Abstract
Background:
Acute myeloblastic leukemia (AML) is a clonal disorder due to bone marrow failure and uncontrolled proliferation of myeloid lineage. Acute promyelocytic leukemia (APL) is a subtype of AML. Heterocyclic compounds, such as indole, are considered as attractive candidates for cancer therapy, due to their abundance in nature and known biological activity. Sal-like protein (SALL4) is a zinc finger transcription factor involving in the multi-potency of stem cells, in the NB4 cell line. This study was aimed to evaluate the effects of basal indole and its new derivative, 2-(1-((2, 4-Aril)imino)-2,2,2-trifluoroethyl) phenyl-1H Indole-3- carbaldehyde (TFPHC), on the expression of SALL4.
Methods:
Cells were cultured and treated with different concentrations (75, 150, and 300 µg/mL) of the new indole derivative and DMSO, as a vehicle control, for 24 and 48 hours. Cell proliferation was evaluated by using Trypan blue exclusion and MTT assays. The percentage of apoptotic cells was determined by flowcytometry analysis using the Annexin V/PI apoptosis detection kit; mRNA expression of SALL4 was studied using absolute quantitative RT-PCR.
Results:
Our findings demonstrated the effects of new indole derivatives on SALL4 mRNA expression. Expression of SALL4 mRNA was significantly decreased at 75, 150, and 300 µg/mL concentrations.
Conclusion:
SALL4 plays a role in the survival of APL cells. SALL4 expression could be suppressed by the novel indole derivative. Additionally, SALL4 gene suppression can serve as a target in APL therapy.
Keywords: SALL4 protein, Indoles, Leukemia, Acute promyelocytic
INTRODUCTION
Acute leukemia, based on the cellular origin, is categorized into two various divisions, including acute lymphoblastic leukemia (ALL) and acute myeloblastic leukemia (AML). ALL and AML are further divided into different subcategories. Acute promyelocytic leukemia (APL) is a subtype of AML and comprises 4.8%-34% of AML[1]. There is no investigation reporting the exact prevalence of AML in Iran; however, a previous study has shown that the prevalence of the disease is high within the country[2]. Although promising advances have been made specifically via chemotherapy and other supportive cares, remission occurs only in 70-90% of patients with AML[3-5]. Chemotherapy is associated with different side effects varying from nausea and mouth sores to vomiting[4]. Indole-3-carbinol (I3C), found in vegetables, especially in the cruciferous family, has been reported to be beneficial to animal models of cancers[6-8]. There are a few studies addressing the anticancer activity of indole derivatives[9,10]. In the present study, a novel indole derivative was synthesized by our group, and its regulatory effects on the expression of Sal-like protein (SALL4) in NB4 cells were further examined.
SALL4 is a zinc finger transcription factor encoded by a member of the spalt-like gene family and regarded as a key factor for the maintenance of pluripotency in embryonic stem cells[11,12]. In normal situation, SALL4 is expressed in bone marrow stem cells but down-regulated in mature blood cells[13]. SALL4, as one of the numerable genes, has been indicated to be able to bridge the unique properties of stem cells and malignancies. Unlike its absence or decreased level in most adult tissues, SALL4 is re-expressed in various human tumors, including hematologic malignancies, as well as liver, gastric, lung, endometrial, and breast cancers[14]. SALL4 also has an antagonistic function in normal hematopoiesis and leukemia and in proliferation and differentiation of normal hemato-poiesis. However, suppression of the SALL4 gene in leukemia leads to the induction of apoptosis without notable effects on differentiation[15]. Altogether, these data confirm the important role of SALL4, as a cell survival factor, in tumor cells[16-18]. Considering the above issues, we aimed to examine the effects of both basal indole and 2-(1-((2,4-Aril)imino)-2,2,2-trifluoroethyl) phenyl-1H Indole-3-carbaldehyde (TFPHC), as a synthetic novel indole derivative on the expression of SALL4 in NB4 cells.
MATERIAL AND METHODS
Preparation of TFPHC
The chemical compound was synthesized using the following procedure. A dry, two-necked, 50-mL round-bottomed flask equipped with a nitrogen inlet was charged with 5 mL dry acetonitrile, 0.145 g (1.0 mmol) I3C, and 0.24 g (1.0 mmol) NaH. The resultant solution was stirred under nitrogen atmosphere at room temperature for 30 min. Afterwards, a solution of 2,2,2-trifluoro-N-(3-(trifluoromethyl)phenyl) acetimidoyl chloride (1.0 mmol; Sigma, USA) was added gently and dropwise via a syringe. The mixture was stirred under the N2 atmosphere at room temperature for 20 h and then was filtered. The products obtained from I3C were purified by recrystallization from ethanol (twice; Fig. 1).
Fig. 1.
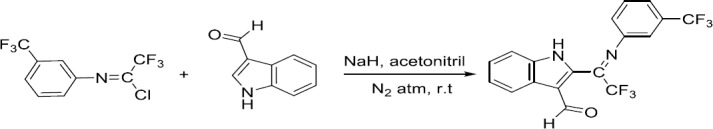
Structure of 2-(1-((2,4-Aril)imino)-2,2,2-trifluoroethyl) phenyl-1H Indole-3-carbaldehyde
The compound was obtained as a white solid, melting point = 114-116 °C, Yield = 82%, Fourier transform infrared spectroscopy (KBr) υmax = 1698, 1673, 1557 cm-1. 1H-NMR (DMSO-d6 500 MHz) δ = 10.03 (s, 1H), 8.65 (s, 1H), 8.01 (s, 1H), 7.39 (m, 2H), 7.31 (m, 2H), 7.16 (m, 3H) ppm. 19F-NMR (CFCl3 475 MHz) δ = -70.852, -62.098 ppm. Anal.Calcd for C18H10F6N2O (384.28): C, 56.26; H, 2.62; N, 7.29%. Found: C, 56.34; H, 2.73; N, 7.14%[1].
Cell culture
The NB4 cell line was purchased from the National Cell Bank of Iran (Pasteur Institute of Iran, Tehran, Iran). Cells were seeded (1 × 106 cells/well) into RPMI-1640 (Gibco Laboratories, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (Gibco Laboratories, Grand Island, NY, USA) supplemented with 100 IU/mL penicillin and 100 μg/mL streptomycin in a 37ºC humidified incubator with 95% O2-5% CO2. With appropriate confluence, cells were subjected to passage and then treated with TFPHC that was already dissolved in cell culture medium.
Cell viability assay
MTT assay
This colorimetric assay determines the MTT [3-(4,5-dimethylthiazolyl)-2,5-diphenyl-tetrazolium bromide] reduction. The MTT technique is based on the mitochondrial dehydrogenase activity to generate blue formazan product, reflecting the normal activities of mitochondria, which facilitates the measurement of upcoming cytotoxicity and cell viability.
NB4 cells were seeded at a density of 1 × 104 cells/well into a 96-well plate. The cells were then treated with different concentrations of the novel indole derivative TFPHC (75, 150, and 300 µg/mL), the vehicle control (DMSO), as well as the similar doses of basal indole. After incubation for 24 and 48 hours, the MTT reagent (5 mg/L) was added to each well and incubated for further 4 h. The supernatant was replaced by DMSO, and the relative absorbance was read at 570 nm using a microplate scanning spectrophotometer (ELISA reader, Bio Tek EIK 808, USA). The numbers of viable cells were calculated using appropriate controls. The mean ± SD values are shown from three independent experiments. The inhibition rates were also calculated according to the following formula: Inhibition rate = absorbance value of control group-absorbance value of test group/absorbance value of control group × 100%
Trypan blue-based cell viability assay
NB4 cells were seeded onto a 6-well plate at a density of 1 × 104 cells/well. Briefly, the cells were treated with different concentrations of the novel indole derivative TFPHC (75, 150, and 300 µg/mL), the vehicle control (DMSO), as well as the similar doses of basal indole. After incubation for 24 and 48 hours, the number of viable cells in each well was counted under a microscope using a hemocytometer.
Flowcytometric-based cell analysis of apoptosis
Annexin V-FITC/PI staining was used to determine the apoptotic rates. Following 24 and 48 hours of incubation with various concentrations of the new indole compound, NB4 cells of different groups were collected and transferred into the 5-mL plastic tubes and washed twice with cold PBS. The apoptosis was detected using FITC Annexin V/PI Apoptosis Detection Kit (eBioscience, USA) according to the manufacturer’s instructions using a flow cytometry machine (BD FACSCalibur, USA).
RNA extraction and cDNA synthesis
The total RNA of cells was extracted using the Trizol™ reagent (Invitrogen, USA) according to the manufacturer’s protocol (Invitrogen, USA). The purity and fidelity of RNA were checked by evaluating optical density (calculation of 260/280 nm ratio) and running onto agarose gel, respectively. The first-strand cDNA was generated using 2 µg total RNA by a high-capacity cDNA Reverse Transcription Kit (Thermo Scientific, USA). The real-time PCR analysis was conducted by application of SYBR Green I Master Mix PCR (GeNet Bio, Korea). Reactions were run using a real-time system (Bio-Rad Company, USA). The software vector NTI was used to design the specific primers for SALL4; ß-actin was used as a house keeping gene. The sequences of primers used in this study were 5’ ATTTGTGGCGGAGAGG3’ (SALL4 forward) and 5’ ACCCCAGCACATCAACTC 3’ (SALL4 reverse) and 5’GGGCATGGGTCAGAAGG ATT 3’ (β-actin forward), 5’ CGCAGCTCATTGTAG AAGGT 3’ (β-actin reverse).
Statistical analysis
Student’s t-test was applied to analyze the data using SPSS software (version 21). All experiments were performed three times, and all of the acquired results were presented as the mean ± SEM. P values less than 0.05 were considered to be significant statistically.
RESULTS
Synthesis of a new indole compound
The structure-activity analysis of I3C indicated that the substituents linked to nitrogen atom in the indole ring, which inhibits dehydration and the formation of the reactive indolenine, can increase the potency. It stabilizes the compound and prevents the formation of oligomeric self-condensation products. In contrast, the C-3 hydroxy methyl substituent on the indole ring was derived for the compound biological functions. Further, the increased number of carbons in the N-alkoxy derivatives led to the considerably increased potency, purposing that the hydrophobic properties of the substituents at this indole ring positions were created to evaluate the potency of the generated molecules. In this investigation, we designed and synthesized a new stable indole-3-carbaldehyde derivative containing two trifluoromethyl groups and one phenyl ring that increases hydrophobic and lipophilicity properties. The elevated stability of carbaldehyde, compared to I3C, may depend on the steric effect of bulky addition of the CF3 groups and substituents, which hampers oligomerization of TFPHC action formed under protonation. Hence, the biological effects of TFPHC are independent of its conversation into a derivative. To compare the efficacies of the anti-proliferative activation of TFPHC and the parental compound (I3C), NB4 cells were treated with increasing concentrations of indole for 24 and 48 hours. Our results suggested that the TFPHC has potent anti-proliferative activities.
Examination of NB4 cell viability by the MTT assay
The MTT assay was used to evaluate the inhibitory effects of new indole derivative on the growth of NB4 cells. NB4 cells were incubated with different concentrations of new indole derivative (0–300 µg/mL) for 24 and 48 hours. The results demonstrated that new indole derivative led to decreased cell viability in NB4 cells in a time and dose-dependent manner (p < 0.05; Fig. 2A).
Fig. 2.
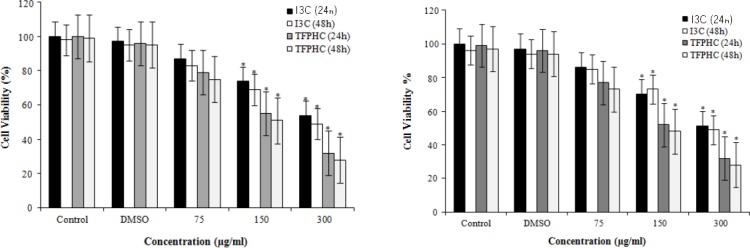
Cytotoxic effects of basal indole and TFPHC plus DMSO on NB4 cell line. (A) Analysis of MTT assay-based viability of cultured NB4 cells. NB4 cells were cultured in the presence of various concentrations of basal indole and TFPHC plus DMSO for 24 and 48 hours. Control cells were remained untreated. Cells were subjected to MTT assay for examination of their viability. (B) Analysis of Trypan-blue based-assay viability of cultured NB4 cells. NB4 cells were cultured in presence of various concentrations of basal indole and TFPHC plus DMSO for 24 and 48 hours. Control cells were remained untreated. At indicated time points, the cells were subjected to staining with Trypan-blue, and the ratio of viable to dead cells was calculated. TFPHC, 2-(1-((2, 4-Aril)imino)-2,2,2-trifluoroethyl) phenyl 1H Indole-3- carbaldehyde; I3C, indole-3-carbaldehyde; * shows significant difference with control DMSO and 75 µg/mL dose.
Examination of NB4 cell viability by Trypan blue exclusion
In Trypan blue analysis, a significant difference in cell viability decline was observed between various concentrations of new indole derivative after 24 and 48 hours (p < 0.05; Fig. 2B).
Examination of NB4 cell viability by flowcytometry
To examine the apoptosis, NB4 cells were treated with novel indole derivatives in IC50 concentrations for 24 and 48 hours. At the indicated time points, the treated cells were harvested and stained with annexin V and propidium iodide, and the apoptotic effect of new indole derivative on NB4 cells was detected by flowcytometry. The flowcytometry analysis revealed that the new indole derivative was able to induce apoptosis in NB4 cells, as compared to the control. The normal viable cells were observed in the LL (lower left) region. The early apoptotic cells were determined in LR (lower right) and the late apoptosis and necrosis cells, appeared mainly in the UR (upper right) and UL (upper left) regions, respectively (Fig. 3).
Fig. 3.
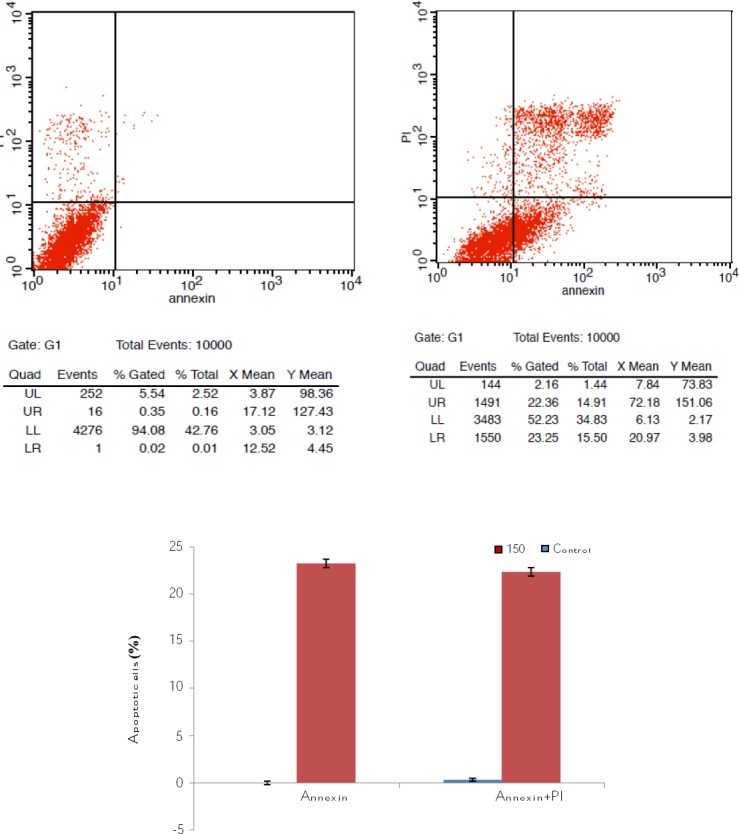
Induction of apoptosis in NB4 cells after treatment with TFPHC in comparison with untreated cells after 24 and 48 at IC50 concentrations. NB4 cells were treated with basal indole and TFPHC for 24 h and 48 h. Cells were stained with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI). Subsequently, apoptotic and necrotic cells were quantified by flow cytometry. Different subpopulations were defined as UL, Annexin V-negative but PI-positive, i.e. necrotic cells; UR, Annexin V/PI-double positive, i.e. late apoptotic cells; LL, Annexin V/PI-double negative, i.e. normal live cells; and LR, Annexin V-positive but PI-negative, i.e. early apoptotic cells. UL, upper left; UR, upper right; LR, lower right; LL, lower left
Evaluation of mRNA expression of SALL4
Results of the present study showed that the expression of SALL4 in I3C and TFPHC derivative-treated cells was down-regulated. The mRNA level of SALL4 was significantly decreased by 1.5fold after 24 hours being exposed to I3C at the dose of 300 µg/ml (p < 0.05; Fig. 4). The mRNA expression of SALL4 was also down-regulated by threefold when the cells were exposed to 15 µg/ml and 300 µg/ml of TFPHC for 24 hours (p < 0.05; Fig. 4). We observed that the expression of SALL4 mRNA was down-regulated after 48 hours in response to I3C and TFPHC (concentrations of 150 and 300 µg/ml) by 2fold and 2.5fold, respectively
Fig. 4.
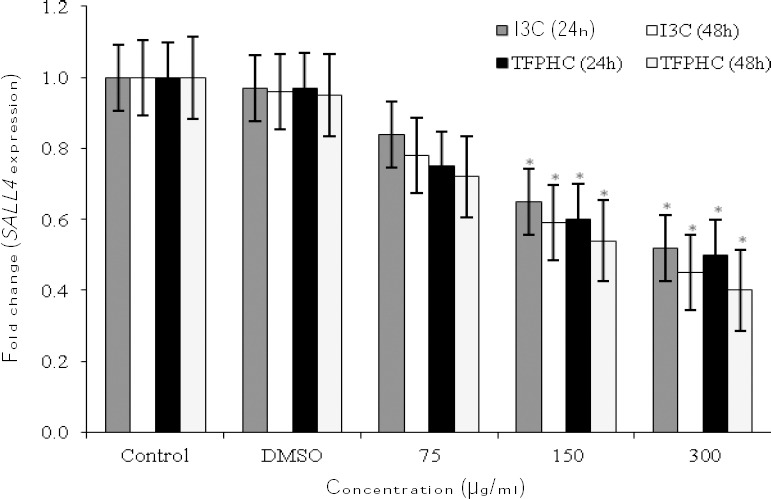
Expression of SALL4 by NB4 cells in the presence of basal indole and TFPHC following 24 h and 48 h of culture. Following 24 h of culture, cells were harvested and subjected to mRNA expression analysis. The cDNA was synthesized and real-time PCR was performed. The mRNA expression of SALL4 was then calculated against β-actin (as the housekeeping gene). TFPHC, 2-(1-((2, 4-Aril)imino)-2,2,2-trifluoroethyl) phenyl 1H Indole-3-carbaldehyde; I3C, indole-3-carbaldehyde
DISCUSSION
SALL4 is expressed in embryonic cells and plays important roles in the maintenance of pluripotency in human embryonic stem cells[12,19-21]. In human normal bone marrow, SALL4 is expressed in germ cells while being almost undetectable in other adult tissues. In other words, in normal bone marrow cells, SALL4 has a key function in hematopoietic differentiation. The present study, for the first time, showed the regulatory effects of both indole and its syntethetic derivative, TFPHC, in vitro on the expression of SALL4, as a transcription factor involving in the pluri-potency of stem cells. SALL4 has been shown to be able to aberrantly expressed in different tumor types and hematological malignancies and is involved in leukemogenesis and to have a role in cell death, cancer,
DNA replication/repair, and cell cycle[22]. SALL4 has also demonstrated to serve as an upstream target for expression of an array of apoptosis genes, including TNF, TP53, PTEN, CARD9, CARD11, CYCS, and LTA, and apoptosis inhibiting genes such as Bmi-1, BCL2, XIAP, DAD1, and TEGT. Cellular apoptosis occurred following the knocking-down of SALL4[23].
AML is amongst the most frequent acute leukemias with an increased incidence with age[24]. The pathogenesis of AML involves mutations in critical genes involved in normal cell development, cellular survival, proliferation, maturation and apoptosis[25]. The complexity of the molecular and cellular structure in AML may explain the reason why advancing the treatment of AML has been an important challenge. Previous studies on different cell lines have shown that the I3C is able to induce potent anti-proliferative activities[26,27]. Our results showed that the expression of SALL4 was significantly down-regulated in the presence of indole and its derivative as compared with DMSO and control in a dose-dependent fashion; however, the indole derivative was shown to be more potent in several folds when compared to indole itself. Because there was no comparable study within literature concerning the expression of SALL4, these data provide novel evidence that SALL4 serves as an upstream target for pro-/anti-apoptosis genes. Therefore, these findings are in agreement with the results of investigators who reported the up-regulated levels of pro-apoptotic genes, including TNF, TP53, PTEN, CARD9, CARD11, CYCS, LTA, as well as Bmi-1, BCL2, XIAP, DAD1, and TEGT[23]. Several studies approached the enhancing potencies of cytotoxicity as well as induction of apoptosis in cancer cells by synthesizing more potent I3C dimerized products[28-30]. This novel I3C derivative is one of the most potent synthetic derivatives of I3C that results in several folds increased tendency of the cell cycle arrest[10] and apoptosis[31] in NB4 cancer cells. Our recent data also demonstrated that this novel compound (TFPHC) has down-regulated both OCT4 and NANOG, the genes which are involved in cellular immortality[32].
As mentioned above, SALL4 is a key regulator of cell growth and apoptosis in leukemic cells[33]; hence, it is able to be targeted as a promising therapeutic target. In the present study, NB4 cells were treated with 75, 150, and 300 µg/mL of TFPHC for 24 hours. Further, RNA was extracted from the cultured cells, and cDNA was synthesized, followed by RNA analysis using real-time PCR. According to our data, the down-regulation of SALL4 in the presence of TFPHC (the new indole derivative) may explain a mechanism by which the compound prevents the proliferation of NB4 cells (via decreasing the SALL4 expression).
The role of SALL4 in APL is unknown, but this study showed that this gene may probably be involved in pathophysiology of this disease, and TFPHC is able to reduce the expression of SALL4. Further studies are required to address the exact role of SALL4 in APL. In addition, this study demonstrated that targeting of SALL4 by TFPHC can possibly be used as a promising treatment for APL.
ACKNOWLEDGEMENTS
The financial support of the present study by Kerman University of Medical Sciences (Kerman, Iran) is acknowledged.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Ribeiro RC, Rego E. Management of APL in developing countries:epidemiology, challenges and opportunities for international collaboration. ASH education program book. 2006;2006(1):162–168. doi: 10.1182/asheducation-2006.1.162. [DOI] [PubMed] [Google Scholar]
- 2.Ziaei JE. High frequency of acute promyelocytic leukemia in northwest Iran. Asian pacific journal of cancer prevention. 2004;5(2):188–189. [PubMed] [Google Scholar]
- 3.Bellm LA, Epstein JB, Rose-Ped A, Martin P, Fuchs HJ. Patient reports of complications of bone marrow transplantation. Supportive care in cancer. 2000;8(1):33–39. doi: 10.1007/s005209900095. [DOI] [PubMed] [Google Scholar]
- 4.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106(4):1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 5.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia:rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 6.Jin L, Qi M, Chen DZ, Anderson A, Yang G-Y, Arbeit JM, Auborn KJ. Indole-3-carbinol prevents cervical cancer in human papillomavirus type 16 (HPV16) transgenic mice. Cancer research. 1999;59(16):3991–3997. [PubMed] [Google Scholar]
- 7.Kojima T, Tanaka T, Mori H. Chemoprevention of spontaneous endometrial cancer in female Donryu rats by dietary indole-3-carbinol. Cancer research. 1994;54(6):1446–1449. [PubMed] [Google Scholar]
- 8.Oganesian A, Hendricks JD, Williams DE. Long term dietary indole-3-carbinol inhibits diethylnitrosamine-initiated hepatocarcinogenesis in the infant mouse model. Cancer letters. 1997;118(1):87–94. doi: 10.1016/s0304-3835(97)00235-8. [DOI] [PubMed] [Google Scholar]
- 9.Fraley ME, Steen JT, Brnardic EJ, Arrington KL, Spencer KL, Hanney BA, Kim Y, Hartman GD, Stirdivant SM, Drakas BA, Rickert K, Walsh ES, Hamilton K, Buser CA, Hardwick J, Tao W, Beck SC, Mao X, Lobell RB, Sepp-Lorenzino L, Yan Y, Ikuta M, Munshi SK, Kuo LC, Kreatsoulas C. 3-(Indol-2-yl) indazoles as Chek1 kinase inhibitors:Optimization of potency and selectivity via substitution at C6. Bioorganic and medicinal chemistry letters. 2006;16(23):6049–6053. doi: 10.1016/j.bmcl.2006.08.118. [DOI] [PubMed] [Google Scholar]
- 10.Karimabad MN, Mahmoodi M, Jafarzadeh A, Darekordi A, Hajizadeh MR, Khorramdelazad H, Sayadi AR, Khanamani Falahati-Pour S, Hassanshahi G. Regulatory effects of the novel synthesized Indole-3-carbaldehyde on expression of cell cycle genes:A study on Cyclin D and P21 expression by acute promylocytic leukemia cell line (NB4) Cellular and molecular biology (Noisy-le-grand) 2017;63(5):60–67. doi: 10.14715/cmb/2017.63.5.12. [DOI] [PubMed] [Google Scholar]
- 11.Kohlhase J, Schuh R, Dowe G, Kühnlein RP, Jäckle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Köhler A, Müller U, Raab-Vetter M, Burkhardt E, Engel W, Stick R. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the drosophila gene spalt. Genomics. 1996;38(3):291–298. doi: 10.1006/geno.1996.0631. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS one. 2010;5(5):e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao C, Kong NR, Li A, Tatetu H, Ueno S, Yang Y, He J, Yang J, Ma Y, Kao GS, Tenen DG, Chai L. SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion. 2013;53(5):1037–1049. doi: 10.1111/j.1537-2995.2012.03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong H-W, Cui W, Yang Y, Lu J, He J, Li A, Song D, Guo Y, Liu BH, Chai L. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PLoS one. 2011;6(4):e18372. doi: 10.1371/journal.pone.0018372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Liu L, Leung L-H, Cooney AJ, Chen C, Rosengart TK, Ma Y, Yang J. Knockdown of SALL4 protein enhances all-trans retinoic acid-induced cellular differentiation in acute myeloid leukemia cells. Journal of biological chemistry. 2015;290(17):10599–10609. doi: 10.1074/jbc.M114.634790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueno S, Lu J, He J, Li A, Zhang X, Ritz J, Silberstein LE1, Chai L. Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia:Mechanism, function, and implication for a potential novel therapeutic target. Experimental hematology. 2014;42(4):307–316. e8. doi: 10.1016/j.exphem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki E, Chiba T, Yokosuka O. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. The new England journal of medicine. 2013;369(12):2266–2276. doi: 10.1056/NEJMc1308785. [DOI] [PubMed] [Google Scholar]
- 18.Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan B, Srivastava S, Lim GS, Tang P, Yang H, Tenen DG, Chai L. SALL4 is a new target in endometrial cancer. Oncogene. 2015;34(1):63–72. doi: 10.1038/onc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darehkordi A, Ramezani M. One-pot synthesis of novel (2R,4S)-N-aryl-4-hydroxy-1-(2,2,2-trifluoroacetyl) pyr-rolidine-2-carboxamides via TiO2TiO2-NPs and Pd (PPh3)2Cl2Pd(PPh3)2Cl2 catalysts and investigation of their biological activities. Molecular diversity. 2017;21(2):305–315. doi: 10.1007/s11030-017-9726-y. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W, Ji X, Zhang F, Li L, Ma L. Embryonic stem cell markers. Molecules. 2012;17(6):6196–6236. doi: 10.3390/molecules17066196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell stem cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nature cell biology. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Zhao W, Kong N, Cui W, Chai L. The next new target in leukemia:The embryonic stem cell gene SALL4. Molecular and cellular oncology. 2014;1(4):e969169. doi: 10.4161/23723548.2014.969169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D, Fink LM, Ward DC, Ma Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112(3):805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto JF, Goodman MT. Patterns of leukemia incidence in the United States by subtype and demographic characteristics 1997–2002. Cancer causes and control. 2008;19(4):379–390. doi: 10.1007/s10552-007-9097-2. [DOI] [PubMed] [Google Scholar]
- 26.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, Huberman K, Cheng J, Viale A, Socci ND, Heguy A, Cherry A, Vance G, Higgins RR, Ketterling RP, Gallagher RE, Litzow M, van den Brink MR, Lazarus HM, Rowe JM, Luger S, Ferrando A, Paietta E, Tallman MS, Melnick A, Abdel-Wahab O, Levine RL. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. The new England journal of medicine. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gali R, Banothu J, Porika M, Velpula R, Hnamte S, Bavantula R, Abbagani S2, Busi S. Indolylmethylene benzo [h] thiazolo [2,3-b] quinazolinones:synthesis, characterization and evaluation of anticancer and antimicrobial activities. Bioorganic and medicinal chemistry letters. 2014;24(17):4239–4242. doi: 10.1016/j.bmcl.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Kim YS, Milner JA. Targets for indole-3-carbinol in cancer prevention. The journal of nutritional biochemistry. 2005;16(2):65–73. doi: 10.1016/j.jnutbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Abdelrahim M, Newman K, Vanderlaag K, Samudio I, Safe S. 3, 3′-diindolylmethane (DIM) and its derivatives induce apoptosis in pancreatic cancer cells through endoplasmic reticulum stress-dependent upregulation of DR5. Carcinogenesis. 2006;27(4):717–728. doi: 10.1093/carcin/bgi270. [DOI] [PubMed] [Google Scholar]
- 30.Maciejewska D, Rasztawicka M, Wolska I, Anuszewska E, Gruber B. Novel 3, 3′-diindolylmethane derivatives:Synthesis and cytotoxicity, structural characterization in solid state. The European journal of medicinal chemistry. 2009;44(10):4136–4147. doi: 10.1016/j.ejmech.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Karimabad MN, Mahmoodi M, Jafarzadeh A, Darehkordi A, Hajizadeh MR, Khorramdelazad H, Falahati-Pour SK, Hassanshahi G. The novel Indole-3-formaldehyde (2-AITFEI-3-F) is involved in processes of apoptosis induction? Life science. 2017;181:31–44. doi: 10.1016/j.lfs.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Karimabad MN, Mahmoodi M, Jafarzadeh A, Darehkordi A, Hajizadeh MR, Khorramdelazad H, Sayadi AR, Rahmani F, Hassanshahi G. Evaluating of OCT-4 and NANOG was differntially regulated by a new derivative indole in leukemia cell line. Immunology letters. 2017;190:7–14. doi: 10.1016/j.imlet.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Ueno S, Lu J, He J, Li A, Zhang X, Ritz J, Silberstein LE, Chai L. Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia:Mechanism, function, and implication for a potential novel therapeutic target. Experimental hematology. 2014;42(4):307–316. doi: 10.1016/j.exphem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


