deleterious mutations per individual (Brandvain and Wright 2016). However, direct assessment of individual genomes in horses (Schubert et al. 2014) and dogs (Marsden et al. 2016) indicates that an increase in the proportion of deleterious variants in domestic populations is associated with an increased burden of deleterious variants per genome, which implies that domestication bottlenecks have typically led to increased mutational load. One important consideration for quantifying mutational load per individual is the extent to which deleterious mutations have additive or recessive effects, which is not well understood (Simons et al. 2014; Do et al. 2015; Henn et al. 2015).
Domestication Histories of Each Species
Domestic rabbits are derived from an ancestral population of wild rabbits (Oryctolagus cuniculus) from two subspecies that were restricted to France and the Iberian peninsula, where they are still found today (Carneiro et al. 2009). Rabbits were domesticated in the monasteries of southern France likely around 1,400 years ago (Clutton-Brock 1999). We compared whole-genome genetic variation in population sampled across this native range of wild rabbits, and compared it with pools of samples of six domestic breeds (Carneiro et al. 2014). There is a consistent reduction in genetic variation and increase in the nonsyn/syn ratio in all pools, which can likely be attributed to the population bottlenecks that occurred when small populations of rabbits were bred in isolation in monasteries. In addition to population bottlenecks, the domestic populations also show evidence of positive selection at loci across the genome, evidenced by a general enrichment of SNPs in functional genomic regions among those with extreme frequency differences between wild and domestic pools (Carneiro et al. 2014).
The domestication of dogs is the most ancient domestication event, which likely occurred over 15,000 years ago when wolves began scavenging for food in human settlements although the number, timing, and location of events are debated (Thalmann et al. 2013; Shannon et al. 2015; Frantz et al. 2016; Wang et al. 2016). Their genetic ancestors, wolves, are spread across most of the northern hemisphere. Previous studies have found that there is higher proportion of nonsynonymous variants in dogs using both mtDNA and nuclear SNPs (Björnerfeldt et al. 2006; Cruz et al. 2008). Most recently, a study of 90 individually barcoded whole-genome sequences was able to estimate that individual dogs have an average of 2–3% increased genetic load compared with gray wolves (Marsden et al. 2016). Interestingly, this study found an increase in the proportion of amino acid changes in both breed dogs, which have experienced strong selective breeding for desirable characteristics, and village dogs, which have not. This and simulation studies indicated that population bottlenecks rather than recent inbreeding and artificial selection are the main cause for the excess of weakly deleterious variants. Our study corroborates these findings using a separate data set of 60 dogs from 14 breeds compared with 12 wolves (Axelsson et al. 2013). We find a lower genetic variation and a higher nonsyn/syn ratio in all dog pools compared with wolf. We also find that genetic variation in pools containing mixed breeds is higher than in single breed pools, but is still lower than in wolves. Mixed-breed pools likely reflect the variation in the ancestral dog population before the creation of breeds (Lindblad-Toh et al. 2005). Hence, it appears that the initial domestication bottleneck had a large effect on genetic variation and the prevalence of weakly deleterious variation.
Pigs are believed to have at least two independent origins of domestication, in western Eurasia and East Asia (Giuffra et al. 2000). Phylogenetic analysis has revealed an ancient split between Asian and European wild boars 1.6–0.8 Ma, prior to pig domestication, with distinct Asian and European domestic pig lineages derived from these two ancestral populations (Frantz et al. 2015). Asian and European wild boars are separate subspecies with distinct demographic histories (Giuffra et al. 2000; Frantz et al. 2015). A previous analysis of the same data set used here indicated that although both populations declined during the last glaciation, a much bigger population size reduction occurred in the European population, which was likely connected to more extreme fragmentation in northern Europe (Groenen et al. 2012). This could explain the generally lower genetic variation observed in European wild boars (fig. 1;Groenen et al. 2012).
In contrast to most domestication events, pigs were only partially isolated from their wild ancestors, and were able to mix and interbreed with wild boars (Groenen et al. 2012; Frantz et al. 2015). Gene flow between wild and domestic pigs was therefore common during their history and this process is inferred to be more common in Europe compared with Asia. We find similar levels of genetic variation in wild and domestic pigs and Asia, and European pigs actually have higher levels of genetic variation than present-day European wild boar populations. Moreover, the European domestic pig populations are the only species we have analyzed here that do not have an elevated nonsyn/syn ratio compared with their wild relatives, indicating that domestic pigs are not enriched for deleterious SNPs as is common in domestic species. European pigs actually have higher genetic variation than European wild boars, an observation that could reflect human-mediated gene flow between Asian and European domestic pig populations (Giuffra et al. 2000; Frantz et al. 2015).
Archeological and genetic evidence suggests that chickens were domesticated in Asia up to 10,000 years ago and that there are likely to have been multiple centers of domestication in distinct origins across Southeast Asia and China (Stevens 2005; Kanginakudru et al. 2008; Xiang et al. 2014). The ancestor of chickens is the red jungle fowl, although at least some genetic variation in chickens is derived from the gray jungle fowl, indicating that hybridization with this species also contributed to chicken domestication (Eriksson et al. 2008). Our finding of reduced variation and elevated nonsyn/syn ratio in domestic chicken compared with red jungle fowl indicates that despite multiple origins and a huge present-day population, population bottlenecks due to domestication are likely to have affected chicken genetic diversity and increased the mutational load and that gene flow from wild populations into domestic ones has not been common. In particular, we observed a particularly elevated nonsyn/syn ratio (and reduced levels of genetic variation) in the experimentally selected chicken lines (high and low growth line and obese strain; fig. 1 and supplementary table S2, Supplementary Material online), which have been maintained with small numbers of breeding individuals. The population size of Obese Strain (OS) is extremely small, which is reflected in patterns of genetic diversity (Dietrich et al. 1999).
A likely history of rice cultivation is that the Oryza sativa japonica strain was originally developed in southern China and that the Oryza sativa indica strain was subsequently developed through crosses between japonica and local wild strains of Oryza sativa (Huang et al. 2012). We infer that the proportion of weakly deleterious variants segregating in all domestic rice strains is higher than wild rice, but that this effect is much more pronounced in japonica compared with indica rice, which is reasonable considering this evolutionary history (Xu et al. 2011; Huang et al. 2012). Our results are consistent with those reported by Liu et al. (2017) using a different data set.
Cultivated soybean was domesticated in China about 5,000 years ago (Hymowitz 1970). Our analysis is consistent with a strong bottleneck that reduced genetic variation and increased the proportion of weakly deleterious variants. Traces of this process are also observed in greater extent of linkage disequilibrium in domestic populations (Lam et al. 2010). Similar results have been presented by Kono et al. (2016) using a smaller set of sequences.
Silkworms were also domesticated over in China over 5,000 years ago from the wild silkworm that is found throughout Asia. Strong selection for desirable traits has had the result that domestic silkworms are now completely dependent on humans for survival (Goldsmith et al. 2005). However, despite substantial phenotypic changes, it has been estimated that the domestication bottleneck was modest, leading to less than 20% reduction in levels of genetic variation (Xia et al. 2009). This could suggest that the initial selected population was large or that there were multiple such originator populations. However, despite a relatively small reduction in population size, there is still a significant increase in the nonsyn/syn ratio, which likely reflects elevated prevalence of weakly deleterious mutations.
Conclusion
A number of studies from various different animal and plant species support the hypothesis that domestication commonly results in an increased proportion of weakly deleterious variants segregating in populations of domestic species. Here, we have tested for signatures of an increased proportion of nonsynonymous variants in new data sets from seven domestic species compared with present-day populations of their wild ancestors. Our results corroborate previous findings in additional species and are consistent with the hypothesis that population bottlenecks associated with the initial domestication events result in a rise in frequency of existing weakly deleterious mutations. We also show that this pattern is observable in conserved noncoding regions as well as protein-coding genes. We do not observe an increase in deleterious mutations in European pig populations, where domestication did not entail a bottleneck and isolation. Ancient domestication events have therefore given rise to an increased proportion of deleterious variants, which persist in domestic species even though their current population sizes are huge.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
Hon-Ming Lam kindly provided soybean SNP data sets. This work was supported by Leading Young Researcher Overseas Visit Program from Tohoku University, KAKENHI (17H03728) from the Japan Society for the Promotion of Science and Environment Research and Technology Development Fund (4-1605) of the Ministry of the Environment, Japan (to T.M.). Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. M.C. was supported by Fundação para a Ciência e Tecnologia (FCT) through POPH-QREN funds from the European Social Fund and Portuguese MCTES (IF/00283/2014/CP1256/CT0012).
Literature Cited
- Auton A, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 912:e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, et al. 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 4957441:360–364.http://dx.doi.org/10.1038/nature11837 [DOI] [PubMed] [Google Scholar]
- Björnerfeldt S, Webster MT, Vila C.. 2006. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 168:990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, et al. 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 45:e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Wright SI.. 2016. The limits of natural selection in a nonequilibrium world. Trends Genet. 324:201–210.http://dx.doi.org/10.1016/j.tig.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Carneiro M, et al. 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 3456200:1074–1079.http://dx.doi.org/10.1126/science.1253714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Ferrand N, Nachman MW.. 2009. Recombination and speciation: loci near centromeres are more differentiated than loci near telomeres between subspecies of the European rabbit (Oryctolagus cuniculus). Genetics 1812:593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 103:195–205.http://dx.doi.org/10.1038/nrg2526 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1999. The genetic basis of inbreeding depression. Genet Res. 743:329–340.http://dx.doi.org/10.1017/S0016672399004152 [DOI] [PubMed] [Google Scholar]
- Choi Y, Chan AP.. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 3116:2745–2747.http://dx.doi.org/10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC.. 2011. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 78:e1002240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J. 1999. A natural history of domesticated mammals. Cambridge: Cambridge University Press. [Google Scholar]
- Crow JF, Kimura M.. 2009. An introduction to population genetics theory. Repr. of 1970 Ed. Caldwell (NJ: ): Blackburn. [Google Scholar]
- Cruz F, Vila C, Webster MT.. 2008. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol Biol Evol. 2511:2331–2336.http://dx.doi.org/10.1093/molbev/msn177 [DOI] [PubMed] [Google Scholar]
- Darwin C. 1876. The variation of animals and plants under domestication. New York: Appleton. [Google Scholar]
- Dietrich HM, Cole RK, Wick G.. 1999. The natural history of the obese strain of chickens–an animal model for spontaneous autoimmune thyroiditis. Poult Sci. 7810:1359–1371.http://dx.doi.org/10.1093/ps/78.10.1359 [DOI] [PubMed] [Google Scholar]
- Dimitrieva S, Bucher P.. 2013. UCNEbase—a database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic Acids Res. 41(Database issue):D101–D109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, et al. 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 472:126–131.http://dx.doi.org/10.1038/ng.3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, et al. 2008. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4:e1000010.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS.. 1998. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl Acad Sci U S A. 958:4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD.. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet. 88:610–618.http://dx.doi.org/10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Frantz LAF, et al. 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 47:1141–1148.http://dx.doi.org/10.1038/ng.3394 [DOI] [PubMed] [Google Scholar]
- Frantz LAF, et al. 2016. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 3526290:1228–1231.http://dx.doi.org/10.1126/science.aaf3161 [DOI] [PubMed] [Google Scholar]
- Freedman AH, et al. 2014. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 101:e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Gittelman RM, Bamshad MJ, Akey JM.. 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet. 954:421–436.http://dx.doi.org/10.1016/j.ajhg.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E, et al. 2000. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 1544:1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith MR, Shimada T, Abe H.. 2005. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 50:71–100. [DOI] [PubMed]
- Groenen MAM, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 4917424:393–398.http://dx.doi.org/10.1038/nature11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfield M, Otto SP.. 2011. Recombination and hitchhiking of deleterious alleles. Evol Int J Org Evol. 659:2421–2434.http://dx.doi.org/10.1111/j.1558-5646.2011.01311.x [DOI] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S.. 2015. Estimating the mutation load in human genomes. Nat Rev Genet. 166:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 1134:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1966. The effect of linkage on limits to artificial selection. Genet Res. 83:269–294.http://dx.doi.org/10.1017/S0016672300010156 [PubMed] [Google Scholar]
- Huang X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 4907421:497–501.http://dx.doi.org/10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz T. 1970. On the domestication of the soybean. Econ Bot. 244:408–421.http://dx.doi.org/10.1007/BF02860745 [Google Scholar]
- Inada DC, et al. 2003. Conserved noncoding sequences in the grasses. Genome Res. 139:2030–2041.http://dx.doi.org/10.1101/gr.1280703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanginakudru S, Metta M, Jakati R, Nagaraju J.. 2008. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol. Biol 8:174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K.. 2008. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 99:713–725.http://dx.doi.org/10.1038/nrg2382 [DOI] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A.. 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 1774:2251–2261.http://dx.doi.org/10.1534/genetics.107.080663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Maruyama T, Crow JF.. 1963. The mutation load in small populations. Genetics 48:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, et al. 2013. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci U S A. 11028:E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, et al. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 61:e15925.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono TJY, et al. 2016. The role of deleterious substitutions in crop genomes. Mol Biol Evol. 339:2307–2317.http://dx.doi.org/10.1093/molbev/msw102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, et al. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 4212:1053–1059.http://dx.doi.org/10.1038/ng.715 [DOI] [PubMed] [Google Scholar]
- Larson G, Burger J.. 2013. A population genetics view of animal domestication. Trends Genet. 294:197–205.http://dx.doi.org/10.1016/j.tig.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Larson G, Fuller DQ.. 2014. The evolution of animal domestication. Annu Rev Ecol Evol Syst. 451:115–136.http://dx.doi.org/10.1146/annurev-ecolsys-110512-135813 [Google Scholar]
- Li H, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2516:2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2514:1754–1760.http://dx.doi.org/10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 4387069:803–819.http://dx.doi.org/10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhou Y, Morrell PL, Gaut BS.. 2017. Deleterious variants in Asian rice and the potential cost of domestication. Mol Biol Evol. 344:908–924. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 4517181:994–997.http://dx.doi.org/10.1038/nature06611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, et al. 2006. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 223:126–131.http://dx.doi.org/10.1016/j.tig.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trends Genet. 268:345–352.http://dx.doi.org/10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, et al. 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci U S A. 1131:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith JM, Haigh J.. 1974. The hitchhiking effect of a favourable gene. Genet Res. 2301:23–35.http://dx.doi.org/10.1017/S0016672300014634 [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD.. 2013. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 1412:840–852.http://dx.doi.org/10.1038/nrg3605 [DOI] [PubMed] [Google Scholar]
- Mulvihill JJ. 1972. Congenital and genetic disease in domestic animals. Science 1764031:132–137.http://dx.doi.org/10.1126/science.176.4031.132 [DOI] [PubMed] [Google Scholar]
- Nabholz B, et al. 2014. Transcriptome population genomics reveals severe bottleneck and domestication cost in the African rice (Oryza glaberrima). Mol Ecol. 239:2210–2227.http://dx.doi.org/10.1111/mec.12738 [DOI] [PubMed] [Google Scholar]
- Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 2465428:96–98.http://dx.doi.org/10.1038/246096a0 [DOI] [PubMed] [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 231:263–286.http://dx.doi.org/10.1146/annurev.es.23.110192.001403 [Google Scholar]
- Ostrander EA, Kruglyak L.. 2000. Unleashing the canine genome. Genome Res. 109:1271–1274.http://dx.doi.org/10.1101/gr.155900 [DOI] [PubMed] [Google Scholar]
- Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L.. 2013. On the accumulation of deleterious mutations during range expansions. Mol Ecol. 2224:5972–5982. [DOI] [PubMed] [Google Scholar]
- Renaut S, Rieseberg LH.. 2015. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other compositae crops. Mol Biol Evol. 329:2273–2283.http://dx.doi.org/10.1093/molbev/msv106 [DOI] [PubMed] [Google Scholar]
- Rubin C-J, et al. 2012. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci U S A. 10948:19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CJ, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 4647288:587–591.http://dx.doi.org/10.1038/nature08832 [DOI] [PubMed] [Google Scholar]
- Schubert M, et al. 2014. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci U S A. 11152:E5661–E5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon LM, et al. 2015. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci U S A. 11244:13639–13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G.. 2014. The deleterious mutation load is insensitive to recent population history. Nat Genet. 463:220–224.http://dx.doi.org/10.1038/ng.2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. 2005. Genetics and evolution of the domestic fowl. Cambridge/New York: Cambridge University Press. [Google Scholar]
- Thalmann O, et al. 2013. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 3426160:871–874.http://dx.doi.org/10.1126/science.1243650 [DOI] [PubMed] [Google Scholar]
- Vaysse A, et al. 2011. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 710:e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-D, et al. 2016. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 261:21–33.http://dx.doi.org/10.1038/cr.2015.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 72:256–276.http://dx.doi.org/10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Xia Q, et al. 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 3265951:433–436.http://dx.doi.org/10.1126/science.1176620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, et al. 2014. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 11149:17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. 2011. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 301:105–111.http://dx.doi.org/10.1038/nbt.2050 [DOI] [PubMed] [Google Scholar]
- Zeder MA. 2012. The domestication of animals. J Anthropol Res. 682:161–190.http://dx.doi.org/10.3998/jar.0521004.0068.201 [Google Scholar]
- Zhang M, Zhou L, Bawa R, Suren H, Holliday JA.. 2016. Recombination rate variation, hitchhiking, and demographic history shape deleterious load in poplar. Mol Biol Evol. 3311:2899–2910.http://dx.doi.org/10.1093/molbev/msw169 [DOI] [PubMed] [Google Scholar]
- Zohary D, Hopf M, Weiss E.. 2012. Domestication of plants in the Old World: the origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin. 4th ed Oxford/New York: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.