Figure 2.
Validation of the HLA-A*02:01 library with the DMF5 TCR
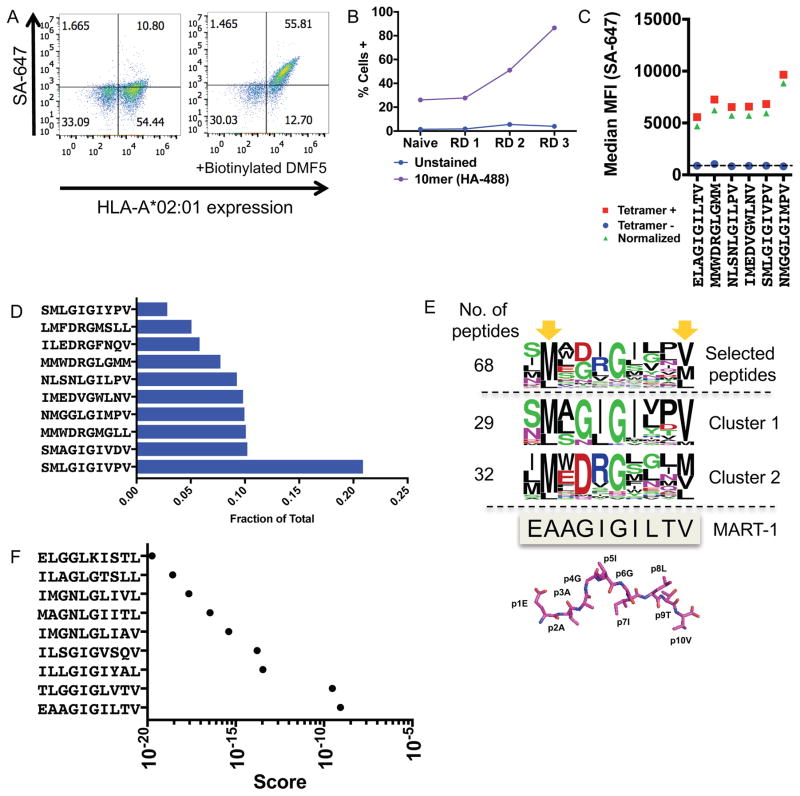
(A) The DMF5 TCR stains yeast displaying the MART-1 peptide (ELAGIGILTV) in complex with HLA-A*02:01 on the surface of yeast. Streptavidin-647 (SA-647) was used to tetramerize and fluorescently label the DMF5 TCR.
(B) Enrichment of the 10mer length HLA-A*02:01 yeast-display library by the DMF5 TCR as measured by anti-HA epitope tag staining by flow cytometry. Three of four rounds of selection shown.
(C) Highly-enriched peptides sequenced from the 10mer selection by the DMF5 TCR are stained by the DMF5 TCR tetramer and measured by flow cytometry.
(D) The fraction of total sequencing read counts of the top 10 peptides according to deep sequencing of round 3 of the 10mer HLA-A*02:01 library selections by the DMF5 TCR.
(E) Unique peptides from round 3 of selection fall into two major clusters that appear similar to the wildtype MART-1 peptide sequence. Clusters are determined by first calculating reverse hamming distance between all peptides present in round 3 of the selection and then clustered by score. The MART-1 decamer structure (PDB: 4L3E) is aligned to the selected peptides.
(F) A substitution matrix (2014PWM) using cluster 1 peptides predicts the MART-1 peptide as the most probable peptide to bind the DMF5 TCR among eight other predicted peptides.