Abstract
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) represent a powerful cellular platform for illuminating mechanisms of human cardiovascular disease and for pharmacological screening. Recent advances in CRISPR/Cas9-mediated genome editing technology underlie this profound utility. We have generated hiPSC-CMs with fluorescently-tagged sarcomeric proteins (CPHG UNIT 21.12), which provide a tool to non-invasively study human sarcomere function and dysfunction. In this unit, we illustrate methods for conducting high-efficiency, small molecule-mediated differentiation of hiPSCs into cardiomyocytes and subsequently, performing non-invasive contractile analysis through direct sarcomere tracking of GFP-sarcomere reporter hiPSC-CMs. We believe that this type of analysis can overcome sensitivity problems found in other forms of contractile assays involving hiPSC-CMs by directly measuring contractility at the fundamental contractile unit of the hiPSC-CM, the sarcomere.
Keywords: cardiomyocytes, pluripotent stem cells, sarcomere, fluorescent reporter, contractility, iPSCs, CRISPR
INTRODUCTION
The development of human induced pluripotent stem cells (hiPSCs) enabled the mass production of patient-specific, human cell types that would otherwise be difficult to obtain, such as human cardiomyocytes, the major contractile cell type found in the heart (Burridge, Sharma, & Wu, 2015; Takahashi et al., 2007). These beating human cardiomyocytes can, on a cellular level, recapitulate genetically-acquired cardiac disorders such as arrhythmias and cardiomyopathies, although hiPSC-CMs do demonstrate some differences from primary human cardiomyocytes in terms of cellular structure, electrophysiology, and metabolism, a reflection of their cellular “immaturity” (Hinson et al., 2015; Itzhaki et al., 2011; Robertson, Tran, & George, 2013). Additionally, recent developments in efficient mammalian genome editing using CRISPR/Cas9 has enabled rapid genetic modification of human pluripotent stem cell lines, allowing for the production of a variety of genetically customized cell types (Mali et al., 2013).
Thanks to improvements in differentiation protocols, human cardiomyocytes can be mass-produced from hiPSCs for the purposes of cardiovascular drug screening, in-vitro disease modeling, and potentially for cellular replacement therapy (Sharma et al., 2017; Sharma et al., 2014; Sharma, Wu, & Wu, 2013). Human pluripotent stem cell differentiation protocols have rapidly progressed over the past two decades. Early human pluripotent stem cell differentiation protocols relied on aggregation-based, embryoid body approaches to promote differentiation into cardiomyocytes (Kehat et al., 2001). These protocols were laborious and result in heterogeneous populations of differentiated cells within embryoid bodies. Initial differentiation approaches also relied on expensive cardiogenic proteins such as Activin A and BMP4, members of the TGF-beta pathway (Kattman et al., 2011). Both issues were solved with the development of pluripotent stem cell monolayer-based cardiomyocyte differentiation protocols utilizing small molecules modulating the Wnt signaling pathway (Lian et al., 2012). This approach allows rapid production of human cardiomyocytes in a highly-efficient and cost-effective manner. Recent refinements have introduced chemically-defined components into the differentiation medium, further reducing the experimental cost and batch-to-batch variability affecting cardiomyocyte differentiation (Burridge et al., 2014).
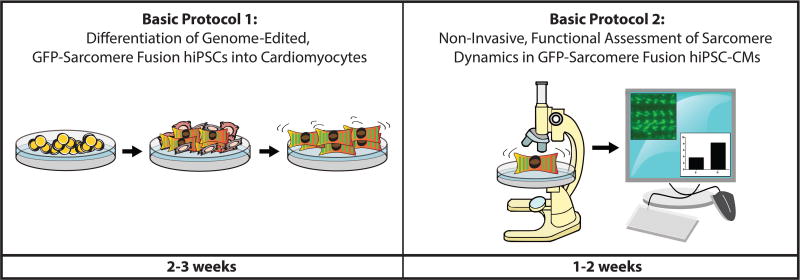
Advances in CRISPR/Cas9 genome editing have allowed researchers to generate genetically customized, patient-specific human cardiomyocytes. CRISPR/Cas9-mediated editing enables fluorescent tagging of proteins in the cardiomyocyte sarcomere, the intracellular contractile unit found in muscle cells. These custom hiPSC-CMs can be sorted with fluorescence activated cell sorting to isolate purified populations of human cardiomyocytes based on fluorescence alone. Existing hiPSC-CM purification protocols largely rely on metabolic selection, which can be inefficient due to hiPSC-CM metabolic immaturity, as well as a lack of highly-specific cell surface markers for hiPSC-CMs (Tohyama et al., 2013). GFP-tagged sarcomeres also allow the visual examination of cardiomyocytes through differentiation, offering a new window into studying the temporal regulation of cardiac development. Finally, since hiPSC-CMs are a dynamic cell type with spontaneous contractility, generation of fluorescently-tagged sarcomere reporter hiPSC-CMs enables non-invasive sarcomere tracking in live cells. This has been developed with the intention to provide a platform for introducing patient-specific sarcomere mutations to examine alterations in contractile function in an otherwise isogenic cell line. In this unit, we provide current protocols for the differentiation and functional characterization of multipurpose, genetically-customized hiPSC-CMs with GFP-tagged sarcomeres, with a focus on high-efficiency differentiation of genome-edited hiPSCs into cardiomyocytes and non-invasive evaluation of cardiomyocyte contractility using fluorescently-tagged sarcomeres (Figure 1).
Figure 1.
Workflow for differentiation and functional analysis of GFP-sarcomere fusion reporter hiPSC-CMs. Expect the entire protocol to take approximately one month.
STRATEGIC PLANNING
Selection of an appropriate GFP-sarcomere fusion reporter hiPSC line can influence the downstream hiPSC-CM contractility analysis. We highly recommend another Current Protocols publication for details about CRISPR-mediated generation of hiPSCs with fluorescently-tagged endogenous proteins (see UNIT 21.11, Sharma et al 2017). [*Copy Editor: Unit 21.11 also publishing in this supplement.] The human sarcomere has many candidate proteins, for tagging with a fluorescent GFP-fusion reporter. When selecting a GFP-sarcomere fusion reporter hiPSC line for differentiation and analysis, consideration should be given to the location of the fluorescently-tagged protein in the sarcomere. In this instance, we have chosen to fluorescently tag onto a protein present in the sarcomeric Z disk (N-terminal Titin), which illuminates typical striated muscle sarcomeric periodicity, as the Z disks demarcate the bounds of a sarcomeric unit. This technique is useful for measuring real time sarcomere length and shortening.
BASIC PROTOCOL 1: DIFFERENTIATION OF GENOME-EDITED, FLUORESCENT SARCOMERE FUSION REPORTER hiPSCs INTO CARDIOMYOCYTES
Human induced pluripotent stem cells can be differentiated into a variety of somatic cell types including beating human cardiomyocytes, the contractile muscle cells of the heart. Small-molecule mediated differentiation protocols can yield human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) in approximately 10 days after application of modulators of the Wnt cell signaling pathway. Cells can begin beating as early as 7 days post-differentiation and express sarcomeric proteins and ion channels found in adult human cardiomyocytes. These hiPSC-CMs can be metabolically purified from the other non-CM cell types produced during differentiation, by removing glucose from the cell culture medium (Tohyama et al., 2013). Cardiomyocytes can shift to utilizing alternative metabolic pathways such as fatty acid-based metabolism in an environment devoid of glucose, whereas non-cardiomyocytes cannot and are selected out by glucose starvation. It should be noted that due to hiPSC-CM immaturity and heterogeneity some hiPSC-CMs will also be lost during glucose starvation. This is one reason why fluorescent sarcomere fusion reporter hiPSC-CMs could be useful. These cells begin expressing GFP upon sarcomeric protein expression, a signal that they are of the intended lineage. Due to this green signal, GFP-sarcomere fusion hiPSC-CMs can be sorted with fluorescence-activated cell sorting to obtain pure cell populations. Additionally, the expression of GFP-tagged sarcomeres enables real-time robust evaluation of sarcomere contractility, which has not been previously possible in live human cardiomyocytes (see Figure 2 for differentiation details).
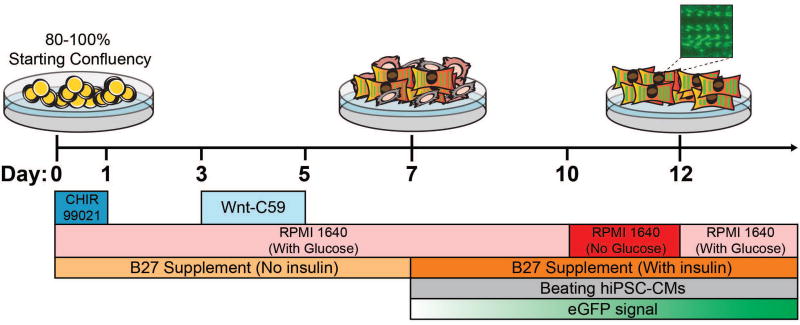
Figure 2.
Protocol for hiPSC-CM differentiation of genome-edited, GFP-sarcomere fusion hiPSCs. This small-molecule mediated differentiation approach involves modulation of the Wnt cell signaling pathway to induce cardiac mesoderm and cardiomyocytes. The earliest GFP-sarcomere signal will coincide with the earliest sarcomeric protein expression and will strengthen over the course of differentiation.
Materials
Genome-edited, GFP-sarcomere fusion hiPSCs
Matrigel (hESC-qualified; BD Sciences, cat. no. 354277)
DMEM/F12 medium (Thermo Fisher Scientific, cat. no. 11320033)
RPMI 1640 medium (Thermo Fisher Scientific, cat. no. 11835055)
Glucose-free RPMI 1640 medium (Thermo Fisher Scientific, cat. no. 11879-020)
B27 Supplement Minus Insulin (Thermo Fisher Scientific, cat. no. A1895601)
B27 Supplement with Insulin (Thermo Fisher Scientific, cat. no. 17504-044)
CHIR99021 (Thermo Fisher Scientific, cat. no. 508306)
Wnt-C59 (Biorbyt, cat. no. orb181132)
6-well tissue culture-treated plates
Standard cell culture laminar flow hood
mTeSR1 medium (StemCell Technologies, cat. no. 05850)
Rho kinase inhibitor (ROCKi; Tocris cat. no. 1254)
0.5M EDTA (Thermo Fisher Scientific, cat. no. 15575020)
DMSO (Sigma-Aldrich, cat. no. D-2650)
Differentiate Genome-Edited, GFP-Sarcomere Fusion hiPSCs to Cardiomyocytes
-
Prepare 6-well plates coated with Matrigel extracellular matrix, on which genome-edited, fluorescent fusion reporter hiPSCs will grow in a feeder cell-free format prior to differentiation. Thaw Matrigel on ice and add 125 µL cold Matrigel to 50 mL of cold DMEM/F12, a 1:400 dilution. Pre-cool pipette tips to 4°C before pipetting Matrigel. 2 mL of Matrigel solution should be added per well of a 6-well dish, leave to coat for at least 1 hour before use.
Matrigel-coated plates can typically be kept at 37°C for 1 month.
For cell culture, remove Matrigel mixture from a 6-well plate, wash once with 2 mL of PBS and add hiPSCs (ideally below passage 30), resuspended hiPSCs in mTeSR1 pluripotent stem cell growth medium containing 10 µM rho kinase inhibitor. Rho kinase inhibitor dramatically improves survival of stem cells during passaging (Watanabe et al., 2007). These hiPSCs can be freshly cultured or obtained from a frozen stock.
-
24 hours after plating cells, replace media with fresh mTeSR1 without rho kinase inhibitor. Allow cells to grow to 80–100% confluency, with daily replacement of mTeSR1.
The optimal cell confluency for differentiation can vary across cell lines, we recommend optimizing confluencies for your cell line.
-
At optimal confluency, passage one well from a 6-well plate of hiPSCs in a 1:12 passaging ratio in mTeSR1 with 10 µM rho kinase inhibitor into a new, Matrigel pre-coated 6-well plate.
This passage maintains the hiPSC line for future experiments and differentiations.
Passaging begins with the removal of mTesR1 and the addition of 1 mL 0.5 mM EDTA in PBS to a well of ~80% confluent hiPSCs. Incubate the plate at 37°C for 5 minutes to partially dissociate hiPSCs.
Gently aspirate the 0.5 mM EDTA. Cells should not be completely dissociated. If examined under the microscope, the hiPSC colonies are breaking apart into single cells and only lightly adhered to the cell culture plate.
Using a 1000 µL manual pipetman, disturb the loosened hiPSCs with 1 mL fresh, prewarmed mTesR1 with 10 µM rho kinase inhibitor. Collect the cell suspension in a new 15 mL tube and add 11 mL mTesR1 with 10 µM rho kinase inhibitor for a 1:12 dilution.
Obtain a new Matrigel-coated 6-well dish and aspirate off the Matrigel solution. In each well of the new 6-well plate, add 1 mL of the 1:12 dilution of hiPSCs suspended in mTesR1 with 10 µM rho kinase inhibitor. Distribute the cells evenly in the new 6-well plate.
Continue culturing this newly-passaged plate by replacing 2 mL mTesR1 (without rho kinase inhibitor) daily for approximately 4 days. The new plate should be ready for passage again around 4 days after a 1:12 passage of an 80% confluent well of hiPSCs.
With the remaining 5 non-passaged wells in the original 6-well dish, begin treatment with 12 µM CHIR99021 GSK3β inhibitor dissolved in DMSO (day 0 of differentiation). To begin CHIR99021 treatment, remove old mTeSR1 medium. Add fresh RPMI 1640 medium, supplemented with B27 supplement (without insulin), and 12 µM CHIR99021. Leave cells in this medium for 24 hours.
After 24 hours of CHIR99021 treatment (day 1 of differentiation), aspirate RPMI 1640 medium (with B27 supplement without insulin) containing CHIR99021 and replace with fresh RPMI 1640 medium (with B27 supplement without insulin). Leave cells in this medium for 48 hours.
After 48 hours, (day 3 of differentiation), begin treatment with Wnt-C59 small molecule Wnt inhibitor. Aspirate RPMI 1640 medium (with B27 supplement without insulin) and replace with fresh RPMI 1640 medium (with B27 supplement without insulin) containing 2 µM Wnt-C59 dissolved in DMSO. Leave cells in this medium for 48 hours.
After 48 hours (day 5 of differentiation), aspirate RPMI 1640 medium (with B27 supplement without insulin) containing Wnt-C59 and replace with fresh RPMI 1640 medium (with B27 supplement without insulin). Leave cells in this medium for 48 hours.
After 48 hours (day 7 of differentiation), aspirate RPMI 1640 medium (with B27 supplement without insulin) and replace with fresh RPMI 1640 medium (with B27 supplement with insulin). Leave cells in this medium for 72 hours. Continue growing cells on RPMI 1640 medium (with B27 supplement with insulin), replacing medium every 48 hours starting at day 10.
-
GFP signal may become visible in the differentiated cells as early as day 7 of differentiation.
This is typically the earliest timepoint that sarcomeric proteins are expressed allowing the differentiated hiPSC-CMs to begin beating. GFP expression will be dependent on the relative expression of the tagged sarcomeric protein, as some proteins are more highly expressed than others.
Once beating begins, metabolically purify the hiPSC-CMs from the non-cardiomyocytes. Remove old RPMI 1640 medium (with B27 supplement with insulin) and add fresh RPMI 1640 medium without glucose (with B27 supplement with insulin). Leave cells on this medium for 48 hours, replenishing every 48 hours as necessary. Non-cardiomyocyte death likely begins after 24 hours. Cardiomyocytes may stop beating and there may be some death in this fraction. Stop glucose starvation and return to RPMI 1640 medium containing glucose (with B27 supplement with insulin) after purification is complete (typically after two days) or if hiPSC-CM death becomes excessive. These purified hiPSC-CMs are now ready for functional contractile analysis.
BASIC PROTOCOL 2: NON-INVASIVE, FUNCTIONAL ASSESSEMENT OF SARCOMERE DYNAMICS IN GENOME-EDITED, GFP-SARCOMERE FUSION REPORTER hiPSC-CMs
Once the genome-edited, GFP-sarcomere fusion reporter hiPSC-CMs have been produced, they can be utilized in a variety of ways. Because the cells glow green upon cardiomyocyte differentiation, they can be used to visually and temporally examine the stages of human cardiac development in an in-vitro setting. Also, because the GFP signal is specific to cardiomyocytes, it can be used to parse the hiPSC-CMs from other cell lineages. Finally, these cells can be used to track individual sarcomeres in live, contracting human cardiomyocytes, which to our knowledge has not been previously possible. This modality allows for phenotypic interrogation of human genetic mutations in genes encoding for sarcomeric proteins, accomplished by introducing patient-specific mutations in hiPSCs with the GFP-sarcomere reporter background. In this protocol, we present a method for non-invasive evaluation of sarcomere contractility in genome-edited, GFP-sarcomere fusion reporter hiPSC-CMs. Specifically, we demonstrate contractility analysis using a custom hiPSC-CM line with a Z disk-localized, N-terminus GFP tag attached to the sarcomeric protein titin (see Figure 3 for details).
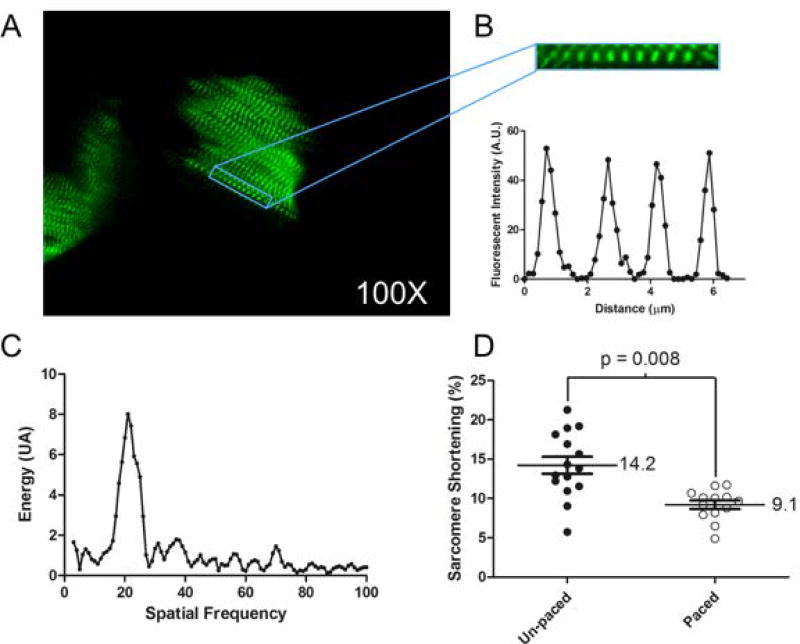
Figure 3.
Outline of sarcomeric tracking for cellular shortening in beating titin-GFP sarcomere reporter hiPSC-CMs. A) Representative 100x frame from an acquired video capture of paced day 30 hiPSC-CMs. See Video 1 for full video. B) Enlarged myofibril running through a single hiPSC-CM, with a representative plot profile of the GFP signal intensity across the N-terminus titin-GFP tag located at the Z disks of each sarcomere. Three sarcomeres shown. C) Fast Fourier Transform (FFT) of enlarged myofibril indicating the signal (large peak) to be tracked through each frame of the vide sequence. D) Measurements of sarcomere shortening for un-paced and 1 Hz paced hiPSC-CMs using this method of acquisition and sarcomere tracking.
Materials
Genome-edited, GFP-sarcomere fusion hiPSC-CMs (from previous steps)
Matrigel (hESC-qualified; BD Sciences, cat. no. 354277)
RPMI 1640 medium (Thermo Fisher Scientific, cat. no. 11835055)
B27 Supplement with Insulin (Thermo Fisher Scientific, cat. no. 17504-044)
6-well tissue culture–treated plates
Standard cell culture laminar flow hood
Rho kinase inhibitor (ROCKi; Tocris cat. no. 1254)
TrypLE™ Express Enzyme (Thermo Fisher Scientific, cat. no. 12605010)
Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, cat. no. 10438026)
Cell culture centrifuge with optional plate adapter
Phosphate-buffered saline (PBS; Life Technologies, cat. no. 20012-050)
Steriflip-GP, 0.22 µm (Millipore, cat. no. SCGP00525)
12-well glass bottom imaging plate (MatTek Part no. P12G-1.0-10-F)
15- and 50-ml conical centrifuge tubes (e.g., BD Falcon)
Fluorescence plate microscope with 40x, 60x, or 100x objectives
SarCoptiM ImageJ analysis software (http://pccv.univ-tours.fr/ImageJ/SarcOptiM/)
Replate GFP-sarcomere fusion hiPSC-CMs into imaging wells
-
Begin with a purified population of genome-edited, GFP-sarcomere fusion reporter hiPSC-CMs. We recommend using hiPSC-CMs that are no older than day 20 for the replating process.
We have noticed that hiPSC-CM dissociation is easier in younger cells.
To re-plate hiPSC-CMs for imaging, start with coating a glass-bottom, imaging-optimized 12-well plate with 1:400 Matrigel in RPMI 1640 medium. Allow this plate to coat for at least 1 hour at 37°C.
While the glass-bottom imaging plate is being coated with Matrigel, prepare hiPSC-CM passaging medium and TrypLE dissociation enzyme. Warm 2 mL TrypLE at 37°C for 1 hour.
Create a solution of RPMI 1640 medium (with B27 supplement with insulin) with 20% fetal bovine serum. Add 10 µM rho kinase inhibitor to this solution, as it improves hiPSC-CM survival post-dissociation (Braam, Nauw, Ward-van Oostwaard, Mummery, & Passier, 2010). Sterile filter this solution using a 50 mL Steriflip-GP with 0.22 µM filter. This filtered solution will be the cardiomyocyte passaging medium to facilitate hiPSC-CM plating after passaging. Warm cardiomyocyte passaging medium at 37°C for 1 hour.
Select a well of genome-edited, GFP-sarcomere fusion hiPSC-CMs for passaging. Wash well with 1 mL PBS three times. Aspirate PBS.
Add 1 mL pre-warmed TrypLE onto the selected well of hiPSC-CMs and leave cells at 37°C in a cell culture incubator for 5 minutes.
Post-incubation, add 1 mL of pre-warmed cardiomyocyte passaging medium to deactivate TrypLE. Immediately manually dissociate hiPSC-CMs from cell culture plate with a 1000 µL pipetman. Vigorously pipette the hiPSC-CMs for approximately 50 repetitions to ensure hiPSC-CMs are in a single-cell format, more repetitions may be necessary for older cells.
Take the dissociated cardiomyocytes and add to a 15-mL conical tube. Centrifuge the tube at 200 x g for 5 minutes to pellet the cardiomyocytes. Aspirate the supernatant and resuspend the hiPSC-CMs in 1 mL pre-warmed cardiomyocyte passaging medium. Vigorously pipette and resuspend the hiPSC-CM pellet to ensure that hiPSC-CMs are in a single-cell format.
Aspirate Matrigel solution from 12-well imaging plate. Add 1 mL of cardiomyocyte passaging medium to each well. This plate will receive the newly-passaged hiPSC-CMs.
-
Take the resuspended hiPSC-CMs and plate them in a serial dilution in the glass-bottom imaging plate. We recommend plating at a dilution of 1:2 down to 1:512, with each well receiving half as many hiPSC-CMs as the well before it. For imaging, individual, isolated hiPSC-CMs are preferred, however this plating density should be optimized to the user’s needs.
It must be noted cell survival post-passaging can be affected by high dilution. Cells replated at higher confluency tend to survive better after passaging.
Return the replated hiPSC-CMs to the cell culture incubator at 37°C. Do not disturb these cells for 48 hours post-plating.
After 48 hours, cardiomyocyte passaging medium can be replaced with RPMI 1640 (with B27 with insulin). Continue replacing RPMI 1640 (with B27 with insulin) every 48 hours. Cardiomyocytes will tend to spread out over the course of cell culture. Image cells at ~day 30 post-differentiation under a fluorescent microscope.
Image GFP-sarcomere fusion hiPSC-CMs and evaluate sarcomere dynamics
Approximately 10 days after replating GFP-sarcomere fusion hiPSC-CMs (ideally at day 30 post-differentiation), prepare for imaging, a 100x oil immersion objective provides optimal magnification and quality for sarcomere tracking. 10 days post-plating, replated cells become flatter, improving sarcomere alignment in the tight focal plane of the 100x objective. This is important for the sarcomere tracking program to best resolve the z-disk periodicity. Use a microscope that will permit imaging of a 12-well plate and be able to maintain hiPSC-CMs at 37°C.
During imaging, place plates for contractile analysis in a temperature and gas controlled chamber (37°C, 5% CO2 and 20% O2). Allow plates to equilibrate temperature for 5 minutes before making measurements.
One can apply a pacing probe to the well to be imaged, this will synchronize the beating of the cells. We have found that a pacing frequency of 1 Hz is maintainable throughout experimentation and mimics the approximate 60 beats per minute of an adult heart. The electrode delivers 20V, which reliably paces all hiPSC-CMs in a well. For imaging, we used, a Keyence BZX microscope with a Nikon 100x 1.4NA oil immersion objective. This objective allows a shallow focal plane to minimize background fluorescence, while providing a high resolution for tracking the Z disk-localized fluorescent signal (Figures 3A and 3B). Similar objectives can be used on other microscopes. We used a pre-built Keyence GFP filter cube to acquire images. Using this methodology, frame acquisition can be as great as 40 frames per second.
-
Once image acquisition is finished, frames can be analyzed using a free ImageJ plug-in called SarcOptiM (Pasqualin et al., 2016). SarcOptiM uses a Fast Fourier Transform (FFT) to localize the Z disk signal and track sarcomere lengths over the course of the experimental acquisition (Figure 3C). This plug-in process provides a time course of sarcomere length over the hiPSC-CM contractile cycle, which can be saved as .txt files for data plotting.
We find that as well as synchronizing cell contraction pacing of hiPSC-CMs also reduces heterogeneity of contractility, in comparison to spontaneous contraction. This reduction in heterogeneity of contractile measures across cells should increase assay sensitivity providing a method to distinguish changes in cellular contractility by genetic mutation mimicking human disease. It is also evident that upon pacing on average contractility is reduced (Figure 3D).
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps, unless otherwise specified. Shelf life for all cell culture media is 1 month at 4°C.
Matrigel-diluted medium
Combine Matrigel (hESC-qualified; BD Sciences, cat. no. 354277) with DMEM/F12 medium (Thermo Fisher Scientific, cat. no. 11320033) in a dilution of 1:400 to coat plates for hiPSC and hiPSC-CM maintenance. Thaw Matrigel on ice at 4°C or in a refrigerator overnight. Dilute Matrigel in cold DMEM/F12 quickly, using cooled pipette tips, to prevent premature solidification. Once plates are coated with Matrigel dilution, store them at 37°C until needed. When ready for use, aspirate old Matrigel dilution from plates and add fresh culture media specific to hiPSCs or hiPSC-CMs.
hiPSC maintenance medium (for daily hiPSC culture)
mTeSR1 feeder-free pluripotent stem cell maintenance medium (StemCell Technologies, cat. no. 05850)
hiPSC passaging medium (for passaging hiPSCs)
mTeSR1 feeder-free pluripotent stem cell maintenance medium containing:
10 µM rho kinase inhibitor
hiPSC-CM differentiation medium (days 0–7 of hiPSC-CM differentiation)
RPMI 1640 medium containing:
1x B27 supplement without insulin
Supplement as necessary with Wnt-regulating small molecules (see Figure 2).
hiPSC-CM maintenance medium (after day 7 of hiPSC-CM differentiation)
RPMI 1640 medium containing:
1x B27 supplement with insulin
hiPSC-CM purification medium (for metabolic selection of differentiated hiPSC-CMs)
RPMI 1640 medium without glucose, containing:
1x B27 supplement with insulin
hiPSC-CM passaging medium (for dissociation and replating of differentiated hiPSC-CMs)
RPMI 1640 medium containing:
1x B27 supplement with insulin
20% fetal bovine serum
10 µM rho kinase inhibitor
COMMENTARY
Background Information
With rapid improvements in human pluripotent stem cell differentiation protocols, cell types such as human cardiomyocytes can now be mass-produced in vitro (Burridge et al., 2015). For the field of cardiac regenerative medicine, the possibility of producing hiPSC-CMs in large quantities has unlocked new opportunities in disease modeling. Genetically-influenced cardiac disorders such as hereditary long-QT syndrome, associated with abnormal heart rhythms, and various cardiomyopathies, associated with thickening or thinning of the heart muscle, can be recapitulated at the cellular level in a dish (Hinson et al., 2015; Itzhaki et al., 2011). Additionally, with the advent of hiPSC-CM technology, drugs can also be screened for their cardiovascular efficacy and toxicity on a patient-specific level, something that was not possible previously due to a scarcity of human cardiac tissue for pharmacological testing (Sharma et al., 2017). Finally, CRISPR/Cas9-mediated genome editing can be readily applied to hiPSCs to make disease mimicking cell lines in the GFP-sarcomere fusion hiPSC-CMs. The GFP reporter line acts as the background in which to engineer subsequent patient specific contractile mutations. These lines are then used to directly track contractility in the fundamental contractile unit of the cell, the sarcomere. This is a far more precise and sensitive measure of contractility in this juvenile cell state. This approach may overcome the problem of sensitivity in the contractile measures from hiPSC-CMs that has slowed progress in the field. This cell line could potentially play an important role in providing a high-throughput pipeline to validate the importance of relevant human genetic variants and their involvement in disease penetrance, thus shedding light on fundamental mechanisms of disease that affect cellular contractility in genetic cardiomyopathies.
Critical Parameters and Troubleshooting
Differentiation and functional characterization of fluorescent sarcomere reporter hiPSC-CMs is possible within one month, if all aspects of this protocol are successful. Here, we list additional parameters that may facilitate the differentiation and analysis of these multipurpose hiPSC-CMs and troubleshooting that may be helpful if problems arise.
hiPSC-CM differentiation of fluorescent sarcomere reporter hiPSCs
For sarcomere contractility assessments, we recommend working with a fluorescent reporter hiPSC-CM cell line with a fluorescently-tagged protein in the Z disk of the sarcomere, as this allows for easy identification of individual sarcomeric units. It is best to use a protein in general that is less likely to be functionally perturbed by the GFP tag and is localized in a specific area of the sarcomere. The sarcomeric gene of interest should also be highly expressed in hiPSC-CMs, preferably beginning at day 7–8 post-differentiation. It may be necessary to conduct qPCR or RNA-sequencing to determine the most highly-expressed sarcomeric genes in differentiated cardiomyocytes. Fluorescent sarcomere reporter hiPSCs can be differentiated into contracting cardiomyocytes within a matter of weeks. However, this differentiation process can be somewhat variable, especially within the first week of differentiation. We recommend testing a range of starting hiPSC confluencies and concentrations of CHIR99021, the GSK3β inhibitor added during the first day of the protocol, to obtain optimal hiPSC-CM differentiation. For example, we have found that hiPSC starting confluencies from 80–100% and CHIR99021 concentrations from 6–24 µM could be effective. An advantage of the GFP-sarcomere reporter hiPSC-CMs is that by day 7–10 of differentiation, you should be able to visibly identify areas of green cells that represent early cardiomyocytes. If no green signal is detected by day 10 of differentiation, start over with a new round of differentiation using different starting confluencies or CHIR99021 concentration. We have also seen that a large amount of cardiomyocyte cell death can occur during the metabolic selection and glucose deprivation step. If this happens, change back to RPMI 1640 media containing glucose with B27 (with insulin) and continue culture. In contrast, we find that in very low efficiency differentiations where cardiomyocytes constitute less than 10% of the total population, metabolic selection may be less effective, and less purification may occur. If efficient hiPSC-CM differentiation is still not obtained, we recommend using fresh Wnt-modulating small molecules and media, properly seeding hiPSCs during monolayer culture through even cell distribution after passaging, preventing hiPSCs from becoming overconfluent during cell maintenance, and scaling up the number of hiPSC wells differentiated to increase the number of technical replicates.
Functional assessment of sarcomere dynamics in GFP-sarcomere reporter hiPSC-CMs
After purified GFP-sarcomere reporter hiPSC-CMs have been created, they can be replated for downstream functional analysis of sarcomere dynamics. These cells can be non-invasively imaged to evaluate how individual sarcomeres shorten and lengthen during hiPSC-CM contractility in both wild type and patient-specific mutant cell lines. Replating purified hiPSC-CMs can be a variable process in terms of cell survival post-plating. If hiPSC-CMs do not survive well after plating, increase the concentration of FBS up to 40% in RPMI 1640 with B27 (with insulin) and 10 µM rho kinase inhibitor. We have also observed that replating cells at higher confluencies tends to improve cell survival. After replating, allow cells to spread out to reduce the level of three-dimensionality for optimal imaging. This allows for a greater proportion of the myofibrilar components to lie within the tight focal plane of the objective used to image these cells, enabling better resolution for tracking sarcomeric shortening as well as eliminating out-of-focus background fluorescence. Using a phenol-red free version of the RPMI 1640 cell culture medium may further reduce background fluorescence. In hiPSC-CMs, sarcomeres can be irregularly arranged, unlike the parallel sarcomeres found in adult cardiomyocytes, so we recommend taking multiple sarcomere measurements per cell. Overlapping sarcomeres can also interfere with the downstream contractility analysis, so we recommend taking videos of multiple cells. Sarcomere tracking by SarCoptiM becomes most robust if a linear track of 10+ sarcomeres can be traced, this should be considered during movie acquisition. We recommend pacing cells at 1 Hz for contractility analysis to provide the best sensitivity in the measures.
Interpreting Results
Differentiation of genome-edited cells into hiPSC-CMs
Variation in differentiation capacity exists between hiPSC lines. Unique hiPSC lines will differentiate best at different confluencies and concentrations of CHIR99021, so we recommend conducting pilot experiments to determine the optimal starting confluency (typically between 80–100%) and CHIR99021 concentration (typically between 6–12 µM for 1 day). Cells should begin beating and expressing GFP in the sarcomeres as early as day 7 post-differentiation, with GFP signal strengthening over the course of differentiation. Note that during subsequent metabolic selection process, cardiomyocytes may die off as well, so monitor hiPSC-CMs daily during the selection process. If hiPSC-CMs begin dying off, end the metabolic selection process.
Functional assessment of sarcomere dynamics in GFP-sarcomere fusion hiPSC-CMs
When replating GFP-sarcomere fusion hiPSC-CMs for functional analysis, expect up to 25% cell loss. Sarcomeres will likely be more visible if GFP-tagged proteins are present in the Z disk of the hiPSC-CMs. While pacing a well of hiPSC-CMs, some cells may not respond to pacing, which is expected. Expect to obtain anywhere between 1–10 sarcomere contraction measurements per cardiomyocyte. As hiPSC-CMs tend to be more irregularly-shaped than rod-shaped adult cardiomyocytes, sarcomere orientation may be radial. However, GFP signal should be highly visible at 100x magnification, at which point individual sarcomeres can be discerned. Localizing contractility measures on linear myofibrils running through hiPSC-CMs allows for a reliable measure of linear contraction, more typical to contraction one would expect to observe in an adult cardiomyocyte. This situation, where individual sarcomeres can be distinguished at high resolution, is usually the best for downstream contractility measurements. Additionally, it is possible that introduction of a fluorescent sarcomere tag could alter hiPSC-CM contractility by sterically altering the function of the sarcomeric protein that it is attached to. Thus, it may be advantageous to use a custom hiPSC line where a large sarcomere protein is fluorescently tagged to reduce the percentage of the total fusion protein that is made up of GFP. Alternatively, it may be possible to computationally identify sarcomere protein domains that are less susceptible to steric hindrance when tagged by GFP.
Time Considerations
Differentiating and analyzing GFP-sarcomere fusion reporter hiPSC-CMs will require a considerable amount of time and effort in cell culture. Work involving hiPSCs requires daily maintenance and replenishment of pluripotency-sustaining media such as mTeSR1. Similarly, differentiation of hiPSCs into cardiomyocytes is a multi-day protocol with regular media exchanges required. See figures for details of time constraints for differentiation protocols and contractility analysis.
Supplementary Material
Video 1: Video of beating day 30 post-differentiation hiPSC-CMs exhibiting an N-terminus titin-GFP fusion tag. The hiPSC-CMs spontaneously contract and express GFP beginning at day 7–10 post-differentiation. Individual sarcomere Z disks can be discerned by the GFP signal. Cells were recorded using a 100x objective.
SIGNIFICANCE STATEMENT.
Human induced pluripotent stem cellss (hiPSC) can be differentiated into a variety of different cell types; here we describe a procedure for differentiating iPSC into cardiomyocytes (hiPSC-CMs). These hiPSC-CMs provide useful models for studying the development and dysfunction of the human heart. Functionally hiPSC-CMs are juvenile cardiomyocytes, or beating heart muscle cells, which can be mass-produced. Additionally, CRISPR/Cas9 genome editing technology has allowed human pluripotent stem cells to be rapidly modified, to generate hiPSC-CMs with fluorescent tags attached to sarcomere proteins (see CPHG UNIT 21.12). These tags can be used to track contraction at the level of the single sarcomere, providing measures of sarcomere length throughout the contractile cycle and cellular shortening, thus potentially enabling new insights into human cardiovascular biology.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH) Grants 1R01HL080494 and 1R01HL084553 (to C.E.S.), the Foundation Leducq (C.E.S.), the Howard Hughes Medical Institute (C.E.S.), the NIH NHLBI Cardiovascular Development Consortium (CVDC) Postdoctoral Research Fellowship (A.S.), Wellcome Trust Grant 206466/Z/17/Z (C.N.T.), and a Sarnoff Cardiovascular Research Foundation Fellowship (A.C.G).
LITERATURE CITED
- Braam SR, Nauw R, Ward-van Oostwaard D, Mummery C, Passier R. Inhibition of ROCK improves survival of human embryonic stem cell-derived cardiomyocytes after dissociation. Ann N Y Acad Sci. 2010;1188:52–57. doi: 10.1111/j.1749-6632.2009.05083.x. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Sharma A, Wu JC. Genetic and Epigenetic Regulation of Human Cardiac Reprogramming and Differentiation in Regenerative Medicine. Annu Rev Genet. 2015;49:461–484. doi: 10.1146/annurev-genet-112414-054911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Seidman CE. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108(3):407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualin C, Gannier F, Yu A, Malecot CO, Bredeloux P, Maupoil V. SarcOptiM for ImageJ: high-frequency online sarcomere length computing on stimulated cardiomyocytes. Am J Physiol Cell Physiol. 2016;311(2):C277–283. doi: 10.1152/ajpcell.00094.2016. [DOI] [PubMed] [Google Scholar]
- Robertson C, Tran DD, George SC. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells. 2013;31(5):829–837. doi: 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Wu JC. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Science translational medicine. 2017;9(377) doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circulation research. 2014;115(6):556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Wu JC, Wu SM. Induced pluripotent stem cell-derived cardiomyocytes for cardiovascular disease modeling and drug screening. Stem Cell Res Ther. 2013;4(6):150. doi: 10.1186/scrt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Fukuda K. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1: Video of beating day 30 post-differentiation hiPSC-CMs exhibiting an N-terminus titin-GFP fusion tag. The hiPSC-CMs spontaneously contract and express GFP beginning at day 7–10 post-differentiation. Individual sarcomere Z disks can be discerned by the GFP signal. Cells were recorded using a 100x objective.