Abstract
Mitochondria are responsible for bioenergetics, metabolism and apoptosis signals in health and disease. The retina being a part of the central nervous system consumes large amounts of glucose and oxygen to generate ATP via the mitochondrial oxidative phosphorylation for its phototransduction and visual function. During ATP generation, electrons leak from the mitochondrial electron transport chain, which is captured by molecular oxygen to produce reactive oxygen species (ROS). These mtROS damage mitochondrial proteins, mtDNA, and membrane lipids and release them in the cytosol. Mitochondrial components are recognized as danger-associated molecular patterns (DAMPS) by cytosolic pattern recognition receptors such as NOD-like receptors, NLRP3 inflammasomes. They process pro-caspase-1 to active caspase-1, which cleaves pro-inflammatory IL-1β o mature IL-1β causing inflammation and cell death by pyroptosis. To counter the damaging action of mtROS and inflammasomes in fully differentiated cells in the retina, the removal of the damaged and dysfunctional mitochondria by a double-membrane autophagic process via lysosomal degradation called mitophagy is critical for mitochondrial homeostasis and cell survival. Nonetheless, under chronic diseases including diabetic retinopathy (DR), mitophagy dysregulation and NLRP3 inflammasome activation exist, which cause premature cell death and disease progression. Recently, the thioredoxin-interacting protein TXNIP, which is strongly induced by diabetes and inhibits anti-oxidant function of thioredoxin, has been implicated in mitochondrial dysfunction, mitophagic dysregulation and NLRP3 inflammasome activation in DR. Therefore, TXNIP silencing or pharmacological inhibition may normalize mitophagic flux and NLRP3 inflammasome activation, which will prevent or slow down the progression of DR.
Keywords: TXNIP, Mitophagy, mt-Keima, NLRP3 inflammasome, Diabetic retinopathy
Introduction
Diabetic retinopathy (DR) is the most common cause of blindness among the working adult population in the developed and developing countries including the US. As obesity increases, the number of people with diabetes and its complications including DR is considered to increase in the coming decades. Therefore, there is tremendous pressure on socio-economic burden on families due to these chronic diseases. DR is generally defined by microvascular dysfunction including endothelial dysfunction, inner blood-retinal-barrier breakdown, pericyte loss and acellular capillary formation, capillary basement membrane thickening, microaneurisms (non-proliferative diabetic retinopathy, NPDR) followed by formation of new fragile and leaky blood vessels, a serious form of proliferative diabetic retinopathy (PDR) and blindness [1,2]. In addition to microvascular complications, recent studies also have demonstrated that retinal neurons undergo degeneration and glial activation occurs, which may lead to chronic low grade neuroinflammation and disease progression [3,4]. In addition to microvascular capillary dysfunction, the tight junction proteins in retinal pigment epithelium in the outer blood-retinal-barrier may also be compromised leading to infiltration of blood components into the subretina space and photoreceptor damage [5,6]. Therefore, DR may involve both microvascular damage and neuronal dysfunction. Nonetheless, whether microvascular damage and/or neuronal dysfunction are interrelated and which one is damaged first in DR is yet to be determined.
TXNIP and Mitochondrial Dysfunction in Diabetic Retinopathy
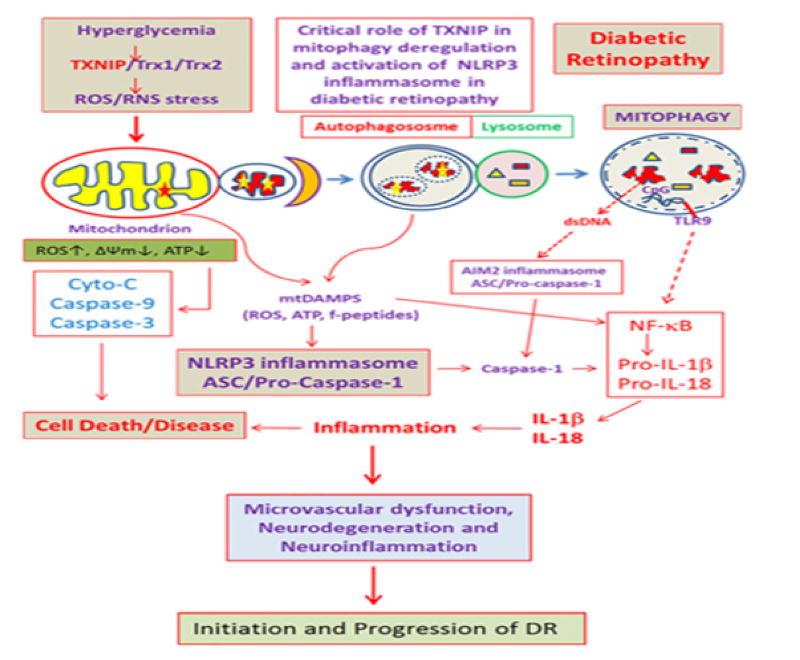
Thioredoxin-interacting protein TXNIP has been defined as a pro-oxidative stress, pro-inflammatory and pro-apoptotic protein, which is strongly induced by high glucose and diabetes in most tissues examined including the pancreatic beta, renal and retinal cells [7–10]. TXNIP binds to and inhibits the thiol-reducing and anti-oxidant capacity of thioredoxin (Trx) and causes cellular oxidative/nitrosative stress [11]. Trx1 is present in the cytosol and nucleus while Trx2 is the mitochondrial iso form. Under cellular stress, TXNIP is known to migrate to the mitochondrion and mediates mitochondrial dysfunction, membrane depolarization and release of pro-apoptotic proteins in the cytosol [12]. In addition, we have recently shown that the level of TXNIP in mitochondria is enhanced under high glucose exposure and induces mitochondrial damage and mitophagy [13]. Knocking out TXNIP by CRISPR/Cas9 and TXNIP gRNA reduces mitochondrial damage and mitophagy, indicating a role for TXNIP in mitophagy regulation [13]. Damaged mitochondria, if not removed via mitophagy, accumulate and release ROS and assemble the cytosolic NOD-like receptor NLPR3 inflammasome (an inflammatory platform composed by NLRP3, ASC, and procaspase-1) and activate caspase-1 [14–16]. Caspase-1 then processes pro-IL-1β and pro-IL-18 to mature forms of IL-1β and IL-18, which are initiators of innate immune response and inflammatory processes. In addition, TXNIP itself is known to involve in high glucose-induced NLPR3 inflammasome assembly under oxidative stress [15]. Therefore, mitophagy regulation is critically important for maintaining mitochondrial homeostasis, bioenergetics and cellular health. In addition to mitophagy, mitochondrial quality control also requires fission, fusion, biogenesis and transport as well as protein quality control [17]. Furthermore, mitochondrial protein quality control under oxidative stress involves mitochondrial unfolded protein response (UPRmt), in which a retrograde signal is generated to synthesize nuclear-encoded mitochondria targeted proteases, chaperones and anti-oxidants [18]. Under chronic hyperglycemia and sustained TXNIP up-regulation, mitochondrial oxidative stress and depolarization may dominate leading to mitochondrial fragmentation (fission) and mitophagy [13], (Figure 1).
Figure 1.
A potential role for TXNIP in mitochondrial damage, mitophagy and inflammasome activation in diabetes and its complications including DR.
Mitophagy Regulation
Autophagy is a catabolic process that degrades oxidative damaged or misfolded/aggregated proteins during starvation or oxidative stress to recycle the macromolecular or organelle components as nutrients [19]. Mitophagy is defined as a specific process of macroautophagy, which removes damaged, old and dysfunctional mitochondria, via the lysosomal degradation pathway. Therefore, mitophagy uses autophagic components such as autophagy-related ATG proteins (~33 ATGs) that are involved in the formation a double-membrane autophagosome [20] and also proteins involved in mitochondrial fission (such as Drp1, fis1, mff), mitochondrial outer membrane protein ubiquitination proteins (E3 ligases) and ubiquitin adaptors (such as Pink1, Parkin, Optineurin, p62/SQSTRM1 and so on [21]. Pink1 is initially imported into the mitochondrial inner membrane and is degraded by mitochondrial protease PARL (presenilin associated rhomboid-like protein) [22]. However, when mitochondria are depolarized and fail to import Pink1, it accumulates in the outer membrane and phosphorylates membrane proteins, which are targeted by Parkin for ubiquitination. Furthermore, proper targeting of mitophagasome (double-membrane enclosed mitochondrial cargo) to lysosomes for degradation are important components of mitophagy. How all these different events are integrated from mitochondrial dysfunction is yet to be elucidated. In a nutshell, ATG8 or LC3BII (Microtubule-associated proteins 1A/1B light chain 3B) is important for double membrane nucleation and elongation of the autophagophore. Initially, a pro-LC3B is processed by ATG4 (multiple isoforms exists such as A, B, C and D), to expose the C-terminus glycine to form LC3BI. Further, LC3BI is lipidated with phosphatidylethanolamine (PE) to form LC3BII. Several proteins are involved in LC3BII phagophore formation including ATG7 and ATG3, as well as ATG12, ATG5, and ATG16L. Among the ATG4s, ATG4B also is involved in delipidation of LC3BII from autophagophores and autophagosomes in order to maintain a sizable pool of LC3BI under basal condition and regulate mitophagy or autophagy. Under oxidative stress, ATG4B is oxidized at cysteine 81 near the protease active site at Cysteine 77 [23]. TXNIP has been shown to interact with REDD1 (regulated in development and DNA damage responses 1) and induces ROS generation and ATG4B inactivation [23]. In addition, ATG4B may also be down regulated under high glucose conditions [13]. These processes will enhance LC3BII and double membrane autophagophore formation. On the other hand, ubiquitinated mitochondrial proteins are recognized by optineurin and other adaptors/receptors such as p62/SQSTM1 and NDP52, which also bind to LC3BII in autophagophore. Phorphorylation of ubiquitin adaptors by TBK1 (TRAF family member-associated NF-κB activator (TANK)-binding kinase1) increases their binding activities and mitophagic flux [24]. Lastly, the autophagosomes carrying the ubiquitinated cargos (damaged mitochondria and aggregated proteins) fuse with the lysosomal membrane and form autolysosomes. The fusion of autophagosome and lysosome may involve SNAREs, Rab GTPase and lysosome membrane-associated proteins 1 and 2A (LAMP1 and LAMP2A). In addition, the Atg17-Atg31-Atg29 complex and Atg11, may also facilitate the fusion process [25]. Acidic lysosomal enzymes digest proteins, lipids, and DNA into their constituent molecules, which are recycled for cellular anabolic metabolism.
We have recently shown that LC3B puncta is increased in the diabetic rat retina than observed in non-diabetic normal rat retina [13]. Furthermore, alterations in mitochondria-targeted gene expression are seen in the diabetic retina suggesting mitochondrial dysfunction and mitophagy. TXNIP knockdown by intravitreal TXNIP siRNA delivery prevents LC3B puncta formation [13]. In cultured rat retinal Muller cell line, rMC1, high glucose also induces TXNIP expression, mitochondrial dysfunction and mitophagy, which are prevented by TXNIP knockout by CRISPR/Cas9 and TXNIP gRNA [13]. Nonetheless, mitophagy is a dynamic process and its flux may alter with the duration of disease and progression. Further, not all retinal cells may have similar mitophagic flux rates. Therefore, an accurate measurement of mitophagic flux in different retinal cell types and the rate of mitophagic flux with disease progression in vivo will be critically important. Towards this goal, newly generated mitophagy probes such as mt-Keima [26,27], which is pH sensitive and emits different colors when present in mitochondria or in lysosome will be helpful in determining the mitophagic states of cells in chronic diseases suchas DR and age-related neurodegenerative diseases.
Nlrp3 Inflammasome
Cellular innate defense mechanisms involve recognition of the invading pathogens and responses to eliminate the danger [28,29]. These pathogen-associated molecular patterns (PAMPS) such as the membrane lipid, protein, double-stranded DNA, single- or double-stranded RNAs are recognized by various pattern recognition receptors (PRRs) such as membrane associated toll-like receptors (TLRs) and cytosolic PRRs including NOD-like receptor NLPR3 and absent in melanoma 2 (AIM2) [30]. Among these PRRs, the cytosolic NLRP3 inflammasome composed by NLRP3, ASC, and pro-caspase-1 is induced by a wide variety of stimulants, toxins and signals, which lead to the activation of caspase-1. Caspase-1 then processes inflammatory pro-IL-1β and pro-IL-18 to their mature forms IL-1β and IL-18. IL-1β initiates the innate immune response by inducing various cytokines including TNF-α, IL-6 and CXC chemokines [31]. The pattern recognition receptors, including TLRs and NLRP3 inflammasomes, also recognize cellular components such as nuclear and mitochondrial constituents released during tissue injuries and evoke innate immune responses [32]. This form of non-invading tissue inflammation is termed as sterile inflammation. The mitochondrion, being a symbiotic bacterium residing inside the cell when damaged, releases various DAMP molecules including mtROS, mtDNA, formyl peptides and lipid components. These mtDAMPs are recognized by TLRs and NLRP3 and other cytosolic immune response platforms, and activate caspase-1 and IL-1β. Hence, if mitophagy is dysregulated, damaged mitochondria accumulate and release mtDAMPs thereby activating NLRP3 inflammasomes. We have observed that high glucose-induced TXNIP expression is associated with NF-kB nuclear translocation and pro-IL-1β expression in retina cells in culture as well as in the diabetic rat retina [10,16]. TXNIP also is known to involve in NLRP3 inflammasomes under oxidative stress [25]. In this case, TXNP bound to Trx is released under excess ROS, and assists in the assembly of NLRP3 with ASC and pro-caspase-1. Furthermore, lysosomal overloading can lead to incomplete processing of the mtDNA, which consists of non-methylated CpG DNA, which is recognized by endosomal and lysosomal membrane-associated TLR9 [33]. In addition, lysosomal membrane destabilization and permeability lead to dsDNA release and activate AIM2 inflammasome and caspase-1. Cathepsins (B, D, K, L) when released to the cytosol degrade mitochondrial outer membrane proteins and render mitochondrial damage, which further generates ROS and NLPR3 inflammasomes [33]. Such events cause a vicious cycle of oxidative stress and mitochondrial and lysosomal dysfunction and NLRP3 inflammasome activation/inflammation. Reducing or blocking TXNIP alone and/or NLRP3 inflammasome may be a potential therapeutic approach to reduce chronic low-grade inflammation associated with diabetes and its complications including DR.
Mt-Keima as a mitophagic probe
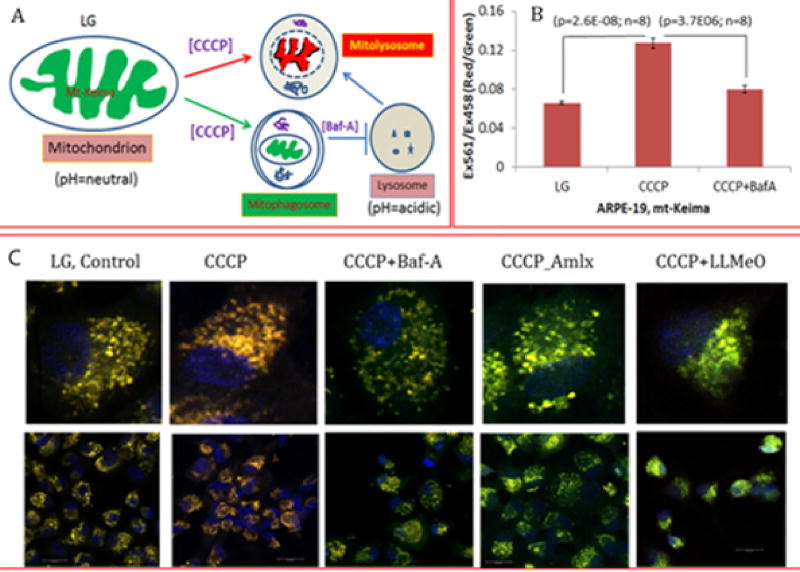
Keima is a coral-derived pH-sensitive fluorophore with dual excitation and single emission properties. This probe therefore has been employed intargeting to mitochondria by tagging the COXVIII mitochondrial target sequence. Mt-Keima emits green when present inside mitochondria at alkaline pH 8.0 (or in neutral pH) whereas in the lysosome during mitophagic flux emits red at acidic pH (4.5pH or <5.0). These properties of mt-Keima make it an excellent mito-probe to investigate mitophagic flux to lysosome both in vitro and in vivo systems. Recently, Sun et al. [27] described a method to measure in vitro and in vivo mitophagic flux using mt-Keima in transgenic mice stably expressing mt-Keima [26]. In this study, confocal microscopy was used to monitor mitophagic flux in living cells and methods to quantify mitophagy based on sequential laser excitation at 561 and 458nm, which emits a red and green at 620nm, respectively. Changes in the ratio of red/green (561/458nm) are used to quantitate mitophagic flux in the presence mitophagy inducers such as FCCP/oligomycin or under hypoxia [26,27]. We also examined mitophagy in human retinal pigment epithelial cell line (ARPE-19) transfected transiently with adenovirus-CMV-mt-Keima in the presence of CCCP with or without bafilomycin A to induce mitophagy or block autophagosome fusion with lysosome (Figure 2). In addition to fluorescence microscopy, fluoremetric measurements can also be applied to quantify mitophagic flux using theEx561/458nm ratio and emission at 620nm. We show that CCCP increases the red/green ratio, which is blocked by bafilomycin A, indicating that this method is applicable at least in in vitro studies.
Figure 2.
Mt-Keima as a molecular probe to study mitophagic flux.
Future Studies
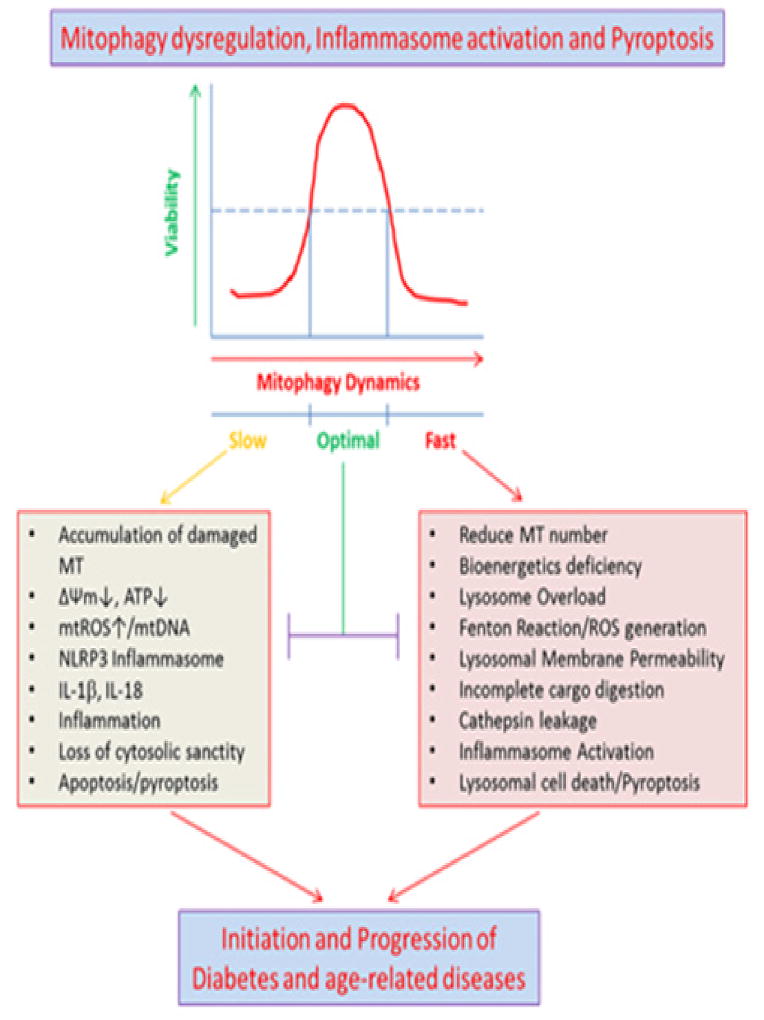
Optimal mitophagy is needed for maintaining mitochondrial homeostasis, bioenergetics, and cell survival. Either a slow or too fast mitophagic flux will cause mitochondrial content imbalance, oxidative stress, inflammation and premature cell death, some of which are depicted in Figure 3. Mt-Keima may be used to investigate mitophagic flux in DR using various vector-mediated intravitreal or subretinal transduction in adult animals, in addition to transgenic models, saving time and resource. The retina being fully differentiated cells, lentiviral-mediated mt-Keima expression vector or adenovirus-associated vectors such as AAV9 and AAV2/8 serotypes can be employed to transduce mt-Keima in the retina in adult animals [34–36]. CMV-promoters or cell type specific promoter-driven expression vectors may be constructed to address cell-specific mitophagic flux in a duration-dependent manner. Various cell types in the retina may have different mitophagic flux rates depending on their metabolic rates and function. For example, the retinal pigment epithelium is a highly phagocytic cell, which recycles photoreceptor outer segments daily, therefore may have a high mitophagic capacity and machinery while neurons with long axons and dendrites may have slower mitophatic rates. These questions may be answered to an extent using mt-Keima as an endogenous mitophagic probe and by manipulating mitophagic genes and pathways such as the Pink1/Parkin, Optineurin/TBK1 and/or TXNIP/Trx2 and ATGs. Understanding accurate rates of mitophagy in vivo under normal and disease conditions will allow drug treatment to optimize mitophagic flux so as to normalize bioenergetics, reduce inflammation and maintain cell viability. Our laboratory aims to develop mt-Keima probes that will target globally or to specific cells in the retina.
Figure 3.
Mitophagic flux dysregulation, inflammation, and cell death.
Conclusion
Mitophagy is a protective mechanism for maintaining mitochondrial homeostasis, bioenergetics and cell survival. The question as to what extent the mitophagic flux is beneficial or detrimental is yet to be defined in various chronic diseases including diabetes and its complications, neurodegenerative diseases and cardiac dysfunction. Maintaining an optimal mitophagic flux will be important to check inflammasome activation and premature cell death. Will manipulation of the mitophagic flux include a therapeutic addition for checking DR is to be established yet? At present, TXNIP represents a potential therapeutic target to limit mitochondrial damage, mitophagy, and inflammasome activation for preventing or slowing down the progression of DR.
Acknowledgments
Funding from NIH/NEI: R01 EY023992 (LPS), NIH Core Grant: P30EY004068 to the Department of Anatomy and Cell Biology, and Research to Prevent Blindness unrestricted grant to the Department of Ophthalmology are acknowledged.
References
- 1.Roy S, Kern TS, Song B, Stuebe C. Mechanistic Insights into Pathological Changes in the Diabetic Retina: Implications for Targeting Diabetic Retinopathy. Am J Pathol. 2017;187(1):9–19. doi: 10.1016/j.ajpath.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong TY, Cheung CM, Larsen M, Sharma S, Simó R. Diabetic retinopathy. Nat Rev Dis Primers. 2016;2:16012. doi: 10.1038/nrdp.2016.12. [DOI] [PubMed] [Google Scholar]
- 3.Abcouwer SF, Gardner TW. Diabetic retinopathy: loss of neuroretinal adaptation to the diabetic metabolic environment. Ann N Y Acad Sci. 2014;1311:174–190. doi: 10.1111/nyas.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran EP, Wang Z, Chen J, Sapieha P, Smith LE, et al. Neurovascular cross talk in diabetic retinopathy: Pathophysiological roles and therapeutic implications. Am J Physiol Heart Circ Physiol. 2016;311(3):H738–H749. doi: 10.1152/ajpheart.00005.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Y, Veenstra A, Palczewski K, Kern TS. Photoreceptor cells are major contributors to diabetes-induced oxidative stress and local inflammation in the retina. Proc Natl Acad Sci USA. 2013;110(41):16586–16591. doi: 10.1073/pnas.1314575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponnalagu M, Subramani M, Jayadev C, Shetty R, Das D. Retinal pigment epithelium-secretome: A diabetic retinopathy perspective. Cytokine. 2017;95:126–135. doi: 10.1016/j.cyto.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Shalev A. Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic β-cell. Mol Endocrinol. 2014;28(8):1211–1220. doi: 10.1210/me.2014-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng DW, Jiang Y, Shalev A, Kowluru R, Crook ED, et al. An analysis of high glucose and glucosamine-induced gene expression and oxidative stress in renal mesangial cells. Arch Physiol Biochem. 2006;112(4–5):189–218. doi: 10.1080/13813450601093518. [DOI] [PubMed] [Google Scholar]
- 9.Singh LP, Perrone L. Thioredoxin Interacting Protein (TXNIP) and Pathogenesis of Diabetic Retinopathy. J Clin Exp Ophthalmol. 2013;4 doi: 10.4172/2155-9570.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrone L, Devi TS, Hosoya K, Terasaki T, Singh LP. Thioredoxin interacting protein (TXNIP) induces inflammation through chromatin modification in retinal capillary endothelial cells under diabetic conditions. J Cell Physiol. 2009;221(1):262–272. doi: 10.1002/jcp.21852. [DOI] [PubMed] [Google Scholar]
- 11.Spindel ON, World C, Berk BC. Thioredoxin interacting protein: redox dependent and independent regulatory mechanisms. Antioxid Redox Signal. 2012;16(6):587–596. doi: 10.1089/ars.2011.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxena G, Chen J, Shalev A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. The Journal of biological chemistry. 2010;285(6):3997–4005. doi: 10.1074/jbc.M109.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devi TS, Somayajulu M, Kowluru RA, Singh LP. TXNIP regulates mitophagy in retinal Müller cells under high-glucose conditions: implications for diabetic retinopathy. Cell Death Dis. 2017;8(5):e2777. doi: 10.1038/cddis.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11(2):136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 15.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 16.Devi TS, Lee I, Hüttemann M, Kumar A, Nantwi KD, et al. TXNIP links innate host defense mechanisms to oxidative stress and inflammation in retinal Muller glia under chronic hyperglycemia: implications for diabetic retinopathy. Exp Diabetes Res. 2012;2012:438238. doi: 10.1155/2012/438238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8(11):870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 18.Fiorese CJ, Haynes CM. Integrating the UPRmt into the mitochondrial maintenance network. Crit Rev Biochem Mol Biol. 2017;52(3):304–313. doi: 10.1080/10409238.2017.1291577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redmann M, Darley UV, Zhang J. The Role of Autophagy, Mitophagy and Lysosomal Functions in Modulating Bioenergetics and Survival in the Context of Redox and Proteotoxic Damage: Implications for Neurodegenerative Diseases. Aging Dis. 2016;7(2):150–162. doi: 10.14336/AD.2015.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111(42):E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meissner C, Lorenz H, Hehn B, Lemberg MK. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy. 2015;11(9):1484–1498. doi: 10.1080/15548627.2015.1063763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, et al. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun. 2015;6:7014. doi: 10.1038/ncomms8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore AS, Holzbaur EL. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci USA. 2016;113(24):E3349–E3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Mao K, Yu AY, Omairi NA, Austin J, et al. The Atg17-Atg31-Atg29 Complex Coordinates with Atg11 to Recruit the Vam7 SNARE and Mediate Autophagosome-Vacuole Fusion. Curr Biol. 2016;26(2):150–160. doi: 10.1016/j.cub.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun N, Yun J, Liu J, Malide D, Liu C, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015;60(4):685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun N, Malide D, Liu J, Rovira II, Combs CA, et al. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc. 2017;12(8):1576–1587. doi: 10.1038/nprot.2017.060. [DOI] [PubMed] [Google Scholar]
- 28.Russo MV, McGavern DB. Immune Surveillance of the CNS following Infection and Injury. Trends Immunol. 2015;36(10):637–650. doi: 10.1016/j.it.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249(1):158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ori D, Murase M, Kawai T. Cytosolic nucleic acid sensors and innate immune regulation. Int Rev Immunol. 2017;36(2):74–88. doi: 10.1080/08830185.2017.1298749. [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265(1):130–142. doi: 10.1111/imr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duann P, Lianos EA, Ma J, Lin PH. Autophagy, Innate Immunity and Tissue Repair in Acute Kidney Injury. Int J Mol Sci. 2016;17(5):E662. doi: 10.3390/ijms17050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repnik U, Turk B. Lysosomal-mitochondrial cross-talk during cell death. Mitochondrion. 2010;10(6):662–669. doi: 10.1016/j.mito.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Bemelmans AP, Kostic C, Crippa SV, Hauswirth WW, Lem J, et al. Lentiviral gene transfer of RPE65 rescues survival and function of cones in a mouse model of Leber congenital amaurosis. PLoS Med. 2006;3(10):e347. doi: 10.1371/journal.pmed.0030347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bemelmans AP, Duqué S, Rivière C, Astord S, Desrosiers M, et al. A single intravenous AAV9 injection mediates bilateral gene transfer to the adult mouse retina. PLoS One. 2013;8(4):e61618. doi: 10.1371/journal.pone.0061618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Da Costa R, Röger C, Segelken J, Barben M, Grimm C, et al. A Novel Method Combining Vitreous Aspiration and Intravitreal AAV2/8 Injection Results in Retina-Wide Transduction in Adult Mice. Invest Ophthalmol Vis Sci. 2016;57(13):5326–5334. doi: 10.1167/iovs.16-19701. [DOI] [PubMed] [Google Scholar]