Abstract
Generating accurate 3D models from cone-beam computed tomography (CBCT) images is an important step in developing treatment plans for patients with craniomaxillofacial (CMF) deformities. This process often involves bone segmentation and landmark digitization. Since anatomical landmarks generally lie on the boundaries of segmented bone regions, the tasks of bone segmentation and landmark digitization could be highly correlated. However, most existing methods simply treat them as two standalone tasks, without considering their inherent association. In addition, these methods usually ignore the spatial context information (i.e., displacements from voxels to landmarks) in CBCT images. To this end, we propose a context-guided fully convolutional network (FCN) for joint bone segmentation and landmark digitization. Specifically, we first train an FCN to learn the displacement maps to capture the spatial context information in CBCT images. Using the learned displacement maps as guidance information, we further develop a multi-task FCN to jointly perform bone segmentation and landmark digitization. Our method has been evaluated on 107 subjects from two centers, and the experimental results show that our method is superior to the state-of-the-art methods in both bone segmentation and landmark digitization.
1 Introduction
Craniomaxillofacial (CMF) deformities include acquired and congenital deformities of the head and the face. It is estimated that approximate 16.8 million Americans require surgical or orthodontic treatment to correct the CMF deformities based on computed tomography (CT) scans. Comparing to the spiral multi-slide CT (MSCT) scan, cone-beam CT (CBCT) scan has the advantages of lower radiation exposure and cost, thus it has been widely used in doctor’s offices. To design accurate treatment plans, it is important to generate a 3D model of CMF structures (e.g., midface, and mandible) and digitize anatomical landmarks for quantitative analysis. However, due to severe image artifacts (e.g., imaging noise, inhomogeneity, and truncation), it is challenging to accurately segment bony structures and digitize anatomical landmarks on CBCT images.
The current clinical gold-standard is to manually perform bone segmentation and landmark digitization, which is very time-consuming and labor-intense. Current published reports regarding automated bone segmentation and landmark digitization can be generally divided into (1) multi-atlas (MA) based methods [1] and (2) learning based methods [2,3]. In the multi-atlas based methods, the segmentation and landmark digitization can be completed by transferring the labeled regions and landmarks from multi-atlas images to the target image via image registration [4]. This process can be computationally expensive (e.g., taking hours) due to the requirement of non-linear registration between multi-atlas images and the target image. In addition, because of the morphological variations across different subjects, it is often difficult to accurately perform bone segmentation and landmark digitization by only using non-linear registration. In the learning based approaches, human-engineered features are often first extracted from CBCT images and then fed into a model for bone segmentation and landmark digitization. Since feature extraction and model training are independent of each other, the learned features and model may not be coordinated with each other, which may lead to sub-optimal performance. Recently, there are reports on using deep learning based methods to incorporate the feature learning and the model training into a unified framework. Ronneberger et al. [5] proposed a U-net framework to perform image segmentation, achieving remarkable performance in biomedical image segmentation. Payer et al. [6] proposed a fully convolutional network (FCN) for landmark heatmap regression, producing good results in landmark localization using limited training data. However, these methods focus on a single task, i.e., either image segmentation or landmark localization, without using the inherent association between two tasks.
It is assumed that the tasks of bone segmentation and landmark digitization are highly associated because the landmarks generally lie on the boundaries of segmented bone regions. Accordingly, previous learning-based approaches have adopted organ segmentation to aid the landmark digitization [7], but still treating bone segmentation and landmark digitization as separate tasks. Motivated by the recent success of multi-task learning [8,9] and deep learning, we propose a joint bone segmentation and landmark digitization (JSD) framework via a context-guided FCN. To our knowledge, this is the first report on integrating bone segmentation and landmark digitization into a unified framework.
Figure 1 illustrates the schematic diagram of our proposed JSD framework. We first develop FCN-1 to learn the displacement maps for multiple landmarks from an input image to model the spatial context information in the whole image. The size of each displacement map is the same size as the input image, and each element in the displacement map records the displacement from the current voxel location to a respective landmark in a specific axis space. We then develop FCN-2 to simultaneously perform bone segmentation and landmark digitization by using both the displacement maps estimated by FCN-1 and the original image as the input. The technical contributions of this proposed method can be summarized as follows. First, we propose to use the displacement maps for explicitly modeling the spatial context information in CBCT images. Second, using the estimated displacement maps as guidance, we introduce a joint learning framework for bone segmentation and landmark digitization via a context-guided FCN.
Fig. 1.
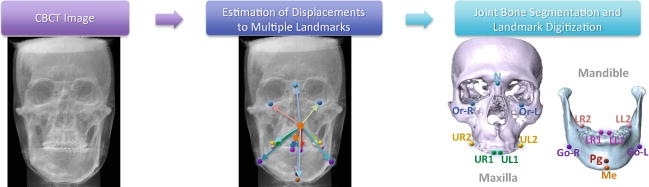
Pipeline of the proposed JSD framework, with 15 anatomical landmarks.
2 Materials and Methods
Data Description
We use 107 CT images acquired from two centers, including 77 CBCT images (0.4 × 0.4 × 0.4 mm3 or 0.3 × 0.3 × 0.3 mm3) of 77 patients with non-syndromic dentofacial deformities, and 30 MSCT images (0.488 × 0.488 × 1.25 mm3) of normal subjects. According to different types of deformity, the patients are categorized into three classes: Skeletal Class I (the mandible is retrognathic caused by mandibular retrusion, maxillary protrusion, or the combination), Skeletal Class II (the mandible is prognathic caused by mandibular protrusion, or maxillary retrusion, or the combination), and Skeletal Class III (the profile is orthognathic by either double-jaw protrusion, retrusion or vertical deformity). Among 77 patients, 20 patients are Skeletal Class I, 21 were Skeletal Class II, and 36 are Skeletal Class III. The study is approved by Institute Review Board prior to the data collection (IRB#Pro00013802). The 30 normal MSCT images, which have been collected in a completely irrelevant study, are used as additional training data. All studied CT images were manually segmented by two experienced CMF surgeons using the Mimics software. As shown in Fig. 1 (right), 15 most clinically relevant anatomical landmarks were also manually digitized by the same CMF surgeons.
Displacement Map Regression via FCN-1
For a 3D image Xn with V vox-els, we represent a displacement map as a 3D volume of the same size as Xn, with each element denoting the displacement from a voxel to a certain landmark in a specific axis space. That is, for the l-th landmark in Xn, there are 3 displacement maps (i.e., , , and ) corresponding to x, y, and z axes, respectively. Given L landmarks, we have 3L displacement maps for each input image. As shown in Fig. 2 (left), the first sub-network (i.e., FCN-1) is developed to learn a non-linear mapping from the input image to the displacement maps. Using a set of training images and their corresponding target displacement maps, FCN-1 adopts a U-net architecture [5] to capture both the global and the local structural information of input images. Specifically, FCN-1 consists of a contracting path and an expanding path. The contracting path follows the typical architecture of a CNN. Every step in the contracting path consist of two 3 × 3 × 3 convolutions, followed by a rectified linear unit (ReLU) and a 2 × 2 × 2 max expanding pooling operation with stride 2 for down-sampling. Each step in the path consists of a 3 × 3 × 3 up-convolution, followed by a concatenation with the corresponding feature map from the contracting path, and two 3 × 3 × 3 convolutions (each followed by a ReLU). Due to the contracting path and the expanding path, such network is able to grasp a large image area using small kernel sizes while still keeping high localization accuracy. Note that the output of the last layer in FCN-1 is normalized into [−1, 1]. Let Xn,v represent the v-th (v = 1, ⋯ , V) voxel of the image Xn. In the a-th (a ε{x, y, z}) axis space, we denote the l-th (l = 1, ⋯, L) displacement map of Xn as and its v-th element as . The target of FCN-1 is to learn a non-linear mapping function to transform the original input image onto its corresponding 3L displacement maps, by minimizing the following loss function:
| (1) |
where f(Xn,v, W1) is the estimated displacement by using the network coefficients W1, and N is the number of training images in a batch.
Fig. 2.
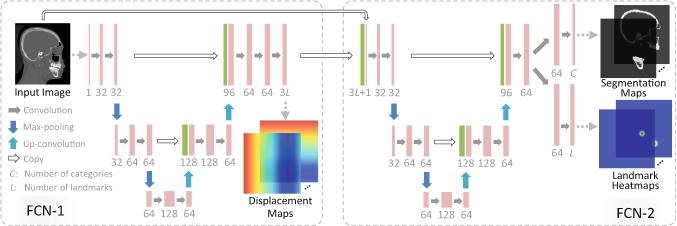
Overview of our context-guided multi-task FCN.
Joint Bone Segmentation and Landmark Digitization via FCN-2
Using the displacement maps learned in FCN-1 as guidance, we further design a sub-network (i.e., FCN-2) with a U-net architecture to jointly perform bone segmentation and landmark digitization. As shown in Fig. 2 (right), FCN-2 uses a stacked representation of displacement maps and the original image as the input, through which the spatial context information is explicitly incorporated into the learning process. In addition, such representation could guide the network to focus on the informative regions, and thus may help alleviate the negative influence of image artifacts. Note that the output of the last layer for bone segmentation is transformed to probability scores by using the softmax function, and that for landmark digitization are normalized to [0, 1]. Denote as the ground-truth segmentation map for the c-th (c = 1, ⋯, C) category, with the v-th element as . Here, a CT image is segmented into C = 3 categories (i.e., midface, mandible, and background). We denote as the ground-truth landmark heatmap for the l-th (l = 1, ⋯, L) landmark in Xn, with its v-th element as . The objective of FCN-2 is to minimize the following loss function:
| (2) |
where the first term is the cross-entropy error for bone segmentation and the second term is the mean squared error for landmark digitization. Here, 1 {·}is an indicator function, with 1 {·} = 1 if {·} is true; and 0, otherwise. indicates the probability of the v-th voxel in the image Xn being correctly classified as the category using the network coefficients W2. The second term in Eq. (2) compute the loss between the estimated value g(Xn,v, W2) and the ground-truth in the l-th landmark heatmap.
Implementation Details
For each landmark, we generate a heatmap using a Gaussian filtering with the standard derivation of 2 mm, and then stretch the values to the range of [0, 1]. For optimizing the network coefficients, we adopt the stochastic gradient descent (SGD) algorithm combined with the backpropagation algorithm. In the training stage, we first train FCN-1 using a CT image and its corresponding target displacement maps as the input and the output, respectively. With FCN-1 frozen, we then train FCN-2 for joint bone segmentation and landmark digitization, by using the stacked representation of the estimated displacement maps from FNC-1 and the original image as the input, while landmark heatmaps and segmentation maps as the output. Finally, using the learned coefficients as initialization, we further train FCN-1 and FCN-2 jointly. In addition, the training process is done in a sliding window fashion (with the fixed window size of 96 × 96 × 96). Then, we can feed a new testing image of any size into the trained model, since FCN only contains the convolutional computation.
3 Experiments
Experimental Settings
Before training the model, all images are spatially normalized to have the same resolution (i.e., 0.4 × 0.4 × 0.4 mm3), and are also intensity-normalized to have similar intensity distributions via a histogram matching technique. For 77 sets of patient CBCT images, we adopt a 5-fold cross-validation strategy. The 30 sets of normal MSCT images are used as additional training samples for model learning in each of 5 folds. To evaluate the accuracy of the bone segmentation (separating the bony structures of the mandible from the midface), we use three evaluation metrics, including (1) Dice similarity coefficient (DSC), (2) sensitivity (SEN), and (3) positive predictive value (PPV). To evaluate the accuracy of landmark digitization (landmark placement on the predetermined anatomical locations), we adopt the detection error as the evaluation measure.
We first compare our JSD method with two state-of-the-art methods: (1) MA based method [1], and (2) random forest (RF) based method with Harr-like features. In the latter method, we use the RF classifier for bone segmentation [10] and the RF regressor for landmark digitization [11]. Note that both MA and RF methods treat the bone segmentation and the landmark digitization as two independent tasks, while our JSD method jointly treats them as highly correlated tasks. To evaluate the specific contributions of two strategies (i.e., using displacement maps as guidance, and joint learning of two tasks) adopt in our JSD method, we further compare JSD with its two variants: JSD1 and JSD2. Specifically, JSD1 only adopts FCN-2 in Fig. 2 to separately perform bone segmentation and landmark digitization without using the joint learning strategy and displacement maps as guidance. JSD2 simply adopts FCN-2 for the jointly learning of two tasks without using displacement maps as guidance.
Results
Table 1 shows the experimental results achieved by the proposed JSD method and the four aforementioned methods in bone segmentation and landmark digitization. In the task of bone segmentation, comparing to the state-of-the-art MA and RF based methods, our JSD method achieves at least a 6.33% and a 5.06% improvement in terms of DSC in the segmentation of mid-face and mandible, respectively. Moreover, JSD generally yields better results than its variants (i.e., JSD1, and JSD2) in bone segmentation. It implies that the proposed two strategies, i.e., using displacement maps as guidance and the joint learning of two tasks, can improve the learning performance of JSD in the task of bone segmentation. In the task of landmark digitization, our JSD method achieves an error of 1.10 mm that is significantly better than the results achieved by the MA and the RF based methods. Also, the digitization performance achieved by JSD is more accurate than that of JSD1 (1.78 mm) and JSD2 (1.33 mm). More specifically, we further report the landmark digitization error for each of 15 anatomical landmarks in Fig. 3. As can be seen from Fig. 3, the proposed JSD method achieves the lowest errors compared with the competing methods, especially for tooth landmarks. It is worth noting that it is very challenging to accurately localize tooth landmarks since there are large variations across subjects in the local appearance of tooth landmarks. We have performed McNemar’s test to compare the landmark detection results achieved by our method and competing methods, and got very small (<0.001) p-values. It suggests the performance difference between our method and each competing method is significant. In addition, note that it is clinically acceptable if the landmark detection error for CBCT images is less than 1.50 mm. The average error achieved by our JSD method is less than 1.50 mm, indicating the great potential of our method in clinical applications.
Table 1.
Results of both bone segmentation and landmark digitization.
| Methods | Bone segmentation | Landmark Digitization Error (mm) |
|||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Midface | Mandible | ||||||
|
| |||||||
| DSC (%) | SEN (%) | PPV (%) | DSC (%) | SEN (%) | PPV (%) | ||
|
| |||||||
| MA | 81.14 ± 2.54 | 80.17 ± 3.27 | 82.48 ± 2.85 | 83.82 ± 2.21 | 84.31 ± 2.21 | 83.29 ± 2.30 | 3.05 ± 1.54 |
| RF | 86.86 ± 1.63 | 87.36 ± 2.98 | 85.92 ± 2.28 | 88.21 ± 1.52 | 88.54 ± 2.77 | 88.01 ± 1.95 | 2.67 ± 1.58 |
| JSD1 | 91.83 ± 1.06 | 90.05 ± 2.35 | 93.72 ± 1.24 | 91.66 ± 1.07 | 91.35 ± 2.13 | 91.99 ± 1.01 | 1.78 ± 1.31 |
| JSD2 | 92.20 ± 1.02 | 92.73 ± 2.50 | 91.78 ± 2.14 | 92.17 ± 0.99 | 93.30 ± 2.29 | 91.13 ± 1.41 | 1.33 ± 0.92 |
| JSD | 93.19 ± 0.89 | 92.82 ± 1.91 | 93.61 ± 1.40 | 93.27 ± 0.97 | 93.63 ± 1.37 | 92.93 ± 1.09 | 1.10 ± 0.71 |
Fig. 3.
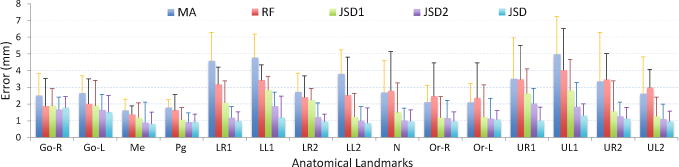
Landmark digitization errors achieved by different methods for 15 landmarks.
We further visually illustrate the results of bone segmentation and landmark digitization achieved by our JSD method for two patients with CMF deformity in Fig. 4. Each row in Fig. 4 reports the results for a specific subject. For the convenience of visualization, we show the 2D probability maps for the segmented midface and mandible in 3 views, given in Fig. 4(a) and (b), respectively. In addition, we overlap the heatmaps of 15 anatomical landmarks onto a single 3D image, and illustrate the results in 3 views in Fig. 4(c). The corresponding 3D rending volumes are provided in the online Supplementary Materials. From Fig. 4(a)–(b), we can see that in the bone segmentation task, our method can accurately separate midface and mandible. From Fig. 4(c), we can see that our method can estimate clear and smooth landmark heatmaps. All these results demonstrate the effectiveness of our proposed method.
Fig. 4.
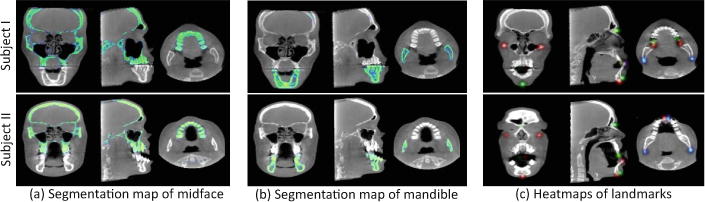
Results of our JSD method on two typical patients with CMF.
4 Discussion and Conclusion
We have proposed a joint CMF bone segmentation and landmark digitization (JSD) framework via a context-guided multi-task FCN. Specifically, to capture the spatial context information of images, we propose to use displacement maps for modeling the displacement information from voxels to anatomical landmarks in the input image. We further develop a context-guided FCN model for jointly performing bone segmentation and landmark digitization. Experimental results suggest that JSD is superior to the state-of-the-art methods.
There are still several limitations in the current study. First, there are only 107 images at hand for model learning. It is interesting to augment the training images by using synthetic data (e.g., by using deformable transformation or Generative Adversarial Networks) to further improve the robustness of our proposed method. Besides, it is reasonable to automatically learn the optimal weights for these different tasks from data. Moreover, we have only 15 landmarks and do not require too much memory. For more landmarks, we can select several salient landmarks to provide the context information, instead of using all landmarks.
Supplementary Material
Footnotes
Electronic supplementary material: The online version of this chapter (doi:10.1007/978-3-319-66185-8 81) contains supplementary material, which is available to authorized users.
References
- 1.Shahidi S, Bahrampour E, Soltanimehr E, Zamani A, Oshagh M, Moattari M, Mehdizadeh A. The accuracy of a designed software for automated localization of craniofacial landmarks on CBCT images. BMC Med Imaging. 2014;14(1):32. doi: 10.1186/1471-2342-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng E, Chen J, Yang J, Deng H, Wu Y, Megalooikonomou V, Gable B, Ling H. 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. EMBC; 2011. Automatic dent-landmark detection in 3-D CBCT dental volumes; pp. 6204–6207. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Liu M, An L, Gao Y, Shen D. Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE J Biomed Health Inform. 2017 doi: 10.1109/JBHI.2017.2704614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao X, Yang J, Gao Y, Guo Y, Wu G, Shen D. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Med Image Anal. 2017 doi: 10.1016/j.media.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015, LNCS. Vol. 9351. Springer; Cham: 2015. pp. 234–241. [DOI] [Google Scholar]
- 6.Payer C, Stern ˇD, Bischof H, Urschler M. Regressing heatmaps for multiple landmark localization using CNNs. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. MICCAI 2016 LNCS. Vol. 9901. Springer; Cham: 2016. pp. 230–238. [DOI] [Google Scholar]
- 7.Zhang J, Gao Y, Wang L, Tang Z, Xia JJ, Shen D. Automatic craniomax-illofacial landmark digitization via segmentation-guided partially-joint regression forest model and multiscale statistical features. IEEE Trans Biomed Eng. 2016;63(9):1820–1829. doi: 10.1109/TBME.2015.2503421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M, Zhang D, Chen S, Xue H. Joint binary classifier learning for ECOC-based multi-class classification. IEEE Trans Pattern Anal Mach Intell. 2016;38(11):2335–2341. [Google Scholar]
- 9.Liu M, Zhang D, Shen D. Relationship induced multi-template learning for diagnosis of Alzheimer’s disease and mild cognitive impairment. IEEE Trans Med Imaging. 2016;35(6):1463–1474. doi: 10.1109/TMI.2016.2515021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroff F, Criminisi A, Zisserman A. Object class segmentation using random forests. BMVC. 2008:1–10. [Google Scholar]
- 11.Criminisi A, Robertson D, Konukoglu E, Shotton J, Pathak S, White S, Siddiqui K. Regression forests for efficient anatomy detection and localization in computed tomography scans. Med Image Anal. 2013;17(8):1293–1303. doi: 10.1016/j.media.2013.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


