Abstract
Background
epigenomes can be influenced by environmental factors leading to the development of diseases.
Objective
To investigate the influence of sun exposure on global DNA methylation and hydroxymethylation status and at specific sites of the miR-9-1, miR-9-3 and MTHFR genes in skin samples of subjects with no history of skin diseases.
Methods
Skin samples were obtained by punch on sun-exposed and sun-protected arm areas from 24 corpses of 16-89 years of age. Genomic DNA was extracted from skin samples that were ranked according to Fitzpatrick's criteria as light, moderate, and dark brown. Global DNA methylation and hydroxymethylation and DNA methylation analyses at specific sites were performed using ELISA and MSP, respectively.
Results
No significant differences in global DNA methylation and hydroxymethylation levels were found among the skin areas, skin types, or age. However, gender-related differences were detected, where women showed higher methylation levels. Global DNA methylation levels were higher than hydroxymethylation levels, and the levels of these DNA modifications correlated in skin tissue. For specific sites, no differences among the areas were detected. Additional analyses showed no differences in the methylation status when age, gender, and skin type were considered; however, the methylation status of the miR-9-1 gene seems to be gender related.
Study limitations
there was no separation of dermis and epidermis and low sample size.
Conclusion
sun exposure does not induce changes in the DNA methylation and hydroxymethylation status or in miR-9-1, miR-9-3 and MTHFR genes for the studied skin types.
Keywords: DNA Methylation, Epigenomics, MicroRNAs, Solar radiation, Skin
INTRODUCTION
Continuous exposure to ultraviolet (UV) radiation leads to lesions in the nuclear and mitochondrial genomic DNA in human skin cells, as well as to the generation of reactive oxygen species, thereby generating various deleterious effects, such as erythema, photoaging, immunosuppression, and cancer.1 Given that skin cancer is associated with epigenetic changes, a current concern of the scientific community is how exposure to solar radiation can affect our epigenome.2 However, studies that address the effects of solar radiation on our epigenome remain scarce and controversial.
Epigenetic changes represent the interface between the genome and the environment, and they are heritable and reversible, and do not alter the primary sequence of the DNA.3 DNA methylation is one of the most common epigenetic modifications, and it is characterised by being able to balance stability with plasticity and tissue-specificity, and potentially varies with age and environmental exposure.4,5 It consists of a covalent modification of DNA in which a methyl group (CH3) is transferred from S-adenosylmethionine (SAM) to a cytosine that precedes a guanine (CpG dinucleotide). It is catalysed by a family of specific enzymes called DNA methyltransferases (DNMTs). Recent studies have reported that aberrant methylation profiles of gene promoters represent a major mechanism associated with gene inactivation, contributing to the initiation and progression of various diseases, including skin cancer.6,7
In addition to DNA methylation, other epigenetic mechanisms are responsible for the integrity and regulation of gene expression such as: DNA hydroxymethylation; methylation, phosphorylation and acetylation of the N-terminal tails of histone proteins and non-coding RNAs.6,8 5-hydroxymethylcytosine (5-hmC) is generated by the oxidation of 5-methylcytosine (5-mC) through the action of TET (Ten-Eleven translocation) enzymes. 5-hmC is responsible for regulating gene transcription over short and long terms for the maintenance of unmethylated DNA in gene promoters, and they are considered one of the key molecules involved in the process of DNA demethylation.9
Cells have developed a system that involves a group of small non-coding endogenous RNAs (known as microRNAs) to negatively regulate gene expression at the post-transcriptional level by degrading or blocking mRNA translation.10 It is estimated that these molecules regulate the expression of more than 50% of human genes, and it was estimated that approximately 10% of miRNAs are epigenetically regulated by DNA methylation.11,12 It was reported that microR-NAs can act as oncogenes or tumour suppressors, depending on the cell type in which they are expressed.13 The epigenetic regulation of microRNAs has been considered a major bet of current research due to their important roles in the silencing of genes involved in cell proliferation, differentiation, apoptosis and carcinogenesis.14,15
In particular, the family of microRNAs called miR-9, presents three independent loci in DNA, including miR-9-1 located on chromosome 1q22, miR-9-2 located on chromosome 5q14.3 and miR9-3 located on chromosome 15q26. They are an interesting family, because their association with skin cancer has been demonstrated.16
The methylene tetrahydrofolate reductase (MTHFR) gene is located on chromosome 1 and encodes the methylenetetrahydrofolate reductase enzyme involved in folate metabolism. MTHFR is essential for metabolic pathways generating S-adenosylmethionine (SAM), which is the most important methyl donor for DNA.17 Most of the studies focusing on MTHFR have considered polymorphisms, some of which have shown an association with DNA methylation changes.18
The mechanisms governing how external factors affect the methylation profiles of the miR and MTHFR genes have rarely been explored. However, studies have shown that a variety of environmental factors, including solar radiation, can influence microRNA expression, suggesting that they are valuable novel biomarkers for exposure.19,20 For the MTHFR gene, it was shown that exposure to arsenic can change their methylation profile.21
In this context, the aim of this study was to investigate the influence of sun exposure on global DNA methylation and hydroxymethylation, as well as at specific sites of the miR-9-1, miR-9-3 and MTFHR genes, in subjects with no history of skin disease. The basis of the study was that changes in epigenetic profiles could be used as markers of changes related to sun exposure.
METHODS
Ethics statement and subject population
This study was performed in accordance with the Ethics Committee in Research from Universidade Federal da Paraíba, Brazil (protocol number 430/2011), and with the Helsinki Declaration of 1975. Twenty-four corpses of both genders of individuals ranging from 16-89 years of age at the time of their death were included. The corpses were obtained from the Service of Death Investigation (SDI-João Pessoa, PB, Brazil). Written informed consent for participation was obtained from all families. The demographic data and general health of each individual were obtained via SDI records and transcripts to our database and did not include individuals with smoking habits or a medical history of skin diseases. Skin samples were collected and ranked according to Fitzpatrick's criteria by two observers and confirmed by skin type declaration in the record of each individual.22
Sample collection and genomic DNA extraction
Skin samples were collected from the 24 corpses by punch (5mm diameter) from the outer forearm (sun-exposed area) and inner arm (sun-protected area) of each corpse, up to 10 hours after death. Immediately after removal, the samples (epidermis and dermis) were stored in a tube containing 800µL of RNAholder (Bioagency, São Paulo, SP, Brazil) and frozen at −20ºC until DNA extraction. After, the genomic DNA was purified using the TRIZOL reagent (Invitrogen, Carlsband, CA, USA) following the manufacturer's recommendation and using a tissue homogeniser. DNA quantification was performed using a NanoDrop spectrophotometer (Thermo Scientific). The samples were then frozen at −20ºC until further analysis.
Global DNA methylation and hydroxymethylation quantification
Global DNA methylation and hydroxymethylation levels were assessed with an ELISA-based technique. Briefly, wells were coated with 50ng of DNA diluted in Coating solution. The methylated and hydroxymethylated fraction of DNA was detected separately, using the respective capture monoclonal antibodies [5-mC-1:125 (Merck Millipore) and 5-hmC-1:2000 (Epigentek)]. Coating, Wash, Stop and Blocking Solutions and detection antibodies were provided by the Protein Detector Kit™ ELISA (KPL Inc.), and used according to the manufacturer's recommendations. The optical density was read at 405nm using a microplate reader Multiskan GO ™ Microplate Spectrophotometer (Thermo Fisher Scientific Inc.). The amount of methylated DNA is proportional to the OD intensity measured. The relative percentage of global DNA methylation and hydroxymethylation was derived by subtracting the optical density of the positive controls for both studied modifications - methylated or hydroxymethylated DNA (5-Methylcytosine & 5-hydroxymethylcytosine DNA Standard Set-Zymo Research) - on the optical density of each sample.23 All analyses were performed in duplicate and the average was reported.
DNA methylation analysis of the promoters of the miR-91, miR-9-3 and MTHFR genes
DNA methylation analysis of specific sites was performed using methylation-specific PCR (MSP). Bisulfite modification of genomic DNA (500ng) was performed with the CpGenome™ Turbo Bisulfite Modification Kit (Merck Millipore), according to the manufacturer's instructions. Each MSP reaction incorporated 100ng of bisulfite-modified DNA, 1µL (10µM) of each primer, and 1 × Go Taq Hot Start Green Master Mix (Promega Corporations, Madison, WI, USA) in a final reaction of 25µL. Fragments were amplified with specific primers for either methylated or unmethylated targets, as previously described.24,25 (Table 1). Methylated DNA (Methylated Control DNA, Sigma Aldrich) and unmethylated DNA (CpGenome Universal Unmethylated DNA, Merck Millipore) were modified, as previously described, and amplified by PCR as control reactions with primers for the methylated and unmethylated conditions, respectively. All reactions were performed in duplicate, and amplified PCR samples (10µL) were loaded in 6% polyacrylamide gels and subjected to electrophoresis. DNA bands were detected after silver staining.
Table 1.
| Primer (5'-3') F/R | Fragment Size (bp) | Anneling Temperature (°C) | Cycles | |
|---|---|---|---|---|
| 110 | 55 (40 sec) | 35 | ||
| miR-9-1 | tataagggtttcgtttcgtttttaac (M) | |||
| acccgtcaccgaaaaaaa (M) | ||||
| 110 | 60 (40 sec) | 35 | ||
| ggttttgttttgtttttaatgt (U) | ||||
| caaccaaaaccctaccttcaac (U) | ||||
| 116 | 58 (1 min) | 35 | ||
| miR-9-3 | attggtcgatttttggattgac (M) | |||
| cgcttaaaaaacctcgaacg (M) | ||||
| 116 | 55 (1 min) | 35 | ||
| gattggttgatttttggattgat (U) | ||||
| caaaacacttaaaaaacctcaaaca (U) | ||||
| 180 | 58 (40 sec) | 40 | ||
| MTHFR | tagatttaggtaggtgaagtagggtagac(M) | |||
| gaaaaactaataaaaaacggacgaa(M) | ||||
| 180 | 58 (40 sec) | 40 | ||
| tttaggtatgtgaaatagggtagatgt (U) | ||||
| caaaaaactaataaaaaaccaacaaa (U) | ||||
* F-Forward, R-Reverse, M-methylated, U-unmethylated, MSP- Methylation Specific PCR.
Statistical analysis
To evaluate the differences in DNA methylation among sun-exposed and sun-protected skin areas, a paired non-parametric McNemar test was used for specific methylation and a paired t-test was used for global methylation and hydroxymethylation. Differences between methylation and hydroxymethylation levels in the skin were analysed with a t-test, while a Pearson correlation test was used for correlation analysis. Additional tests to evaluate the influence on age, gender and skin type were performed using t-tests and ANOVA. All analyses were performed using Bioestat 5.0 software (Pará-Brazil) at a 5% level of significance.
RESULTS
Demographic and skin type data are shown in table 2.
Table 2.
Demographic and skin type data in studied population
| DNA modification /Gene | n | Age (Mean ± SD) | Gender (Male/Female) | Skin Type (number of individuals for each skin type) |
|---|---|---|---|---|
| Global DNA methylation and hydroxymethylation | 24 | 57,9 ± 21,6 | 11/13 | 09-type III; 10-type IV; 05-type V |
| miR-9-1 | 17 | 57,8 ± 20,9 | 7/10 | 06-type III; 07-type IV; 04-type V |
| miR-9-3 | 21 | 56,1 ± 21,3 | 10/11 | 08-type III; 09-type IV; 04-type V |
| MTHFR | 14 | 58,5 ± 21,8 | 5/9 | 06-type III; 04-type IV; 04-type V |
Type III: light brown, type IV: moderate brown and type V: dark brown. We collected samples from 24 corpses, but some samples are not available for amplification or showed no fragments after PCR amplification, and were excluded from the study.
Global DNA methylation and hydroxymethylation
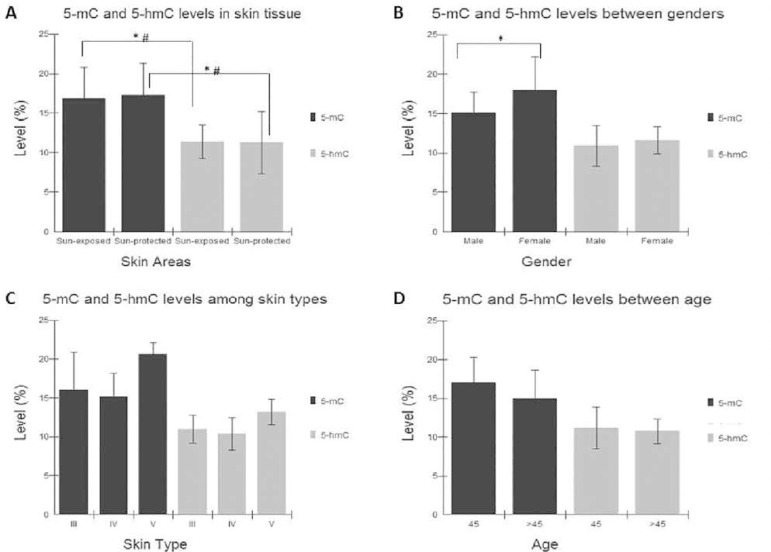
Quantification of 5-mC and 5-hmC levels showed no significant difference between the sun-exposed and sun-protected skin areas (5-mC = 16.9 % and 17.3%; 5-hmC = 11.4% and 11.3%; p > 0.05). However, 5-mC levels were higher in comparison to 5-hmC levels (p < 0.0001) in skin cells. In addition, a significant correlation was found between %methylation and %hydroxymethylation (p < 0.0001) (Figure 1A). Females presented a higher methylation level (18.4%) in comparison to that of men (15.1%) (p = 0.03), while no differences were observed for DNA hydroxymethylation (p > 0.05) (Figure 1B). In relation to age and skin type, no differences were observed for both DNA methylation and hydroxymethylation (p > 0.05) (Figure 1C and D).
Figure 1.
Global DNA methylation and hydroxymethylation levels in skin tissues. Data are presented as mean and standard deviation. A - Methylation and hydroxymethylation among sun-exposed and sun-protected skin areas (p>0.05; Paired t test). Methylation versus hydroxymethylation in the skin (*p<0.0001; T test). Correlation of methylation and hydroxymethylation (#p<0.0001; Pearson Correlation). B - Global DNA methylation and hydroxymethylation between genders (p=0.03; T test). C - Global DNA methylation and hydroxymethylation among skin types (p>0.05; ANOVA). D - Global DNA methylation and hydroxymethylation between age (p>0.05; T test). n=24, SE - sun-exposed, SP - sun-protected, III-light brown, IV- moderate brown, V-dark brown.
DNA methylation at specific sites
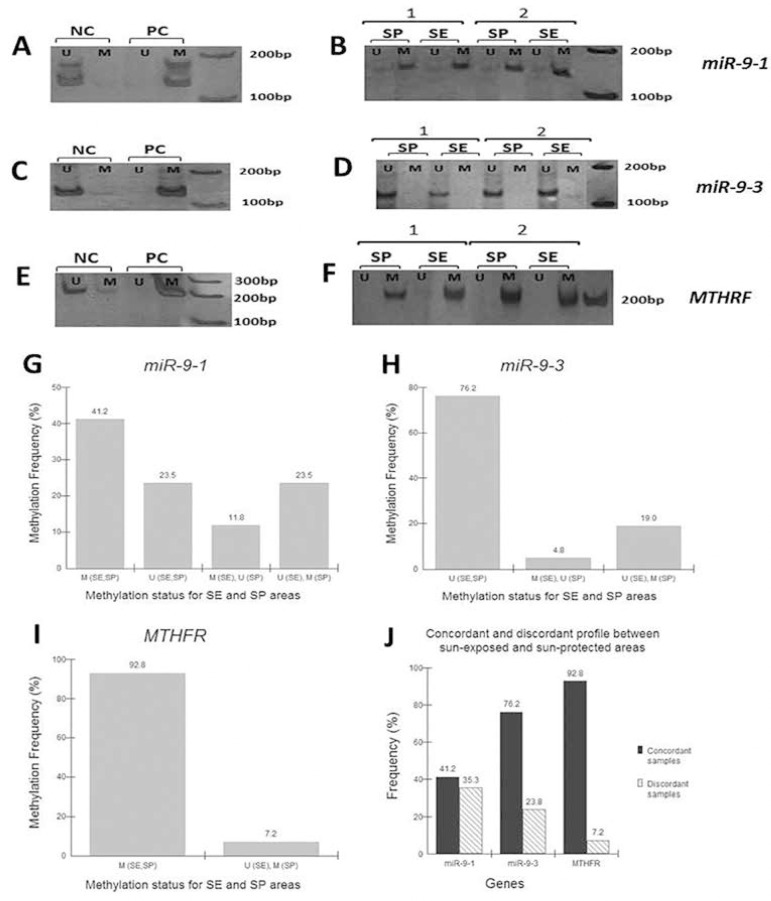
miR-9-1- no significant differences were observed between areas, given that, out of a total of 17 samples, only six (35.3%) showed differences between sun-exposed and sun-protected areas [2 methylated in SE (11.8%) and 4 methylated in SP (23.5%)], and most (64.7%) had the same condition for both areas [7 methylated (41.2%) and 4 unmethylated (23.5%); p = 0.68] (Figure 2A, B and G).
Figure 2.
DNA methylation analysis of miR-9-, miR-9-3 and MTHFR promoters of skin tissues. A - Bands of positive (methylated) and negative (unmethylated) control DNA, showing specific amplification for methylated and unmethylated conditions for miR-9-1. B - Bands of two representative samples (1 and 2) obtained after polymerase chain reaction (110 bp). C - Bands of positive (methylated) and negative (unmethylated) control DNA, showing specific amplification for methylated and unmethylated conditions for miR-9-3. D - Bands of two representative samples (1 and 2) obtained after polymerase chain reaction (116 bp). E - Bands of positive (methylated) and negative (unmethylated) control DNA, showing specific amplification for methylated and unmethylated conditions for MTHFR. F - Bands of two representative samples obtained after polymerase chain reaction (180 bp). G - Methylation frequency for miR9-1 (n=17, p>0.05; McNemar). H - Methylation frequency for miR-9-3 (n=21, p>0.05; McNemar). I - Methylation frequency for MTHFR (n=14, p>0.05; McNemar). J - Methylation frequency of concordant samples (samples with the most frequent profile in both sun-exposed and sun-protected areas: methylated for miR-9-1 and MTHFR genes and unmethylated for miR-9-3 gene) and discordant samples (samples with different profile in sun-exposed and sun-protected areas). NC-negative control, PC-positive control, M -methylated, U -unmethylated, SE -sun-exposed, SP - sun-protected.
miR-9-3- no significant differences were observed between areas, given that, out of a total of 21 samples, only five (23.8%) showed differences between sun-exposed and sun-protected areas [1 methylated in SE (4.8%) and 4 methylated in SP (19%)], and most had the same condition for both areas [16 unmethylated (76.2%) (p = 0.37)] (Fig. 2C, D and H).
MTHFR- no significant differences were found between areas, given that, out of a total of 14 samples, only one (7.2%) showed differences between sun-exposed and sun-protected areas (1 methylated in SP and unmethylated in SE), and most had the same condition for both areas [13 methylated (92.8%) (p = 1)] (Fig. 2E, F and I).
Figure 2 shows the analysis of the three genes, emphasizing the frequency of discordant samples (samples with different profiles in sun-exposed and sun-protected areas) and concordant samples (samples with same profile in sun-exposed and sun-protected areas).
DISCUSSION
Although UV radiation is crucial for the synthesis of vitamin D in the skin and other physiological aspects of human life, it has currently been considered one of the most potent carcinogens to which the human population is exposed.26 Furthermore, the intensity of ultraviolet UV sun radiation on the Earth's surface has been consistently increasing and correlates with an increased incidence of skin cancer.27 Therefore, the detection of genomic changes in non-tumoural cells may be a powerful marker to predict a malignant state.
Global DNA methylation and hydroxymethylation
In the present study, differences in global DNA methylation levels between sun-exposed and sun-protected skin areas were not observed (Figure 1). Studies investigating the influence of sun radiation on skin cells are scarce and data are still controversial. It has also been shown that the human keratinocytes chronically exposed to UV radiation have no recognisable global effect on DNA methylation patterns.28 In contrast, in samples obtained from human sun-exposed arm areas, the global DNA hypomethylated condition appeared to be more frequent in comparison to that of sun-protected skin areas, which was more pronounced in the epidermis.29,30 In the present study, it was not possible to separate dermis and epidermis and, therefore, the levels of global DNA 5-mC and 5-hmc for keratinocytes and fibroblasts were analysed together. In contrast, we analysed samples of individuals that represent the Brazilian population, classified as light, moderate and dark brown (types III, IV and V) according to Fitzpatrick's criteria, while the studies above cited analysed Caucasian sample as type II. 22 This type of skin is more sensitive to the sun and, therefore, more likely to undergo epigenetic changes.
We detected differences in relation to gender with women, showing higher global methylation levels than men (Figure 1). Godderis et al.31 observed no significant differences in global DNA methylation levels in the blood and saliva between males and females; however, the sample size was limited (4 males and 10 females). In relation to skin tissue, the hypermethylation of CpG sites associated with female gender in women over 50 years was observed. It was suggested that menopause would be involved in this process, as it causes accelerated aging, related to estrogen signaling.32-34 In this study, the proportion of young women (<45) was lower (n=3) than older women (>45) (n=10), which could explain the observed hypermethylation, although in this case in a global scale.
Furthermore, there were no detectable differences in global methylation levels in relation to age and skin type (Figure 1). It was already shown on a genome-scale level that aging is more associated with methylation changes at specific sites than on the global DNAprofile, which was more pronounced in the epidermis.29,32,35 In contrast, Vandiver et al.30 identify changes across large block regions encompassing ~ 20% of the genome; however, their finding of hypomethylation in epidermal tissue occurs only with chronic exposure, not aging alone. Some authors suggested that methylation variability in aging were accompanied by a reduced connectivity of transcriptional networks. In the present study, the small sample size, especially of young people, and the inseparability of the dermis and epidermis may have influenced the results, thus preventing the confirmation of the above assumptions for the epigenetics of skin aging.
In relation to hydroxymethylation, to the best of our knowledge, this is the first study to report levels of global DNA hydroxymethylation in sun-exposed and sun-protected skin areas and to reveal that there are no differences between them (Figure 1). Studies focussing on the influence of environmental factors on the global DNA hydroxymethylation profile are relatively unexplored; however, a recent study showed that exposure to arsenic is associated with changes at these levels.21 The levels of 5-hmC have previously been shown to be an important hallmark of human melanoma, in which there is a reduction of 5-hmC levels in comparison to that in mature melanocytes.36 In addition, hydroxymethylcytosine is a useful marker to differentiate between malignant melanomas and benign melanocytic nevi.37
In the present study, it was shown that global DNA methylation levels are higher in comparison to hydroxymethylation levels (p < 0.0001; Figure 1). In fact, DNA methylation is the predominant epigenetic modification and, much like 5-mC and the 5-hmC, it is also tissue-specific.38 We also detected a correlation between 5-mC and 5-hmC levels in the skin tissue (p < 0.0001; Figure 1). Godderis et al.31 observed that global DNA methylation and hydroxymethylation levels were correlated in saliva but not in blood. It was previously proposed that the balance between 5-mC and 5-hmC levels is involved in the balance between cell pluripotency and lineage commitment.39 The correlation of the two bases support the hypothesis that the 5-hmC acts as an intermediary of active demethylation of 5-mC.
It is known that both global and site-specific DNA methylation profiles are influenced by aging.40,41 Currently, global DNA hydroxymethylation is being characterised as a strong and reproducible trademark of chronological age, potentially acting on the assessment of health and disease prevention.42 Truong et al.43 recently reported that the level of hydroxymethylation in peripheral blood T cells from humans is dependent on age, and is associated with a gradual decrease of TET3 expression levels during aging. However, in our study, we detected no influence of age on DNA global methylation and hydroxymethylation levels in skin tissue (Figure 1). In relation to skin tissue, it was previously suggested that age-related changes are more pronounced at specific sites than on the global DNA profile, which may also be true for hydroxymethylation levels.29,32
DNA methylation at specific sites
For the miR-9-1 gene, our data show that the methylated condition for both sun-exposed and sun-protected skin areas was the most frequent (41.2%); however, 23.5% of the samples showed an unmethylated condition. Given the lack of influence of sun exposure on this profile, the differences could be explained by age, skin type or gender. However, it was also observed that age and skin type did not influence this profile, because, for the methylated samples (41.2%) in individuals ranging from 30 to 89 years of age, 17.6% presented skin type III, 11.8% skin type IV and 11.8% skin type V.
However, gender may be a factor that influences the DNA methylation profile of this gene, as we observed that the methylated profile included 29.4% of women out of a total of 41.2% of individuals. However, a study with a larger sample size is required to validate this hypothesis. Our group has previously demonstrated that the methylated profile of miR-137 in skin samples is also associated with the female gender.44 In fact, the female gender has proven to be positively associated with DNA methylation for some genes. Here, we also detected hypermethylation in global methylation, which could be associated with aging (menopause), as discussed above.45,46 For unmethylated samples, we observed that aging, gender or skin type did not influence this profile. Other intrinsic factors, such as genetic polymorphisms or even a combination of several intrinsic factors, could explain the variability in methylated and unmethylated miR-9-1 profiles among the skin samples of individuals.18,47
For samples with different methylation profiles between areas (35.3%), both methylated (11.8%) and unmethylated (23.5%) profiles were observed in sun-exposed skin areas. This suggests that factors in addition to sun exposure, such as exposure to chemical agents, occupation and habits, may have influenced the methylation profile. However, there is insufficient information to validate this hypothesis.
Cheng & Cho48 suggest that dermatologists are practiced skeptics of the dogma "one genome per individual", given that genetic diversity within individuals is routinely revealed in skin cells. For example, there are mosaic presentation conditions, such as incontinentia pigmenti, McCune-Albright disease, localized lesions of neurofibromatosis, Darier disease, and even the abundant nevi harbouring BRAF mutations.48 We dare to say "one epigenome per individual" is even more pronounced, since the epigenome is influenced by both intrinsic and extrinsic factors, since every individual and each tissue/cell is exposed to different conditions, and since the combination of these factors can lead to a variety of epigenome profiles. We suggest this is the basis for the miR-9-1 epigenetic profile, given the presence of four observed methylation profiles, indicating that other environmental factors in addition to sun exposure can influence the methylation profile of this gene (Figure 2). Our experience in studying the epigenome of skin cells shows that the methylation status of a particular microRNA can be influenced by a variety of factors in addition to sun exposure and gender. This is based on evidence that in addition to the four different miR-9-1 methylation profiles among the individuals analysed in the present study, there were five different miR-137 methylation profiles previously observed in a sample of 30 individuals.44 Therefore, epigenome profiles can vary both among various cell types and within a particular individual, which may then lead to unique epigenome profiles associated with a particular disease state. This finding reinforces the issue of the significant influence of intrinsic and extrinsic factors on epigenetic profiles, and it should be used as a basis for the individualization of drug therapies.
Our data shows that sunlight exposure does not influence the DNA methylation profiles of the miR-9-3 and MTHFR genes based on the similar frequencies of the unmethylated and methylated conditions for both skin areas for miR-9-3 and MTHFR, respectively (Figure 2).The high frequencies of those profiles show that age, gender and skin type also do not influence the methylation profile of these genes. Although some studies show a correlation with age and gender44 in skin tissue, it is known that these changes occur in a very specific way, that is, some genes are more susceptible than others to these factors.29,32,40,44
A comparison of the frequency of concordant and discordant profiles among skin areas for the three genes revealed that miR-9-1 may be the most susceptible to intrinsic or extrinsic factors. Although miR-9-1 showed no difference between skin type areas, it showed the highest frequency of differences among sun-exposed and sun-protected skin areas (35.3%), and its methylation status was distributed in four profiles, suggesting that this gene may be suscep-tible to intrinsic or extrinsic factors, including gender.
Furthermore, the methylation analysis for the miR-9-3 and MTHFR genes showed that unmethylated and methylated DNA is a common profile of skin cells for these genes (Fig. 2H and I), respectively, because it was observed in both areas and in more than 70% of the cases, in contrast to miR-9-1 (Figure 2). In contrast to miR9-1 profiles, those of miR-9-3 and MTHFR showed a homogeneity among cells in the dermis and epidermis. Therefore, the separation of the epidermis and dermis could elucidate whether or not sun exposure influences the methylation profile of miR-9-1.
According to controversial data that UV radiation can lead to global methylation changes, this is also true for specific sites, since some studies have shown differences among sun-exposed and sun-protected skin areas, while others have not.28,29,30 In relation to specific sites, the literature indicates that changes in the DNA methylation status in sun-exposed areas occur in a specific way in the genome, as it has already been shown that some genes seem to change with sun exposure (CDH1 and KRT75), and other genes are not influenced (cell adhesion, cell cycle, cytokeratin, micro-RNAs and matrix metalloproteinase genes).29,44,49 In fact, genes are not susceptible to environmental factors of the same intensity and even the genes evaluated in this study may present others CpG sites that are more susceptible to solar radiation, as already shown in the case of smoking for example.50
Inconsistencies among studies can be explained by the studied skin types/ethnicity (Grönniger et al.29 and Vandiver et al.30 studied Caucasians (skin type II), in contrast with our studies (previous41 and present), which were performed with skin type III, IV and V); population exposure levels (we do not know the dress habits of the studied population); study designs (Grönniger et al.29 and Vandiver et al.30 separated dermis and epidermis, while our research group did not; Grönniger et al. and Vandiver et al.30 performed Array-based methylation analysis and Bisulfite sequencing, as compared to ELI-SA and MSP performed by our group).
CONCLUSION
It can therefore be concluded that sun exposure does not influence the DNA methylation status at specific sites of the miR-9-1, miR-9-3 and MTHFR genes, as well as the global DNA methylation and hydroxymethylation levels in individuals with light, moderate or brown skin types. However, female-related global methylation levels were detected. In addition, global methylation levels were higher in comparison to hydroxymethylation levels, and the 5-mC and 5-hmC levels are correlated in skin tissues.
Footnotes
Work performed at Universidade Federal da Paraíba (UFPB), João Pessoa, PB, Brazil.
Financial support: Post-graduate program in Cellular and Molecular Biology (PPGBCM), Universidade Federal da Paraíba (UFPB), and Coordination for the Improvement of Higher Education Personnel (CAPES).
Conflict of interest: None.
REFERENCES
- 1.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg ES, Chong KK, Huynh KT, Tanaka R, Hoon DS. Epigenetic Biomarkers in Skin Cancer. Cancer Lett. 2014;342:170–177. doi: 10.1016/j.canlet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer Sci. 2010;101:300–305. doi: 10.1111/j.1349-7006.2009.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farré P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin. 2015 May 09;8:19–19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 7.Schinke C, Mo Y, Yu Y, Amiri K, Sosman J, Greally J, et al. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–265. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davalos V, Esteller M. MicroRNAs and cancer epigenetics a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 11.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 12.Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 14.Meola N, Gennarino VA, Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2:7–7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 16.Shiiyama R, Fukushima S, Jinnin M, Yamashita J, Miyashita A, Nakahara S, et al. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013;23:366–372. doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 17.Matthews RG, Sheppard C, Goulding C. Methylenetetrahydrofolate reductase and methionine synthase: biochemistry and molecular biology. Eur J Pediatr. 1998;157:S54–S59. doi: 10.1007/pl00014305. [DOI] [PubMed] [Google Scholar]
- 18.Weiner AS, Boyarskikh UA, Voronina EN, Mishukova OV, Filipenko ML. Methylenetetrahydro-folate reductase C677T and methionine synthase A2756G polymorphisms influence on leukocyte genomic DNA methylation level. Gene. 2014;533:168–172. doi: 10.1016/j.gene.2013.09.098. [DOI] [PubMed] [Google Scholar]
- 19.Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015;123:399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syed DN, Khan MI, Shabbir M, Mukhtar H. MicroRNAs in Skin Response to UV Radiation. Curr Drug Targets. 2013;14:1128–1134. doi: 10.2174/13894501113149990184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, et al. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ Health Perspect. 2014;122:946–954. doi: 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Arch Dermatol. 1988;124:869–871. doi: 10.1001/archderm.124.6.869. [DOI] [PubMed] [Google Scholar]
- 23.de Arruda IT, Persuhn DC, de Oliveira NF. The MTHFR C677T polymorphism and global DNA methylation in oral epithelial cells. Genet Mol Biol. 2013;36:490–493. doi: 10.1590/S1415-47572013005000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khazamipour N, Noruzinia M, Fatehmanesh P, Keyhanee M, Pujol P. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role epigenetics in male infertility. Hum Reprod. 2009;24:2361–2364. doi: 10.1093/humrep/dep194. [DOI] [PubMed] [Google Scholar]
- 25.Wang LQ, Kwong YL, Kho CS, Wong KF, Wong KY, Ferracin M, et al. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia-implications on constitutiveactivation of NFENTIDADEB pathway. Mol Cancer. 2013;12:173–173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsilimigaki SI, Messini-Nikolaki N, Kanariou M, Piperakis SM. A estudy on the effects of seasonal solar radiation on exposed populations. Mutagenesis. 2003;18:139–143. doi: 10.1093/mutage/18.2.139. [DOI] [PubMed] [Google Scholar]
- 27.Bharath AK, Turner RJ. Impact of climate change on skin cancer. J R Soc Med. 2009;102:215–218. doi: 10.1258/jrsm.2009.080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahtz C, Kim SI, Bates SE, Li AX, Wu X, Pfeifer GP. UVB irradiation does not directly induce detectable changes of DNA methylation in human keratinocytes. F1000Res. 2013 Feb 13;2:45–45. doi: 10.12688/f1000research.2-45.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grönniger E, Weber B, Heil O, Peters N, Stäb F, Wenck H, et al. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandiver AR, Irizarry RA, Hansen KD, Garza LA, Runarsson A, Li X. Age and sun exposurerelated widespread genomic blocks of hypomethylation in nonmalignant skin. Genome Biol. 2015;16:80–80. doi: 10.1186/s13059-015-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godderis L, Schouteden C, Tabish A, Poels K, Hoet P, Baccarelli AA, et al. Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. Biomed Res Int. 2015;2015:845041–845041. doi: 10.1155/2015/845041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bormann F, Rodríguez-Paredes M, Hagemann S, Manchanda H, Kristof B, Gutekunst J, et al. Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell. 2016 Jun;15(3):563–571. doi: 10.1111/acel.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornton MJ. Estrogens and aging skin. Dermatoendocrinol. 2013 Apr 01;5(2):264–270. doi: 10.4161/derm.23872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmerson E, Hardman MJ. The role of estrogen deficiency in skin ageing and wound healing. Biogerontology. 2012;13:3–20. doi: 10.1007/s10522-011-9322-y. [DOI] [PubMed] [Google Scholar]
- 35.Raddatz G, Hagemann S, Aran D, Söhle J, Kulkarni PP, Kaderali L. Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics Chromatin. 2013;6:36–36. doi: 10.1186/1756-8935-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchiyama R, Uhara H, Uchiyama A, Ogawa E, Takazawa Y, Ashida A. 5-Hydroxymethylcytosine as a useful marker to differentiate between malignant melanomas and benign melanocytic nevi. J Dermatol Sci. 2014;73:161–163. doi: 10.1016/j.jdermsci.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Estève PO. Tissuespecific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 40.Bezerra SFO, Costa LA, De Freitas PAN, Oliveira NFP. age-related changes in dna methylation status of HTERT gene promoter of oral epithelial cells. Braz Arch Biol Technol. 2015;58:82–89. [Google Scholar]
- 41.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14:924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong J, Jiang HP, Peng CY, Deng QY, Lan MD, Zeng H, et al. DNA hydroxymethylation age of human blood determined by capillary hydrophilic-interaction liquid chromatography/mass spectrometry. Clin Epigenetics. 2015;7:72–72. doi: 10.1186/s13148-015-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truong TP, Sakata-Yanagimoto M, Yamada M, Nagae G, Enami T, Nakamoto-Matsubara R, et al. Age-dependent decrease of DNA hydroxymethylation in human T cells. J Clin Exp Hematop. 2015;55:1–6. doi: 10.3960/jslrt.55.1. [DOI] [PubMed] [Google Scholar]
- 44.Da Silva Melo AR, Barroso H, Uchôa De Araújo D, Ruidomar Pereira F, De Oliveira NF. The influence of sun exposure on the DNA methylation status of MMP9, miR137, KRT14 and KRT19 genes in human skin. Eur J Dermatol. 2015;25:436–443. doi: 10.1684/ejd.2015.2598. [DOI] [PubMed] [Google Scholar]
- 45.D'Aquila P, Giordano M, Montesanto A, De Rango F, Passarino G, Bellizzi D. Ageand gender-related pattern of methylation in the MT-RNR1 gene. Epigenomics. 2015;7:707–716. doi: 10.2217/epi.15.30. [DOI] [PubMed] [Google Scholar]
- 46.Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31:864–870. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arakawa Y, Watanabe M, Inoue N, Sarumaru M, Hidaka Y, Iwatani Y. Association of polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR genes with global DNA methylation levels and prognosis of autoimmune thyroid disease. Clin Exp Immunol. 2012;170:194–201. doi: 10.1111/j.1365-2249.2012.04646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng JB, Cho RJ. Genetics and epigenetics of the skin meet deep sequence. J Invest Dermatol. 2012;132:923–932. doi: 10.1038/jid.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sathyanarayana UG, Moore AY, Li L, Padar A, Majmudar K, Stastny V, et al. Sun exposure related methylation in malignant and non-malignant skin lesions. Cancer Lett. 2007;245:112–120. doi: 10.1016/j.canlet.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 50.De Oliveira SR, Da Silva IC, Mariz BA, Pereira AM, De Oliveira NF. DNA methylation analysis of cancer-related genes in oral epithelial cells of healthy smokers. Arch Oral Biol. 2015;60:825–833. doi: 10.1016/j.archoralbio.2015.02.022. [DOI] [PubMed] [Google Scholar]