Abstract
Purpose
Fatigue is the most common symptom associated with cancer and its treatment. Investigation of molecular mechanisms associated with fatigue in oncology patients may identify new therapeutic targets. The objectives of this study were to evaluate the relationships between gene expression and perturbations in biological pathways and evening fatigue severity in oncology patients who received chemotherapy (CTX).
Methods
The Lee Fatigue Scale (LFS) and latent class analysis were used to identify evening fatigue phenotypes. We measured 47,214 ribonucleic acid transcripts from whole blood collected prior to a cycle of CTX. Perturbations in biological pathways associated with differential gene expression were identified from public datasets (i.e., Kyoto Encyclopedia Gene and Genomes, BioCarta).
Results
Patients were classified into Moderate (n=65, mean LFS score 3.1) or Very High (n=195, mean LFS score 6.4) evening fatigue groups. Compared to patients with Moderate fatigue, patients with Very High fatigue exhibited differential expression of 29 genes. A number of the perturbed pathways identified validated prior mechanistic hypotheses for fatigue, including alterations in: immune function, inflammation, neurotransmission, energy metabolism, and circadian rhythms. Based on our findings, energy metabolism was further divided into alterations in carbohydrate metabolism and skeletal muscle energy. Alterations in renal function-related pathways were identified as a potential new mechanism.
Conclusions
This study identified differential gene expression and perturbed biological pathways that provide new insights into the multiple and likely inter-related mechanisms associated with evening fatigue in oncology patients.
Keywords: fatigue, cancer, gene expression, chemotherapy, pathway analysis
INTRODUCTION
Fatigue is the most common symptom associated with cancer and its treatments [1]. In addition, severe fatigue persists in approximately 30% of cancer survivors [2]. Severe fatigue has a negative impact on patients’ ability to tolerate treatments as well as on their quality of life [3]. Given the high occurrence rates and significant negative impact of fatigue on patients’ lives, it is imperative that effective treatments be developed for this devastating symptom.
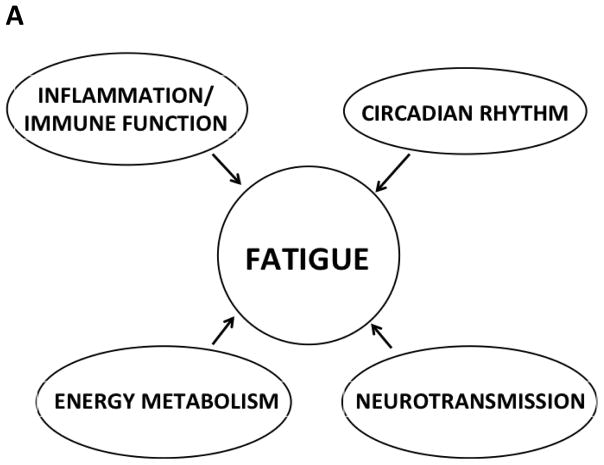
To date, exercise is the only intervention that has demonstrated efficacy for the management of fatigue in oncology patients [4,5]. One of the major reasons for the paucity of efficacious interventions is the lack of knowledge of the underlying mechanisms for fatigue [6–10]. Findings from several studies suggest that fatigue is associated with alterations in: inflammation/immune function, energy metabolism, neurotransmission, and circadian rhythm (Figure 1A). Of note, most of the work on fatigue mechanisms has focused on an evaluation of changes in serum levels of inflammatory mediators and mediators of immune function.
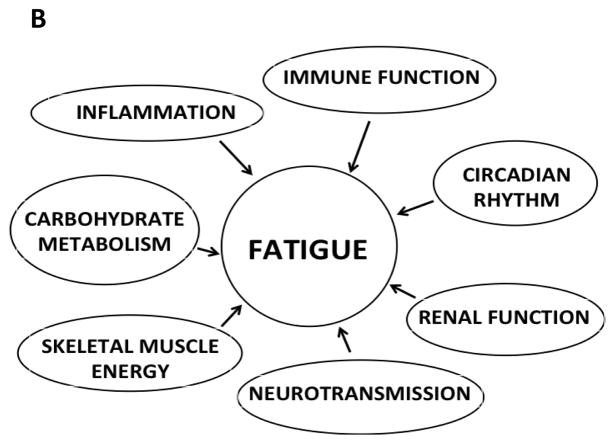
Figure 1.
Potential mechanisms for fatigue. Panel A depicts previously hypothesized mechanisms for fatigue. Panel B depicts a revised model with additional mechanisms for fatigue.
One approach that can be used to identify the mechanisms for fatigue is to evaluate for changes in gene expression using a refined phenotype. An improved understanding of these mechanisms has the potential to identify targets that can be used to develop therapeutic interventions to alleviate this devastating symptom. In a pilot study from our research group [11], changes in gene expression profiling of evening fatigue were evaluated in a sample of breast cancer patients who were categorized into low (n=19) and high (n=25) fatigue groups. This categorization was done using evening LFS scores and an established clinically meaningful cut-point for evening fatigue (i.e., >5.6 on a 0 to 10 scale) [11]. Findings from this pilot study were consistent with previous reports that evaluated the relationship between fatigue severity and changes in gene expression in oncology patients [12–14] and suggest that alterations in inflammation, neurotransmission regulation, and energy metabolism are potential mechanisms for fatigue. In the current study, we used an extreme phenotype approach to evaluate for changes in gene expression in a larger sample of patients who were classified into Moderate versus Very High evening fatigue latent classes in order to further elucidate the molecular mechanisms underlying fatigue.
METHODS
Patients and settings
This study is part of a longitudinal study of the symptom experience of oncology outpatients receiving CTX [15]. Eligible patients were ≥18 years of age; had a diagnosis of breast, gastrointestinal, gynecological, or lung cancer; had received CTX within the preceding four weeks; were scheduled to receive at least two additional cycles of CTX; were able to read, write, and understand English; and gave written informed consent. A total of 969 patients were approached and 582 consented to participate (60.1% response rate). The major reason for refusal was being overwhelmed with their cancer treatment. Of the 582 participants, a subset of 333 had complete gene expression data. For this analysis, we compared the extreme phenotypes in a subset of patients who were not included in our pilot study (i.e., 65 patients with Moderate and 195 patients with Very High levels of evening fatigue) [16].
Instruments
A demographic questionnaire obtained information on age, gender, ethnicity, marital status, living arrangements, education, employment status, income, and past medical history. Karnofsky Performance Status (KPS) scale was used to evaluate patients’ functional status [17]. Self-administered Comorbidity Questionnaire (SCQ) evaluated the occurrence, treatment, and functional impact of common comorbid conditions (e.g., diabetes, osteoarthritis, renal disease) [18]. Total SCQ score can range from 0 to 39.
The 13 item LFS assesses physical fatigue [19]. Patients rated each item using a 0 to 10 numeric rating scale based on how they felt prior to going to bed each night over the previous week (i.e., evening fatigue). Higher LFS scores indicate greater fatigue severity. A score of ≥ 5.6 indicates a clinically meaningful level of evening fatigue [20]. The LFS has well-established validity and reliability [19].
Study Procedures
The study was approved by the Committee on Human Research at the University of California, San Francisco and by each of the study sites. A research staff member approached patients in the infusion unit to discuss participation in the study. All patients signed written informed consent. Depending on the length of their CTX cycle, patients completed questionnaires in their homes, a total of six times over two cycles of CTX (i.e., prior to the next CTX administration, approximately 1 week after CTX administration, approximately 2 weeks after CTX administration). Blood for ribonucleic acid (RNA) was collected at a single time point prior to the administration of the next cycle of CTX. Medical records were reviewed for disease and treatment information.
Procedures for RNA Processing
Sample processing
Total RNA was extracted from whole blood collected into PAXgene RNA stabilization vacutainers and processed using a standard protocol (Qiagen, USA). RNA concentration was measured by NanoDrop UV spectrophotometry (ThermoScientific, USA). Using the RNA 6000 Nano Assay (Agilent, USA), all of the RNA samples were determined to be of good quality (i.e., RNA Integrity Number ≥ 8).
Microarray hybridization
Approximately 100 nanograms of total RNA was labeled using the Illumina Total Prep RNA Amplification Kit (Ambion, USA) and hybridized to the HumanHT-12 v4.0 Expression BeadChip (47,214 transcripts) (Illumina, USA). BeadChips were scanned using the iScan system (Illumina, USA) at the University of California, San Francisco Genomics Core Facility. Each HumanHT-12 BeadChip contained 12 sample BeadArrays.
Microarray preprocessing and normalization
Summary level data were calculated from the uncorrected, non-normalized, and non-transformed summary intensities at the probe level. Data preparation and analyses were done using two well-established protocols [21]. The quality control procedures were described in detail previously [11]. Two arrays were excluded because of poor hybridization performance for positive, background negative, and biotin controls assays. The remaining arrays displayed no unusual distance array signal intensity distributions with ArrayQualityMetrics. Background correction, quantile normalization, and log2 transformation were performed using limma [22]. Probes with insufficient expression measurements were excluded (n=2,619), leaving 43,923 probes spanning 16,980 genes for analysis.
Data Analyses
Latent Profile Analysis (LPA)
The method used to obtain the evening fatigue latent classes is described in detail elsewhere. [16] Briefly, LPA was used to identify subgroups of patients (i.e., latent classes) with distinct evening fatigue trajectories over the two cycles of CTX. Of the 582 patients included in the LPA, 20.0% (n=116) were in the Moderate, 21.8% (n=127) were in the High, and 58.2% (n=339) were in the Very High evening fatigue classes. For this paper, comparisons were made between the extreme phenotypes (i.e., Moderate (n=65) versus Very High (n=195) classes) for whom gene expression data were available.
Demographic and clinical data
Demographic and clinical data were analyzed using SPSS version 23 (IBM, Armonk, NY) and Stata version 13.0 (StataCorp, College Station, TX). Descriptive statistics and frequency distributions were calculated for demographic and clinical characteristics as well as for fatigue severity. Independent sample t-tests, Mann-Whitney U tests, Chi square tests, and Fisher’s Exact tests were used to evaluate for differences in demographic and clinical characteristics between the Moderate and Very High evening fatigue classes.
Differential gene expression
Differential expression (DE) of genes was examined with an estimation of gene-by-gene variance using limma [23]. Phenotypic characteristics that differed between the two classes (i.e., age, gender, body mass index (BMI), KPS score, cancer diagnosis) were included as covariates in the model. To control for batch effects, the BeadChip was included as a covariate in the model. To control for differences in signal intensities, array weights were assessed and applied to the intensity values. Adjustment for multiple comparisons was performed using the Benjamini and Hochberg method [24] and significance was assessed at a false-discovery rate (FDR) of 10. Finally, 488 genes functioning as transcription factors (TF) in humans were identified using the TRANSFAC database [25] obtained from the CIS-BP database [26] based on the gene ontology annotation with term GO:0003700 “sequence-specific DNA binding transcription factor activity” [27] (Supplementary Table 1).
Differential pathway perturbation
A summary signal estimate of expression was calculated from all valid probes spanning each gene [28]. Summary signal intensities were obtained for 16,980 genes. Differential pathway perturbation was performed using competitive analysis with the Generally Applicable Gene Set Enrichment (GAGE) R package [29]. Genes with insufficient background expression levels and a whole genome gene expression microarray were utilized to decrease spurious results associated with alternative approaches [30,11].
Pathways and gene sets were defined using the 229 Kyoto Encyclopedia of Genes (KEGG) [31], 259 BioCarta [32], and 17,202 Gene Ontology (GO) [27] annotated sets provided by the gageData R package. Pathways model the complex interactions between genes in a biological setting and are not expected to be solely all up- or down-regulated. Therefore, differential perturbations were tested under three models: up-regulation, down-regulation, and simultaneous up/down (2D) regulation. While all of the genes in each pathway were included in this analysis, only a subset of these genes had discernible expression changes above background (i.e., essential contributing (EC) genes).
Objective query of publicly available transcriptome experiments
To better categorize and understand the biological significance of the molecular signatures associated with evening fatigue, we employed a data driven approach to leverage the collection of over 1,800 data sets that are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO). Specifically, we employed ProfileChaser (http://profilechaser.stanford.edu) to identify whole transcriptome DE profiles that existed in GEO similar to the ones that we identified [33]. Ten sets containing 5000 randomly selected genes with similar levels of expression observed in this study were evaluated separately. Then, the ten sets were combined and all profiles identified as significant were considered for interrogation.
The “factor” (i.e., unit of analysis) comparison identified by ProfileChaser was used to exclude findings that were un-interpretable. Due to the content-agnostic nature of this analysis (i.e., all pairwise comparisons of all categories are included), many of the significantly similar profiles that were identified were not appropriate for this study (e.g., sample ID) or were not easily interpretable in the context of the current study (e.g., a factor that split samples based on DE at different time points of a yeast colony’s development) were excluded. Results were reviewed for relevance to hypothesized mechanisms for fatigue by two authors (i.e., EF, KK). For all of the matches identified in the GEO database, manuscripts were obtained and findings were reviewed for relevance to the current study.
RESULTS
Demographic and Clinical Characteristics
As shown in Table 1, compared to patients in the Moderate class, patients in the Very High evening fatigue class were significantly younger, had a higher BMI, a lower KPS score, and a higher evening fatigue score at enrollment. A higher percentage of patients in the Very High evening fatigue class were female or had a breast cancer diagnosis. A smaller percentage of the Very High evening fatigue class had a gastrointestinal cancer diagnosis.
Table 1.
Differences in Demographic and Clinical Characteristics Between the Moderate and Very High Evening Fatigue Classes
| Characteristic | Moderate Evening Fatigue (n=65) | Very High Evening Fatigue (n=195) | Statistics |
|---|---|---|---|
|
| |||
| Mean ± SD | Mean ± SD | ||
| Age (years) | 60.0 ± 12.0 | 56.0 ± 12.0 | t=2.39, p=0.018 |
| Education (years) | 16.4 ± 3.1 | 16.4 ± 3.0 | t=−0.01, p=0.992 |
| Body mass index (kg/m2) | 25.3 ± 4.9 | 27.2 ± 6.4 | t=−2.17, p=0.032 |
| Lee Fatigue Scale evening fatigue score | 3.1 ± 1.5 | 6.4 ± 1.5 | t=−15.81, p<0.000 |
| Karnofsky Performance Status score | 84.0 ± 11.2 | 77.9 ± 11.5 | t=3.45, p=0.001 |
| Self Administered Comorbidity Questionnaire score | 2.2 ± 1.3 | 2.5 ± 1.4 | t=−1.57, p=0.116 |
| Time since diagnosis | 1.7 ± 2.8 | 2.5 ± 4.1 | t=−1.46, p=0.146 |
| Hemoglobin (g/dL) | 11.9 ± 1.4 | 11.7 ± 1.4 | t=1.13, p=0.261 |
| Hematocrit (%) | 35.3 ± 4.1 | 34.8 ± 4.0 | t=0.91, p=0.363 |
| n (%) | n (%) | ||
| Gender (female) | 45 (69.2) | 166 (85.1) | FE, p=0.006 |
| Self-reported ethnicity | X2=1.58, p=0.664 | ||
| White | 44 (68.8) | 137 (71.7) | |
| Asian/Pacific Islander | 11 (17.2) | 22 (11.5) | |
| Black | 2 (3.1) | 9 (4.7) | |
| Hispanic/Mixed/Other | 7 (10.9) | 23 (12.0) | |
| Married or partnered (%, yes) | 44 (67.7) | 129 (66.2) | FE, p=0.880 |
| Lives alone (%, yes) | 11 (17.2) | 40 (20.7) | FE, p=0.592 |
| Currently employed (%, yes) | 22 (33.8) | 70 (36.1) | FE, p=0.767 |
| Annual household income | U, p=0.625 | ||
| <$30,000 | 13 (22.4) | 36 (20.1) | |
| $30,000–$70,000 | 12 (20.7) | 34 (19.3) | |
| $70,000–$100,000 | 9 (15.5) | 27 (15.3) | |
| >$100,000 | 24 (41.4) | 79 (44.9) | |
| Childcare responsibilities (% yes) | 11 (16.9) | 56 (29.0) | FE, p=0.071 |
| Eldercare responsibilities (% yes) | 5 (8.3) | 14 (7.9) | FE, p=1.000 |
| Exercise on a regular basis (% yes) | 47 (72.3) | 135 (69.6) | FE, p=0.755 |
| Cancer diagnosis | X2=9.23, p=0.026 | ||
| Breast cancer | 18 (27.7) | 86 (44.1) | p=0.020 |
| Gastrointestinal cancer | 24 (36.9) | 41 (21.0) | p=0.013 |
| Gynecological cancer | 14 (21.5) | 49 (25.1) | p=0.619 |
| Lung cancer | 9 (13.9) | 19 (9.7) | p=0.3610 |
| Prior cancer treatment | U, p=0.219 | ||
| None | 15 (23.1) | 23 (11.9) | |
| Only CTX, surgery, or RT | 24 (36.9) | 96 (49.7) | |
| CTX and surgery, or surgery and RT, or CTX and RT | 19 (29.2) | 34 (17.6) | |
| CTX, surgery, and RT | 7 (10.8) | 40 (20.7) | |
| Metastatic sites | X2=1.22, p=0.748 | ||
| No metastasis | 20 (30.8) | 72 (37.3) | |
| Only one lymph node metastasis | 16 (24.6) | 38 (19.7) | |
| Only metastatic disease in other sites | 15 (23.1) | 41 (21.2) | |
| Metastatic disease in lymph nodes and other sites | 14 (21.5) | 42 (21.8) | |
Abbreviations: CTX – chemotherapy; dL – deciliter; FE – Fisher’s Exact test; g – grams; kg – kilograms; m2 – meters squared; RT – radiation therapy; U – Mann Whitney U test
Differences in Gene Expression
After controlling for significant differences in demographic and clinical characteristics, 29 DE genes were identified (Table 2). Seventeen genes were up-regulated and 12 genes were down-regulated in the Very High compared to Moderate evening fatigue classes. Three loci (i.e., LOC644537, LOC100131792, LOC647747) are not predicted in the current annotation release and their functions are unknown. Of the remaining 26 genes, seven were assigned one or more functional roles that can be associated with hypothesized mechanisms for fatigue (i.e., immune function/inflammation (i.e., ubiquitin D (UBD), ubiquitin conjugating enzyme E2 V1 (UBE2V1), complement factor H related 5 (CFHR5), mitogen activated protein kinase 4 (MAPK14)); energy metabolism (i.e., calcium voltage-gated channel subunit alpha1 E (CACNA1E), potassium voltage-gated channel subfamily Q member 5 (KCNQ5)); and renal function (i.e., CFHR5, matrix metallopeptidase 24 (MMP24)).
Table 2.
Differentially Expressed Genes in Very High Evening Compared to Moderate Fatigue Classes
| Probe | Gene Symbol | Gene Name | Entrez Gene ID | Log Fold Change |
|---|---|---|---|---|
| ILMN_1719206 | EDARADD | EDAR associated death domain | 128178 | 0.042 |
| ILMN_1682100 | IGSF10 | immunoglobulin superfamily member 10 | 285313 | −0.044 |
| ILMN_1797974 | AIG1 | androgen-induced 1 | 51390 | 0.133 |
| ILMN_1678841 | UBD | ubiquitin D | 10537 | 0.052 |
| ILMN_2350183 | ST5 | suppression of tumorigenicity 5 | 6764 | 0.054 |
| ILMN_1810852 | LAMC1 | laminin subunit gamma 1 | 3915 | 0.068 |
| ILMN_1664047 | CACNA1E | calcium voltage-gated channel subunit alpha1 E | 777 | 0.161 |
| ILMN_2318869 | UBE2V1 | ubiquitin conjugating enzyme E2 V1 | 7335 | −0.051 |
| ILMN_1689246 | PIEZO1P1 | piezo type mechanosensitive ion channel component 1 pseudogene 1 | 128615 | 0.051 |
| ILMN_1778333 | MMP24 | matrix metallopeptidase 24 | 10893 | −0.068 |
| ILMN_1704029 | LOC653453 | fibroblast growth factor 7 pseudogene | 653453 | 0.040 |
| ILMN_3188342 | HPN-AS1 | HPN antisense RNA 1 | 100128675 | −0.042 |
| ILMN_1735719 | UNG | uracil DNA glycosylase | 7374 | −0.039 |
| ILMN_1771172 | LOC644537 | ^ | 0.045 | |
| ILMN_3214384 | LOC100131792 | ^ | 0.035 | |
| ILMN_1718248 | CFHR5 | complement factor H related 5 | 81494 | −0.048 |
| ILMN_1689866 | KIF24 | kinesin family member 24 | 347240 | −0.043 |
| ILMN_1745223 | CDC42EP4 | CDC42 effector protein 4 | 23580 | 0.131 |
| ILMN_1741586 | LOC647747 | ^ | 0.045 | |
| ILMN_2388090 | MAPK14 | mitogen activated protein kinase 4 | 1432 | 0.189 |
| ILMN_1690833 | KCNQ5 | potassium voltage-gated channel subfamily Q member 5 | 56479 | −0.032 |
| ILMN_3236231 | MBD3L5 | methyl-CpG binding domain protein 3 like 5 | 284428 | 0.032 |
| ILMN_2335198 | NCOA1 | nuclear receptor coactivator 1 | 8648 | 0.046 |
| ILMN_1663718 | LHFPL3 | lipoma HMGIC fusion partner-like 3 | 375612 | −0.033 |
| ILMN_1714327 | SETD1A | SET domain containing 1A | 9739 | −0.117 |
| ILMN_2259966 | TGM5 | transglutaminase 5 | 9333 | −0.073 |
| ILMN_2400922 | OPRL1 | opioid related nociception receptor 1 | 4987 | 0.195 |
| ILMN_3305466 | ZNF788 | zinc finger family member 788 | 388507 | 0.052 |
| ILMN_1735905 | FLJ36144/GOLGA6L2 | Golgin A6 family-like 2 | 283685 | −0.063 |
Locus not predicted in recent annotation release.
Benjamini-Hochberg false discovery rate of 10
CHO – carbohydrate metabolism; IF – immune function; IN – inflammation; NE – not established; RF – renal function; SME – skeletal muscle energy
Differentially Perturbed Pathways
Differentially perturbed pathways were evaluated using these databases (i.e., KEGG, BioCarta, GO). Using KEGG, 39 down-regulated, 162 2D perturbed, and no up-regulated pathways were identified. Using BioCarta, 84 2D and no up- or down-regulated pathways were identified. Using GO, six up-regulated, 241 down-regulated, and 3,517 2D pathways were identified (Supplementary Table 2). Table 3 summarizes the differentially perturbed pathways that were identified for evening fatigue.
Table 3.
Differentially Perturbed Pathways in the KEGG and BioCarta Databases
| KEGG | Biocarta | ||
|---|---|---|---|
| Inflammation | |||
|
| |||
| hsa04060 Cytokine-cytokine receptor interaction | 2D | fibrinonlysispathway | 2D |
| hsa04610 Complement and coagulation cascades | 2D | Il7pathway | 2D |
| Il3pathway | 2D | ||
| Il10pathway | 2D | ||
| Il2pathway | 2D | ||
| Il12pathway | 2D | ||
| Il22bpathway | 2D | ||
| Il1rpathway | 2D | ||
| Il4pathway | 2D | ||
| Infapathway | 2D | ||
| Il6pathway | 2D | ||
| il2rbpathway | 2D | ||
|
| |||
| Immune Function | |||
|
| |||
| hsa04612 Antigen processing and presentation | 2D | nkcellspathway | 2D |
| hsa05330 Allograft rejection | 2D/↓ | nfkbpathway | 2D |
| hsa05332 Graft-versus-host disease | 2D/↓ | ||
| hsa04640 Hematopoietic cell lineage | 2D | ||
| hsa04650 Natural killer cell mediated cytotoxicity | 2D/↓ | ||
| hsa05323 Rheumatoid arthritis | 2D/↓ | ||
| hsa05310 Asthma | 2D/↓ | ||
| hsa04660 T cell receptor signaling pathway | 2D | ||
| hsa04662 B cell receptor signaling pathway | 2D | ||
| hsa05322 Systemic lupus erythematosus | 2D | ||
| hsa05320 Autoimmune thyroid disease | 2D/↓ | ||
| hsa04672 Intestinal immune network for IgA production | 2D/↓ | ||
| hsa05340 Primary immunodeficiency | 2D | ||
| hsa05220 Chronic myeloid leukemia | 2D | ||
| hsa04062 Chemokine signaling pathway | 2D | ||
| hsa04621 NOD-like receptor signaling pathway | 2D/↓ | ||
| hsa04612 Antigen processing and presentation | 2D | ||
|
| |||
| Circadian Rhythm | |||
|
| |||
| hsa04710 Circadian rhythm - mammal | 2D | ||
|
| |||
| Neurotransmitters | |||
|
| |||
| hsa04722 Neurotrophin signaling pathway | 2D | ||
| hsa04130 SNARE interactions in vesicular transport | 2D | gabapathway | 2D |
|
| |||
| Carbohydrate Metabolism | |||
|
| |||
| hsa04940 Type I diabetes mellitus | 2D/↓ | insulinpathway | 2D |
| hsa04910 Insulin signaling pathway | 2D | ||
| hsa04920 Adipocytokine signaling pathway | 2D | ||
| hsa00010 Glycolysis/Gluconeogenesis | 2D/↓ | ||
| hsa04973 Carbohydrate digestion and absorption | 2D | ||
| hsa00020 Citrate cycle (TCA cycle) | 2D/↓ | ||
| hsa04930 Type II diabetes mellitus | 2D | ||
| hsa00620 Pyruvate metabolism | 2D | ||
| hsa00052 Galactose metabolism | 2D | ||
|
| |||
| Skeletal Muscle Energy | |||
|
| |||
| hsa00190 Oxidative phosphorylation | 2D/↓ | igf1rpathway | 2D |
| igf1mtorpathway | 2D | ||
|
| |||
| Renal Function | |||
|
| |||
| hsa04964 Proximal tubule bicarbonate reclamation | 2D | ||
| hsa04962 Vasopressin-regulated water reabsorption | 2D | ||
| hsa04614 Renin-angiotensin system | 2D | ||
Abbreviations: 2D – two dimensional perturbation; KEGG – Kyoto Encyclopedia of Genes and Genomes; ↓ – down-regulation
Identification of Similar Whole Transcriptome Gene Expression Patterns from Publicly Available Data
ProfileChaser was used to identify publicly available gene expression studies that shared whole-transcriptome patterns of DE that were similar to those patterns observed in this study. A total of 592 unique comparisons spanning 155 GEO sets were identified as significant in the ten split analyses (Supplementary Table 3).
DISCUSSION
Differential Gene Expression and Perturbed Pathways
While the exact mechanisms underlying fatigue in oncology patients are not well understood, the most commonly cited interacting mechanisms include alterations in: immune function/inflammation, energy metabolism, neurotransmission, and circadian rhythm (Figure 1A). In the current study, all of these previously identified mechanisms [11] were confirmed. We expand the discussion to address alterations in immune function and inflammation separately. In addition, we provide evidence that energy metabolism can be divided into carbohydrate metabolism and skeletal muscle energy. We also provide new evidence that suggests that alterations in renal function are involved in the development of fatigue. Based on the findings from this study, a new mechanistic model for fatigue is illustrated in Figure 1B. The remainder of the discussion focuses on the refined and potential new mechanisms underlying fatigue.
Inflammation and Immune Function
We identified discrete genes and pathways that suggest alterations in both inflammation and immune function influence evening fatigue severity. In the current study, two new genes associated with inflammation (i.e., UBE2V1, CFHR5) were down-regulated in patients in the Very High compared to the Moderate evening fatigue class. UBE2V1 activates nuclear factor kappa-light-chain-enhancer of activated B cells (NFκ-β), [32] which controls cytokine production. Alterations in UBE2V1 appear to inhibit stress-induced apoptosis through NF-κβ [34]. CFHR5, which activates complement in the inflammatory cascade, particularly in renal tissues, including the glomeruli, [35] was also down-regulated. Polymorphisms in this gene were associated with changes in complement function associated with renal disease [36]. In addition, risk loci in this gene were associated with immunoglobin A-induced nephropathy [37]. From the KEGG database, the complement and coagulation cascade (hsa04610) was 2D perturbed. From the BioCarta database, ten interleukin pathways and the interferon-α (INF-α) pathway exhibited 2D perturbations. One of the findings from the ProfileChaser query (i.e., GDS1407), that described high and low levels of cytokine responses in leukocytes following lipopolysaccharide treatment, had a similar pattern of whole transcriptome changes as in the current study. Taken together, these findings suggest that alterations in inflammation contribute to the development of fatigue.
Additional findings support the role of immune function as both independent from and overlapping with inflammation. UBD plays a role in the processing of major histocompatibility (MHC) class I and II antigens [38]. A large number of differentially perturbed pathways related to immune function were identified (Table 3), including several found in our prior study (i.e., antigen processing and presentation (hsa04612), natural killer cell mediated cytotoxicity (hsa04650), chemokine signaling pathway (hsa04062), and NOD-like receptor signaling pathway (hsa04621)) [11]. Finally, we found differential perturbation in the NFκ-β pathway (nfkbpathway).
Energy Metabolism
Alterations in energy metabolism may contribute to the development of fatigue [6–10]. In the current study, we identified perturbations in pathways broadly related to energy metabolism and separated these pathways into two distinct categories.
Carbohydrate Metabolism
Insulin-mediated glucose uptake is a process by which carbohydrates and other nutrients are metabolized. Insulin resistance, which occurs in the setting of excessive intake of carbohydrates and other nutrients, is a major cause of inflammation. Patients treated with CTX are at increased risk for impaired insulin sensitivity and increased inflammation [39]. CACNA1E, which is associated with type 2 diabetes and insulin sensitivity [40] and is hypothesized to regulate insulin secretion [41], was one of the DE genes that was up-regulated in the current study. In addition, DE of MAPK14, which up-regulates inflammation in skeletal muscle in insulin resistant individuals with type 2 diabetes, [42] was up-regulated in the current study. In terms of perturbed pathways, nine pathways related to carbohydrate metabolism in the KEGG database and the insulinpathway from the BioCarta database were identified. These data suggest that changes in insulin sensitivity and carbohydrate metabolism may influence inflammatory pathways that result in fatigue in oncology patients receiving CTX. Additional studies are needed to determine whether improving insulin sensitivity in patients receiving CTX could decrease inflammation and fatigue severity.
Skeletal Muscle Energy
KCNQ5, which was DE in the current study, is a highly abundant sodium channel in skeletal muscle [43]. How alterations in this gene’s expression may be related to fatigue severity is not known. From the KEGG database, perturbations in the oxidative phosphorylation pathway and from the BioCarta database perturbations in the igf1rpathway and igf1mtorpathway were identified. Using ProfileChaser, studies of the skeletal muscle system with similar whole transcriptome differential expression patterns to the current study were found (Supplemental Table 3). For example, in one study (i.e., GDS1541), changes in gene expression in whole blood from healthy males both immediately following exercise and after 60 minutes of rest were similar to those observed in our study. The similarity in patterns of whole transcriptome changes found between exercise-induced fatigue and Very High versus Moderate fatigue in oncology patients receiving CTX suggests that some mechanisms that underlie fatigue may be ubiquitous.
Renal Function
CTX can result in renal damage and impaired renal function. Impaired renal function can lead to the accumulation of toxins that increase inflammation and immune responses. In addition, fatigue is a common symptom reported by patients with renal disease [44]. In a previous study of oncology patients, [45] higher levels of blood urea nitrogen were associated with worse fatigue. In the current study, no differences in the occurrence of renal disease were found between the Very High (1%) and Moderate (0%) evening fatigue classes. However, we identified DE of MMP24, which had increased expression in kidney biopsies of patients with diabetes and is hypothesized to play a role in diabetic renal tubular atrophy [46]. As described above, CFHR5 is a component of the inflammation-related complement protein. Variations in this gene are known to be associated with perturbations in the complement pathway, which results in renal damage and disease [35,36]. From the KEGG database, three pathways related to renal function were perturbed. Our study provides evidence that impairment in renal function may be related to fatigue severity in patients receiving CTX. Additional studies are needed to confirm or refute these findings.
Complex Etiology
Based on the evidence from a number of reviews [6–10] and the findings from this study, fatigue is likely to occur as a result of epistasis at single gene and pathway levels, as well as through gene-environment interactions. Two of the DE genes identified in the current study are examples of these complex interactions. Impaired glucose metabolism is a cause of renal damage and this study and others found that MMP24 is DE in individuals with diabetes and renal impairment [46]. Another example is CFHR5. This gene codes for a subunit of the inflammation-related complement protein in renal tissue. Evidence from this study and others [47,6] supports the role of inflammation in the development of fatigue. In addition, we found evidence for perturbations in renal function including DE expression of CFHR5. Finally, the patterns of DE across multiple pathways may be regulated by upstream driver genes. Changes in these genes (e.g., TF) may regulate large number of genes with diverse functions and explain some of the variation observed across functional categories observed in this study. However, no putative TF were found to be significantly DE. Additional studies are needed to understand the complex interactions among the hypothesized mechanisms for fatigue that are illustrated in Figure 1B.
Limitations
Several limitations warrant consideration. Gene expression was measured from whole blood, which may not reflect the same biologic activity that occurs across the blood brain barrier or within discrete organs and tissues. Despite controlling for significant covariates, the sample was heterogenous in terms of the type of CTX received, stage of disease, and number of previous CTX cycles. While no differences were found in caregiver responsibilities, additional measures of caregiver burden should be evaluated in future studies. While all of the gene expression studies of fatigue in oncology patients, including our own, identified differences in genes involved in inflammation, the specific genes and pathways identified differed, which may be due to differences in the classification of the patients who were evaluated and/or the fatigue phenotype.
Conclusions
Findings from this study support the main hypothesized mechanisms for fatigue (i.e., inflammation, immune function, energy metabolism, neurotransmission, circadian rhythm). In addition, we differentiated inflammation and immune function as discrete categories; further specified energy metabolism into insulin metabolism and skeletal muscle energy; and provided new evidence for renal function. Through the use of data-mining approaches, similar whole transcriptome changes were found in non-cancer systems for each of the hypothesized molecular mechanisms of fatigue found in this study. These findings suggest that a number of common mechanisms contribute to the development of fatigue in oncology patients. This study provides novel information about the complex epistatic mechanisms that underlie the development of fatigue in oncology patients receiving CTX. Confirmation of these findings may help to identify patients who are at greater risk for more severe fatigue during CTX. Once confirmed in larger studies that are adequately powered to include additional covariates, new interventions may be developed that may ameliorate this devastating symptom.
Supplementary Material
Acknowledgments
Funding: Research reported in this publication was supported by grants from the National Cancer Institute [grant numbers R01CA134900, K05CA168960], the National Center for Advancing Translational Sciences of the National Institutes of Health [grant number KL2TR000143], and the National Institute for Nursing Research [grant number T32NR008346]. Dr. Miaskowski is an American Cancer Society Clinical Research Professor.
Footnotes
Conflicts of Interest: None
References
- 1.Berger AM, Gerber LH, Mayer DK. Cancer-related fatigue: implications for breast cancer survivors. Cancer. 2012;118(8 Suppl):2261–2269. doi: 10.1002/cncr.27475. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams Y, Laguette MJ, Prince S, Collins M. Polymorphisms within the COL5A1 3-UTR That Alters mRNA Structure and the MIR608 Gene are Associated with Achilles Tendinopathy. Annals of Human Genetics. 2013;77:204–214. doi: 10.1111/ahg.12013. [DOI] [PubMed] [Google Scholar]
- 3.Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain, behavior, and immunity. 2011;25(1):147–150. doi: 10.1016/j.bbi.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meneses-Echavez JF, Gonzalez-Jimenez E, Ramirez-Velez R. Effects of Supervised Multimodal Exercise Interventions on Cancer-Related Fatigue: Systematic Review and Meta-Analysis of Randomized Controlled Trials. Biomed Res Int. 2015;2015:328636. doi: 10.1155/2015/328636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson D, Diorio C, Beyene J, Sung L. Effect of exercise on cancer-related fatigue: a meta-analysis. Am J Phys Med Rehabil. 2014;93(8):675–686. doi: 10.1097/PHM.0000000000000083. [DOI] [PubMed] [Google Scholar]
- 6.Saligan LN, Kim HS. A systematic review of the association between immunogenomic markers and cancer-related fatigue. Brain, behavior, and immunity. 2012;26(6):830–848. doi: 10.1016/j.bbi.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower JE. Cancer-related fatigue--mechanisms, risk factors, and treatments. Nat Rev Clin Oncol. 2014;11(10):597–609. doi: 10.1038/nrclinonc.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barsevick A, Frost M, Zwinderman A, Hall P, Halyard M Consortium G. I’m so tired: biological and genetic mechanisms of cancer-related fatigue. Quality of life research: an international journal of quality of life aspects of treatment, care and rehabilitation. 2010;19(10):1419–1427. doi: 10.1007/s11136-010-9757-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landmark-Hoyvik H, Reinertsen KV, Loge JH, Kristensen VN, Dumeaux V, Fossa SD, Borresen-Dale AL, Edvardsen H. The genetics and epigenetics of fatigue. PM R. 2010;2(5):456–465. doi: 10.1016/j.pmrj.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Saligan LN, Olson K, Filler K, Larkin D, Cramp F, Yennurajalingam S, Escalante CP, del Giglio A, Kober KM, Kamath J, Palesh O, Mustian K Multinational Association of Supportive Care in Cancer Fatigue Study Group-Biomarker Working G. The biology of cancer-related fatigue: a review of the literature. Supportive care in cancer: official journal of the Multinational Association of Supportive Care in Cancer. 2015;23(8):2461–2478. doi: 10.1007/s00520-015-2763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kober KM, Dunn L, Mastick J, Cooper B, Langford D, Melisko M, Venook A, Chen LM, Wright F, Hammer M, Schmidt BL, Levine J, Miaskowski C, Aouizerat BE. Gene Expression Profiling of Evening Fatigue in Women Undergoing Chemotherapy for Breast Cancer. Biological research for nursing. 2016 doi: 10.1177/1099800416629209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiao CP, Reddy SY, Chen MK, Saligan LN. Genomic Profile of Fatigued Men Receiving Localized Radiation Therapy. Biological research for nursing. 2016;18(3):281–289. doi: 10.1177/1099800415618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landmark-Hoyvik H, Reinertsen KV, Loge JH, Fossa SD, Borresen-Dale AL, Dumeaux V. Alterations of gene expression in blood cells associated with chronic fatigue in breast cancer survivors. Pharmacogenomics J. 2009;9(5):333–340. doi: 10.1038/tpj.2009.27. [DOI] [PubMed] [Google Scholar]
- 14.Saligan LN, Hsiao CP, Wang D, Wang XM, St John L, Kaushal A, Citrin D, Barb JJ, Munson PJ, Dionne RA. Upregulation of alpha-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain, behavior, and immunity. 2013;27(1):63–70. doi: 10.1016/j.bbi.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miaskowski C, Dunn L, Ritchie C, Paul SM, Cooper B, Aouizerat BE, Alexander K, Skerman H, Yates P. Latent Class Analysis Reveals Distinct Subgroups of Patients Based on Symptom Occurrence and Demographic and Clinical Characteristics. Journal of pain and symptom management. 2015;50(1):28–37. doi: 10.1016/j.jpainsymman.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kober KM, Cooper BA, Paul SM, Dunn LB, Levine JD, Wright F, Hammer MJ, Mastick J, Venook A, Aouizerat BE, Miaskowski C. Subgroups of chemotherapy patients with distinct morning and evening fatigue trajectories. Supportive care in cancer: official journal of the Multinational Association of Supportive Care in Cancer. 2016;24(4):1473–1485. doi: 10.1007/s00520-015-2895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnofsky DA, WH, Craver LV, Burchenal JH. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–656. [Google Scholar]
- 18.Sangha O, Stucki G, Liang MH, Fossel AH, Katz JN. The Self-Administered Comorbidity Questionnaire: a new method to assess comorbidity for clinical and health services research. Arthritis Rheum. 2003;49(2):156–163. doi: 10.1002/art.10993. [DOI] [PubMed] [Google Scholar]
- 19.Lee KA, Hicks G, Nino-Murcia G. Validity and reliability of a scale to assess fatigue. Psychiatry research. 1991;36(3):291–298. doi: 10.1016/0165-1781(91)90027-m. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher BS, Paul SM, Dodd MJ, Schumacher K, West C, Cooper B, Lee K, Aouizerat B, Swift P, Wara W, Miaskowski CA. Prevalence, severity, and impact of symptoms on female family caregivers of patients at the initiation of radiation therapy for prostate cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26(4):599–605. doi: 10.1200/JCO.2007.12.2838. [DOI] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth G. Limma: Linear Models for Microarray Data. Springer; New York: 2005. [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 25.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJ, Bouget FY, Ratsch G, Larrondo LF, Ecker JR, Hughes TR. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R Gene Ontology C. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimers M. Making informed choices about microarray data analysis. PLoS computational biology. 2010;6(5):e1000786. doi: 10.1371/journal.pcbi.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi S, Glazko GV, Emmert-Streib F. Ensuring the statistical soundness of competitive gene set approaches: gene filtering and genome-scale coverage are essential. Nucleic Acids Res. 2013;41(7):e82. doi: 10.1093/nar/gkt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol. 2007;396:71–91. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2:117–120. [Google Scholar]
- 33.Engreitz JM, Morgan AA, Dudley JT, Chen R, Thathoo R, Altman RB, Butte AJ. Content-based microarray search using differential expression profiles. BMC Bioinformatics. 2010;11:603. doi: 10.1186/1471-2105-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syed NA, Andersen PL, Warrington RC, Xiao W. Uev1A, a ubiquitin conjugating enzyme variant, inhibits stress-induced apoptosis through NF-kappaB activation. Apoptosis: an international journal on programmed cell death. 2006;11(12):2147–2157. doi: 10.1007/s10495-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 35.Gale DP, Pickering MC. Regulating complement in the kidney: insights from CFHR5 nephropathy. Disease models & mechanisms. 2011;4(6):721–726. doi: 10.1242/dmm.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrera-Abeleda MA, Nishimura C, Smith JL, Sethi S, McRae JL, Murphy BF, Silvestri G, Skerka C, Jozsi M, Zipfel PF, Hageman GS, Smith RJ. Variations in the complement regulatory genes factor H (CFH) and factor H related 5 (CFHR5) are associated with membranoproliferative glomerulonephritis type II (dense deposit disease) J Med Genet. 2006;43(7):582–589. doi: 10.1136/jmg.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43(4):321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basler M, Buerger S, Groettrup M. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Molecular immunology. 2015;68(2 Pt A):129–132. doi: 10.1016/j.molimm.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 39.Makari-Judson G, Katz D, Barham R, Mertens W. Deleterious Effect of Chemotherapy on Measures of Insulin Resistance in Patients with Newly-Diagnosed Invasive Breast Cancer. Cancer research. 2009;69(24 Supplement):1054. doi: 10.1158/0008-5472.sabcs-09-1054. [DOI] [Google Scholar]
- 40.Trombetta M, Bonetti S, Boselli M, Turrini F, Malerba G, Trabetti E, Pignatti P, Bonora E, Bonadonna RC. CACNA1E variants affect beta cell function in patients with newly diagnosed type 2 diabetes. the Verona newly diagnosed type 2 diabetes study (VNDS) 3. PLoS One. 2012;7(3):e32755. doi: 10.1371/journal.pone.0032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmkvist J, Tojjar D, Almgren P, Lyssenko V, Lindgren CM, Isomaa B, Tuomi T, Berglund G, Renstrom E, Groop L. Polymorphisms in the gene encoding the voltage-dependent Ca(2+) channel Ca (V)2.3 (CACNA1E) are associated with type 2 diabetes and impaired insulin secretion. Diabetologia. 2007;50(12):2467–2475. doi: 10.1007/s00125-007-0846-2. [DOI] [PubMed] [Google Scholar]
- 42.Brown AE, Palsgaard J, Borup R, Avery P, Gunn DA, De Meyts P, Yeaman SJ, Walker M. p38 MAPK activation upregulates proinflammatory pathways in skeletal muscle cells from insulin-resistant type 2 diabetic patients. American journal of physiology Endocrinology and metabolism. 2015;308(1):E63–70. doi: 10.1152/ajpendo.00115.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roura-Ferrer M, Etxebarria A, Sole L, Oliveras A, Comes N, Villarroel A, Felipe A. Functional implications of KCNE subunit expression for the Kv7.5 (KCNQ5) channel. Cell Physiol Biochem. 2009;24(5–6):325–334. doi: 10.1159/000257425. [DOI] [PubMed] [Google Scholar]
- 44.Amro A, Waldum-Grevbo B, von der Lippe N, Brekke FB, Miaskowski C, Os I. Symptom Clusters From Dialysis to Renal Transplantation: A Five-Year Longitudinal Study. Journal of pain and symptom management. 2016;51(3):512–519. doi: 10.1016/j.jpainsymman.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Kwak SM, Choi YS, Yoon HM, Kim DG, Song SH, Lee YJ, Yeom CH, Koh SJ, Park J, Lee MA, Suh SY. The relationship between interleukin-6, tumor necrosis factor-alpha, and fatigue in terminally ill cancer patients. Palliative medicine. 2012;26(3):275–282. doi: 10.1177/0269216311406991. [DOI] [PubMed] [Google Scholar]
- 46.Romanic AM, Burns-Kurtis CL, Ao Z, Arleth AJ, Ohlstein EH. Upregulated expression of human membrane type-5 matrix metalloproteinase in kidneys from diabetic patients. Am J Physiol Renal Physiol. 2001;281(2):F309–317. doi: 10.1152/ajprenal.2001.281.2.F309. [DOI] [PubMed] [Google Scholar]
- 47.Bower JE, Lamkin DM. Inflammation and cancer-related fatigue: mechanisms, contributing factors, and treatment implications. Brain, behavior, and immunity. 2013;30(Suppl):S48–57. doi: 10.1016/j.bbi.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.