ABSTRACT
Candida auris has emerged globally as a multidrug-resistant health care-associated fungal pathogen. Recent reports highlight ongoing challenges due to organism misidentification, high rates of antifungal drug resistance, and significant patient mortality. The predilection for transmission within and between health care facilities possibly promoted by virulence factors that facilitate skin colonization and environmental persistence is unique among Candida species. This minireview details the global emergence of C. auris and discusses issues relevant to clinical microbiology laboratories, hospital infection control, and antimicrobial stewardship efforts.
KEYWORDS: antifungal susceptibility, Candida auris, epidemiology, infection control
INTRODUCTION
Candida bloodstream infections (BSIs) are the third to fourth most common cause of health care-associated BSIs (1, 2). Candida albicans has historically been considered the most pathogenic species in the genus and remains the most common cause of candidemia. In 2009, a new fluconazole-resistant species, Candida auris, was identified in East Asia and has now been isolated on five continents. This global emergence has been attributed to the near-simultaneous appearance of at least 4 geographically restricted clades, with clonal transmission identified both within and across health care facilities. C. auris can be challenging to identify in the clinical laboratory, and this species frequently exhibits multidrug resistance. Alarmingly, invasive infections have been difficult to treat and are associated with high rates of treatment failure. This minireview summarizes what is currently known about C. auris infection epidemiology, pathogenesis, diagnosis, and management.
EPIDEMIOLOGY
C. auris was first isolated in 2009 from the external ear canal of a patient in Japan, with ribosomal DNA (rDNA) sequencing and biochemical analyses indicating identification of a novel Candida species (3). C. auris was subsequently reported the same year from 15 patients with chronic otitis media in South Korea (4). The first three cases of C. auris bloodstream infection (BSI) were also reported in South Korea in 2011 (5). A retrospective review of South Korean Candida isolates, however, showed that the earliest strain of C. auris actually dated back to 1996 and was identified from a BSI in a child (4). The majority of the South Korean isolates displayed elevated fluconazole MICs, which suggested that this new species is largely resistant to fluconazole.
Ensuing reports of C. auris infections have come from multiple countries around the world, including India, South Africa, Kuwait, Malaysia, the United Kingdom, Pakistan, Kenya, Norway, Germany, Oman, Spain, Israel, Venezuela, Columbia, Panama, Brazil, the United States, and Canada (6–18). The U.S. Centers for Disease Control and Prevention (CDC) continues to map new case reports and transmission globally (https://www.cdc.gov/fungal/diseases/candidiasis/tracking-c-auris.html). Given the flurry of recent reports, a review of >15,000 Candida isolates deposited in the international SENTRY repository was conducted to look for previously misidentified strains (12). SENTRY began cataloguing Candida isolates in 2004, yet only 4 previously misidentified C. auris cultures were found in the search, implying that this pathogen has recently emerged.
Clinical C. auris isolates have been recovered from a variety of specimen types, including normally sterile body fluids, respiratory sections, urine, bile, tissues, wounds, and mucocutaneous swabs. BSI is the most commonly observed invasive infection, with in-hospital mortality rates reported on the order of 30 to 60% (5, 8, 19). Cases have mainly involved patients with underlying medical comorbidities and significant health care exposure, with infection typically occurring weeks after hospital admission. Overlapping contact with long-term-care facilities or acute-care hospitals in the same geographic region was also common among cases (18, 20). In a recent report from India, the risk for the development of C. auris BSI compared to other Candida spp. was associated with longer intensive care unit (ICU) stays, underlying respiratory illness, vascular surgery, prior antifungal drug exposure, and lower acute physiology and chronic health evaluation II (APACHE II) scores (21). The majority of the C. auris isolates in this study were clonal and came from public hospitals in the northern portion of the country.
Beginning in 2016, the CDC, the European Centre for Disease Prevention and Control (ECDC), and Public Health England, among others, released a series of alerts to inform clinicians, laboratorians, infection prevention practitioners, and public health officials about the emerging health threat posed by C. auris and to request that all cases be reported to local, state, or national health departments (see https://www.cdc.gov/fungal/diseases/candidiasis/c-auris-alert-09-17.html, https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/Candida-in-healthcare-settings_19-Dec-2016.pdf, and https://www.gov.uk/government/publications/candida-auris-emergence-in-england). As of September 2017, a total of 127 confirmed and 27 probable cases from 10 states have been reported to the CDC. New York and New Jersey have identified the majority of the confirmed cases, reporting 86 and 26, respectively (https://www.cdc.gov/fungal/diseases/candidiasis/tracking-c-auris.html). Increased numbers of isolates from the New Jersey and New York area are possibly related to population density, geographic health care facility interconnectedness, and/or heightened surveillance efforts. Whole-genome sequencing (WGS) analysis with comparison to previously identified international strains found that isolates within reporting states were highly related (18). Investigation of the first isolates from New York found that they were similar to isolates from South Asia, New Jersey isolates were also similar to South Asian isolates but distinct from those identified in New York, and isolates from Illinois were grouped with isolates from South America. Available epidemiologic information suggested that many of these infections were acquired in the United States. Additionally, three of the four existing lineages have been detected in the United Kingdom (7). The observation that different clonal lineages are now distributed over large geographic distances supports the idea of multiple independent introductions of C. auris into the United States and U.K., with subsequent local spread.
MICROBIOLOGY
C. auris isolates have been analyzed using a variety of laboratory methods in an attempt to (i) establish phylogenetic relationships with other pathogenic yeasts, (ii) evaluate potential relatedness across strains, (iii) determine virulence, and (iv) characterize antifungal susceptibility profiles.
TAXONOMY
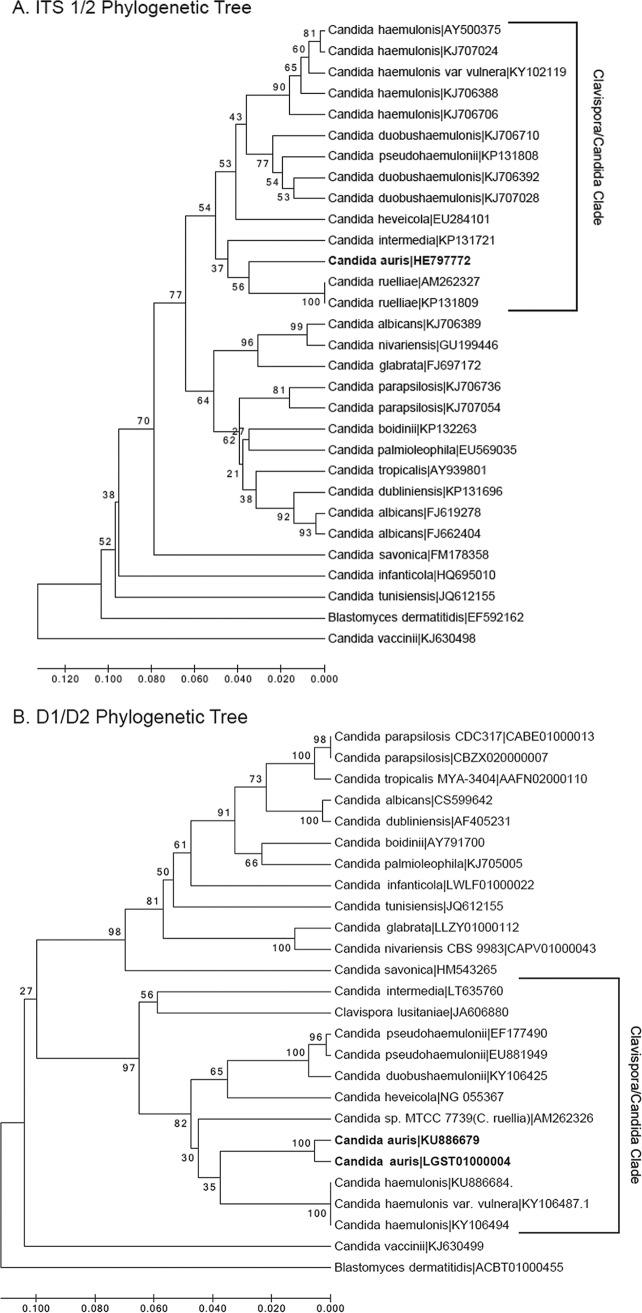
rDNA sequencing of the 26S D1/D2 and 18S internal transcribed spacer (ITS) regions placed the initial Japanese isolate within the Metschnikowiaceae family, most closely related to species of the Clavispora clade (3, 22). D1/D2 sequences from the initial isolate clustered most closely with Candida haemulonii, Candida pseudohaemulonii, and Candida ruelliae (85.7, 83.0%, and 82.4% similarity, respectively), and the ITS 1/2 region most closely matched C. haemulonii, C. pseudohaemulonii, and C. heveicola (87.5, 81.4, and 81.3% sequence similarity, respectively). A phylogenetic tree based on ITS 1/2 and D1/D2 sequences deposited in publically available databases illustrates the position of C. auris within the Clavispora/Candida clade (Fig. 1).
FIG 1.
Dendrograms of Candida auris with other Candida species. Trimmed internal transcribed spacer 1/2 (A) and the long subunit (LSU) D1/D2 sequences (B) were aligned using MEGA7 software. The same software was used to draw the bootstrap (500 replicates) unweighted pair group method using average linkages (UPGMA) dendrograms constructed using a Kimura 2-parameter genetic distances model. Also of note, C. haemulonii (synonym C. haemulonis) is now considered a species complex composed of two species and one variety: C. haemulonii sensu stricto, C. duobushaemulonii, and C. haemulonis var. vulnera (45), and Clavispora lusitaniae is the current name for Candida lusitaniae. Accession numbers are shown to the right of the organism names.
C. auris isolates from India, South Africa, Brazil, and/or South Korea have also been analyzed with classical typing methods, including pulsed-field gel electrophoresis, M13 phage typing, multilocus sequence typing (MLST), and/or amplified fragment length polymorphism (AFLP) fingerprinting (10, 23–25). These were the first studies to suggest geographical strain clustering, but the inherently low discriminatory power of traditional molecular typing methods precluded comparisons of relatedness among isolates with identical fingerprints.
WGS has also been applied to an international collection of isolates in an effort to better assess the relatedness to other Candida species and evaluate clonality among strains (12, 26). The assembled C. auris genome is diploid and has a size range between 12.3 and 12.5 Mb, a G+C content of 44.5 to 44.8%, and approximately 6,600 to 8,300 coding sequences (27, 28). Draft genome comparisons revealed that the majority (≥99.5%) of C. auris genomic reads did not align with previously sequenced C. albicans, Candida glabrata, Candida lusitaniae, and Saccharomyces cerevisiae strains, which suggests that C. auris is highly divergent at the whole-genome level (27). However, compared to each other, C. auris isolates can be grouped into geographic-specific clusters, with unique South Asian (India and Pakistan), South African, South American (Venezuela), and East Asian (Japan) clades identified. Each of the 4 geographically distinct clades was separated by tens of thousands of single-nucleotide polymorphisms (SNPs), while the isolates that grouped within each geospatial clade appeared to be clonal (12).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI TOF MS) has also been used to examine strain relatedness. Protein profiling with cluster analysis also showed evidence of geographically distinct groups, but this did not correlate perfectly with the results of standard genotyping methods (24). It should be noted that MALDI TOF MS has not necessarily been validated or optimized as a tool for strain typing and is limited in the number of proteins that can be detected for strain characterization relative to higher-resolution MS methods.
VIRULENCE FACTORS
Several studies have been conducted to characterize C. auris virulence determinants. Germination, adherence, biofilm formation, phospholipase, and proteinase production are all known to contribute to Candida pathogenesis (27). C. auris does not appear to form germ tubes, pseudohyphae, or chlamydospores in vitro (5, 9, 28) but may produce phospholipase and proteinase in a strain-dependent manner (29, 30). Although C. auris is able to adhere to plastics and form biofilms, these capabilities are significantly reduced relative to those of C. albicans (30, 31). An additional intriguing observation is that some C. auris isolates grow in clumps (i.e., budding occurs but daughter cells are not released), which results in large aggregates of organisms that cannot be easily disrupted in vitro (28). In the Galleria mellonella insect model, aggregate-forming strains were found to be significantly less pathogenic than nonaggregating isolates (28). Interestingly, the most virulent nonaggregative forms exhibited pathogenicity on par with C. albicans, despite the fact they did not form hyphal or pseudohyphal elements in G. mellonella larvae. In a murine model of hematogenous disseminated candidiasis, C. auris cell aggregates were also observed in the kidneys of infected animals compared to the invasive hyphal forms of C. albicans (6). Additionally, inoculation with C. auris was associated with longer median survival than with C. albicans (6). Whether these growth characteristics affect the course of infection in other animal models or humans is not known.
IDENTIFICATION IN THE CLINICAL LABORATORY
Candida isolates from normally sterile body sites should be identified to the species level in order to guide initial therapy based on predictable species-specific susceptibility. Additionally, the CDC recommends considering identifying Candida isolates from nonsterile sites in select situations, such as when a case of C. auris has been identified in a health care facility or when a patient has health care exposure in a location outside the United States where C. auris has been reported. Laboratories should also review past records to identify confirmed or suspected cases as well as conduct prospective surveillance. However, accurate identification of C. auris infection in the clinical laboratory can be problematic especially when relying on phenotypic characteristics.
PHENOTYPIC CHARACTERISTICS
C. auris strains have been described as nondescript white- to cream-colored colonies on Sabouraud dextrose agar (SDA) and pink to beige on CHROMagar. They form ovoid to elongated budding yeast cells without pseudohyphae on corn meal or rice Tween 80 agar, grow well at 37°C and 42°C, and assimilate N-acetylglucosamine, succinate, and gluconate (3, 32). In contrast, C. haemulonii and C. duobushaemulonii strains produce pseudohyphae, do not grow well at 42°C, and do not assimilate the same sugars. Based on these observations, a small study proposed using temperature tolerance and CHROMagar supplemented with Pal's medium as a means to morphologically separate C. auris from members of the closely related C. haemulonii complex (33).
BIOCHEMICAL METHODS
Misidentifications of C. auris as other Candida species by commercial biochemical methods have been widely reported, likely because of a lack of representative organisms in currently available databases. Clinical laboratories should be especially alert to the possibility of C. auris when an isolate is identified by biochemical methods as C. haemulonii. Table 1 summarizes possible C. auris misidentifications grouped by identification method.
TABLE 1.
Common Candida auris misidentifications by commercial biochemical test methoda
| Biochemical method | Misidentification |
|---|---|
| All methods | Candida haemulonii |
| Candida spp. not otherwise specified | |
| API 20C AUX | Candida sake |
| Rhodotorula glutinisb | |
| BD Phoenix | Candida catenulata |
| MicroScan | Candida catenulata |
| Candida famata | |
| Candida guilliermondiic | |
| Candida lusitaniaec | |
| Candida parapsilosisc | |
| Vitek2 | Candida duobushaemulonii |
| Candida famata |
When the characteristic red/pink pigment is absent and urease reaction is negative.
When no blastoconidia/pseudohyphae are present on cornmeal agar.
MALDI-TOF MS
MALDI-TOF MS is a potentially accurate and rapid method for the identification C. auris from pure culture. However, current FDA-cleared libraries for use with the MALDI Biotyper (Bruker, Billerica, MA) and Vitek MS (bioMérieux, Marcy l'Etoile, France) platforms do not include C. auris reference spectra. Identification requires the research use only (RUO) databases for these instruments or, alternatively, the creation of custom libraries. C. auris was added to the MALDI Biotyper RUO library in 2013 (database version 5627 and later). The Saramis version 4.14 RUO database with the Saccharomycetaceae patch is required for a higher confidence that C. auris will not be missed when using the Vitek MS.
A recent multicenter study evaluated the accuracy of current RUO databases for the identification of C. auris versus closely related species (34). Both platforms correctly identified 100% of the test isolates; the Vitek MS accomplished this using the direct on-slide extraction method, whereas the MALDI Biotyper required a separate tube extraction to obtain high-confidence scores. In contrast, another report that included only 2 C. auris strains observed that the Bruker RUO system correctly identified both isolates, while the Vitek MS RUO reported “no species ID” calls (35). Supplementation of the Vitek MS RUO database with in-house generated spectra subsequently resulted in both C. auris isolates being correctly identified to the species level. The variable Vitek MS RUO results may be related to the RUO database employed. Mizusawa et al. did not specify which Vitek MS RUO database version was used in their study (34), while Grenfell et al. reported running version 4.12, which may be why their two C. auris isolates were missed (35).
MOLECULAR METHODS
rDNA sequencing of the 28s D1/D2 region or ITS 1/2 will accurately identify C. auris to the species level. Additionally, PCR-based methods have also been developed. Kordalewska et al. designed a real-time PCR assay to detect C. auris and to differentiate it from C. haemulonii, C. duobushaemulonii, or C. lusitaniae via melt curve analysis (36). The C. auris primers showed excellent selectivity versus common fungal pathogens as well as human DNA, but relatively few members of the Clavispora clade known to share similar ITS sequences were evaluated. The development of a PCR-based test is of particular interest, especially because this approach may be useful for patient and environmental screening from swabs or for direct detection from other clinical specimens.
ANTIFUNGAL DRUG RESISTANCE
One of the reasons the emergence of C. auris has been so alarming is the potential for these organisms to harbor or develop multidrug resistance. In fact, some isolates have demonstrated elevated MICs to all available drug classes, indicating that treatment options for panresistant isolates would be extremely challenging if not impossible.
PHENOTYPIC SUSCEPTIBILITY
Currently, there are no species-specific antifungal susceptibility breakpoints established for C. auris. The isolates tested to date have uniformly displayed elevated fluconazole MICs (i.e., ≥16 μg/ml), along with varied susceptibilities to other azoles, amphotericin, and the echinocandins (9, 12, 24, 30, 32, 37). Table 2 summarizes several of the larger drug susceptibility surveys. Posaconazole followed by isavuconazole have generally been the most potent azoles in vitro, and fewer isolates have had elevated amphotericin or echinocandin MICs. Of note, overrepresentation of clonal strains in any one study has the potential to skew susceptibility patterns, and several of the larger reports have included a significant proportion of isolates from India. It should also be recognized that falsely elevated amphotericin and caspofungin MICs have been reported using the Vitek 2 susceptibility test method (13, 32).
TABLE 2.
Candida auris antifungal MIC ranges determined by the CLSI broth microdilution reference method
| Drug | Tentativea resistance breakpoints | MIC range (μg/ml)b |
||
|---|---|---|---|---|
| MIC | MIC50 | MIC90 | ||
| Fluconazole | ≥32 | 0.12 to ≥64 | ≥64 | ≥64 |
| Voriconazole | NA | 0.032 to 16 | 0.5 to 2 | 2 to 8 |
| Itraconazole | NA | 0.032 to 2 | 0.06 to 0.5 | 0.25 to 1 |
| Posaconazole | NA | 0.015 to 16 | 0.016 to 0.5 | 0.125 to 2 |
| Isavuconazole | NA | 0.015 to 4 | 0.125 to 0.25 | 0.5 to 2 |
| Anidulafungin | ≥4 | 0.015 to 16 | 0.125 to 0.5 | 0.5 to 1 |
| Caspofungin | ≥2 | 0.03 to 16 | 0.25 to 1 | 1 to 2 |
| Micafungin | ≥4 | 0.015 to 8 | 0.125 to 0.25 | 0.25 to 2 |
| Amphotericin | ≥2 | 0.06 to 8 | 0.5 to 1 | 2 to 4 |
Tentative breakpoints proposed by the CDC (https://www.cdc.gov/fungal/diseases/candidiasis/recommendations.html). NA, the CDC has recommended using fluconazole as a surrogate.
MOLECULAR RESISTANCE DETERMINANTS
Molecular mechanisms for azole resistance in C. albicans have been well defined and include: overexpression of multidrug transporters, mutations in lanosterol demethylase (encoded by ERG11), increased expression of ERG11, and an alteration in the sterol biosynthesis pathway which results in the replacement of ergosterol by other sterols in the cytoplasmic membrane (encoded by ERG3). Additionally, mutations in Candida FKS genes result in elevated echinocandin MICs and have been linked to treatment failures. WGS analyses have shown that ERG3, ERG11, FKS1, FKS2, and FKS3 homologs were present as a single copy in the C. auris genome and that these loci share 78% to 85% similarity with C. albicans and C. glabrata genes (26, 38). Additionally, a significant proportion of the C. auris genome is predicted to encode ATP-binding cassette (ABC) and major facilitator superfamily (MFS) efflux pumps (26, 38).
Molecular characterization of drug resistance mechanisms in C. auris has mostly focused on alterations in the lanosterol demethylase enzyme. Lockhart et al. evaluated ERG11 sequences in a global collection of 54 C. auris isolates and determined that substitutions linked to fluconazole resistance in C. albicans were also present in C. auris, and these mutations were strongly associated with geographic clades (i.e., F126T in South Africa, Y132F in Venezuela, and Y132F or K143R in India/Pakistan) (12). Ben-Ami et al. also observed increased ABC-type transporter activity in C. auris compared to that in C. glabrata and C. haemulonii, which suggests that efflux pumps may additionally be involved in antifungal drug resistance (6).
TREATMENT
The optimal treatment regimen for C. auris has not been defined. Because the majority of C. auris isolates identified in the United States have been susceptible to echinocandins, treatment with a drug from this class, along with infectious diseases consultation, is suggested for initial therapy. Alternatively, the treatment of colonization without evidence of active infection is strongly discouraged (https://www.cdc.gov/fungal/diseases/candidiasis/c-auris-treatment.html). Close monitoring for treatment failure and the emergence of resistance upon treatment is required. Preventing C. auris infection altogether is ideal. Many patients with reported C. auris infection or colonization received antecedent broad-spectrum antibiotics and/or antifungals. The implementation of antimicrobial stewardship programs is warranted to minimize unnecessary antibiotic use. Stewardship teams can also assist with rapid case identification, appropriate management, and coordinate with infection prevention to minimize transmission.
INFECTION PREVENTION AND CONTROL
The transmissibility of C. auris within hospitals, especially critical-care settings, has been established (17). Although the precise mode of transmission has yet to be defined, spread from the patient or their environment to the hands of health care workers seems highly plausible. Skin or mucosal colonization of affected patients appears to be common, and the organism has been recovered from a variety of patient contact points, such as mattresses, furniture, sinks, and medical equipment (17, 18). Candida spp. can be transferred from environmental surfaces to hands (39, 40), and C. auris has been shown to persist on plastics ex vivo for at least 14 days, with viability testing indicating that cells are also capable of entering a metabolically active, but nonculturable, state that persisted for 4 weeks (41).
Given the propensity for C. auris to cause outbreaks, the CDC has emphasized adherence to good hand hygiene combined with standard and contact precautions, housing of infected patients in private rooms, performance of thorough daily cleaning, and terminal room disinfection with an agent that is effective against Clostridium difficile (https://www.cdc.gov/fungal/diseases/candidiasis/c-auris-infection-control.html#disinfection). Recent evaluations suggest that nonsporocidal hydrogen peroxide and chlorine-based disinfectants are also effective against C. auris (42–44), but the widely used quaternary ammonium products have relatively poor activity (43). Iodine-based skin cleansers and chlorhexidine (depending on the formulation) also have demonstrated effective C. auris killing (42, 44). However, patients continued to be colonized with C. auris despite daily chlorhexidine washes in a prolonged outbreak setting, potentially as a result of reinfection from the environment or due to reduced chlorhexidine susceptibility (17). Additional work is required to determine the optimal approach to reducing skin colonization.
SUMMARY
C. auris infections continue to be reported across the globe, often with high associated morbidity and mortality. Accurate species-level identification is crucial for case detection and the implementation of appropriate infection prevention measures. Clinical laboratories should have a high suspicion for C. auris when certain yeasts (e.g., C. haemulonii or Candida spp.) are identified by FDA-cleared biochemical or MALDI-TOF MS methods. rDNA sequencing remains the reference method for C. auris species-level identification, but RUO MALDI-TOF MS databases and/or well-designed PCR assays are potentially more rapid alternatives. Suspected and confirmed cases should be reported to public health authorities, with antifungal susceptibility testing performed for invasive infections even though there are no established species-specific breakpoints. Based on available MIC data, the echinocandins appear to be the best choice for initial treatment. Partnerships between clinical laboratories, antimicrobial stewardship, and hospital infection prevention and control are essential for optimizing diagnosis and management and curtailing nosocomial spread.
ACKNOWLEDGMENT
We give special thanks to Keith Simmons for assistance creating the rRNA phylogenetic trees.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 41:1232–1239. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 3.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 5.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 8.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 10.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R. 2015. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohsin J, Hagen F, Al-Balushi ZAM, de Hoog GS, Chowdhary A, Meis JF, Al-Hatmi AMS. 2017. The first cases of Candida auris candidaemia in Oman. Mycoses 60:569–575. doi: 10.1111/myc.12647. [DOI] [PubMed] [Google Scholar]
- 15.Morales-López SE, Parra-Giraldo CM, Ceballos-Garzon A, Martinez HP, Rodriguez GJ, Alvarez-Moreno CA, Rodriguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz Gaitán AC, Moret A, Lopez Hontangas JL, Molina JM, Aleixandre Lopez AI, Cabezas AH, Mollar Maseres J, Arcas RC, Gomez Ruiz MD, Chiveli MA, Canton E, Peman J. 2017. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34:23–27. doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2017. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013–August 2016. Am J Transplant 17:296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 20.Tsay S, Welsh RM, Adams EH, Chow NA, Gade L, Berkow EL, Poirot E, Lutterloh E, Quinn M, Chaturvedi S, Kerins J, Black SR, Kemble SK, Barrett PM, Barton K, Shannon DJ, Bradley K, Lockhart SR, Litvintseva AP, Moulton-Meissner H, Shugart A, Kallen A, Vallabhaneni S, Chiller TM, Jackson BR. 2017. Notes from the field: ongoing transmission of Candida auris in health care facilities—United States, June 2016–May 2017. MMWR Morb Mortal Wkly Rep 66:514–515. doi: 10.15585/mmwr.mm6619a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, Chhina D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 22.Diezmann S, Cox CJ, Schönian G, Vilgalys RJ, Mitchell TG. 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol 42:5624–5635. doi: 10.1128/JCM.42.12.5624-5635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh BJ, Shin JH, Kim MN, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW. 2011. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 49:98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 24.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e271–277.e279. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 26.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US. 2015. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 16:686. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borman AM, Szekely A, Johnson EM. 2016. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere 1:e00189-16. doi: 10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar D, Banerjee T, Pratap CB, Tilak R. 2015. Itraconazole-resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis. J Infect Dev Ctries 9:435–437. doi: 10.3855/jidc.4582. [DOI] [PubMed] [Google Scholar]
- 30.Larkin E, Hager C, Chandra J, Mukherjee PK, Retuerto M, Salem I, Long L, Isham N, Kovanda L, Borroto-Esoda K, Wring S, Angulo D, Ghannoum M. 2017. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother 61:e02396-16. doi: 10.1128/AAC.02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherry L, Ramage G, Kean R, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R. 2017. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis 23:328–331. doi: 10.3201/eid2302.161320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Sachu A, Mohan K, Vinod V, Dinesh K, Karim S. 2017. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal's medium. Rev Iberoam Micol 34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grenfell RC, da Silva Junior AR, Del Negro GM, Munhoz RB, Gimenes VM, Assis DM, Rockstroh AC, Motta AL, Rossi F, Juliano L, Benard G, de Almeida Junior JN. 2016. Identification of Candida haemulonii complex species: use of ClinProTools to overcome limitations of the Bruker Biotyper, VITEK MS IVD, and VITEK MS RUO databases. Front Microbiol 7:940. doi: 10.3389/fmicb.2016.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Candida auris: comparison of the EUCAST and CLSI reference microdilution MICs for eight antifungal compounds and associated tentative ECOFFs. Antimicrob Agents Chemother 61:e00485-17. doi: 10.1128/aac.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. 2015. Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc 3:e00722-15. doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangel-Frausto MS, Houston AK, Bale MJ, Fu C, Wenzel RP. 1994. An experimental model for study of Candida survival and transmission in human volunteers. Eur J Clin Microbiol Infect Dis 13:590–595. doi: 10.1007/BF01971311. [DOI] [PubMed] [Google Scholar]
- 40.van Asbeck EC, Huang YC, Markham AN, Clemons KV, Stevens DA. 2007. Candida parapsilosis fungemia in neonates: genotyping results suggest healthcare workers hands as source, and review of published studies. Mycopathologia 164:287–293. doi: 10.1007/s11046-007-9054-3. [DOI] [PubMed] [Google Scholar]
- 41.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic healthcare surface. J Clin Microbiol 55:2996–3005. doi: 10.1128/jcm.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdolrasouli A, Armstrong-James D, Ryan L, Schelenz S. 2017. In vitro efficacy of disinfectants utilised for skin decolonization and environmental decontamination during a hospital outbreak with Candida auris. Mycoses 60:758–763. doi: 10.1111/myc.12699. [DOI] [PubMed] [Google Scholar]
- 43.Cadnum JL, Shaikh AA, Piedrahita CT, Sankar T, Jencson AL, Larkin EL, Ghannoum MA, Donskey CJ. 2017. Effectiveness of disinfectants against Candida auris and other Candida species. Infect Control Hosp Epidemiol 38:1240–1243. doi: 10.1017/ice.2017.162. [DOI] [PubMed] [Google Scholar]
- 44.Moore G, Schelenz S, Borman AM, Johnson EM, Brown CS. 2017. Yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J Hosp Infect 97:371–375. doi: 10.1016/j.jhin.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 45.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gómez-López A, Boekhout T. 2012. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol 50:3641–3651. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]