ABSTRACT
In patients with hematological malignancies, bronchoalveolar lavage fluid (BALF) specimens are commonly used for the diagnosis of mold infections. However, it is not clear whether the cell pellet (P) or the supernatant fraction (S) of the BALF specimen is optimal for molecular diagnostic testing. Thus, 99 BALF specimens were collected from 96 hematology patients with or without allogeneic hematopoietic stem cell transplant. The cell pellets and supernatants were processed alone and in combination (S/P) for testing by two fungus-specific real-time PCR assays compliant with international recommendations. The results achieved with S/P were revealed to be superior in comparison to those achieved with S and P alone, with the use of each single fraction showing a reduced sensitivity for the detection of Aspergillus DNA (82% and 43% for S and P, respectively). In 57% of the samples, testing of the combination of S and P generated a lower quantification cycle value than testing of S or P alone. Molds would have been missed in 5 and 16 out of 28 samples if only S or P, respectively, was analyzed. No sample was positive by testing of S or P only. Similar results were obtained for the detection of Mucorales DNA in BALF specimens (reduced sensitivity of 67% and 50% for S and P, respectively). Study patients were categorized according to the current European Organization for the Research and Treatment of Cancer/Mycoses Study Group classification for invasive fungal disease (IFD), revealing that 35 patients had proven/probable IFD (36%), 47 patients had possible IFD (49%), and 14 patients had undetermined IFD (15%).
KEYWORDS: Aspergillus, bronchoalveolar lavage fluid, Mucorales, PCR, mold, pellet, supernatant
INTRODUCTION
Invasive mold disease (IMD) is among the most serious and most frequent lung infections in immunocompromised patients who receive solid organ or allogeneic stem cell transplantation (SCT) and who have hematological malignancies (HM). Over the past 3 decades, the incidence of IMD has steadily increased and the spectrum of fungi causing IMD has broadened. As a consequence, over 100,000 cases of IMD occur each year, and these are associated with high morbidity and mortality rates and tremendous health care costs (1–3). Sensitive and specific biomarkers for the diagnosis of IMD are required.
In recent years, substantial efforts have been made to develop better tools and protocols for the diagnosis of IMD. Different blood fractions (whole blood [4, 5], serum [6], plasma [7], or a combination of them [8]) have prospectively been used to both confirm and exclude IMD using real-time PCR assays amplifying multicopy gene regions. To achieve an early confirmatory diagnosis of pulmonary IMD, PCR testing of bronchoalveolar lavage fluid (BALF) was shown to be useful, as the focus of disease is the lung alveoli, where inhaled fungal spores are deposited due to their small size (9, 10). BALF thus represents a clinical sample type originating from the site of infection. Unfortunately, the bronchoscopy process is invasive, limiting it use in hematology settings; furthermore, the procedure used to obtain BALF may vary between centers. In respect to molecular testing of BALF, there has been limited standardization, particularly in relation to the nucleic acid extraction technique to be used and the optimal target to be identified. While it seems obvious that BALF could contain intact fungal cells if such cells were inhaled, an examination of the literature identifies extraction methods that lack the efficiency required to lyse fungal cells but nevertheless provide good performance. The alternative target is likely free DNA released from the organism through the actions of the host's immune response or, potentially, as a result of antifungal therapy. The question remains whether the cell pellet (P), which contains a large amount of intracellular mold DNA, or the supernatant fraction (S) of the BALF specimen, which mostly contains free DNA, is most appropriate for use for subsequent molecular diagnostics.
The objective of this study was to evaluate whether the BALF supernatant, the cell pellet, or a combination of both fractions (S/P) processed separately or combined provides superior sensitivity. A multicenter (n = 8) study was performed using 99 BALF specimens prospectively collected from HM patients with suspected IMD. Patients were categorized according to the current European Organization for the Research and Treatment of Cancer (EORTC)/Mycoses Study Group (MSG) criteria for IMD (11). Additionally, a control PCR assay was performed in parallel to quantify the amount of human DNA in the samples.
(Data from this study were partially presented at Trends in Medical Mycology 2017, Belgrade, Serbia.)
MATERIALS AND METHODS
Study design.
Over a period of 15 months from July 2016 to September 2017, BALF samples taken for routine clinical investigation of chest infection (new or progressive lung infiltrates) in adult (age, ≥18 years) HM/SCT patients across eight hospitals, including seven centers from Germany and one from the United Kingdom, were retained for this anonymous performance evaluation. For each sample, the supernatant and pellet were separated by centrifugation, with each fraction being tested separately or in combination for the presence of fungal DNA (Fig. 1). The analysis was performed in Würzburg, Germany (University Hospital Würzburg [UKW]), or in Cardiff, United Kingdom (CAR), by using European Aspergillus PCR Initiative (EAPCRI)-compliant real-time PCR assays specific for Aspergillus (12) and Mucorales (6, 13). EAPCRI recommendations for the Mucorales PCR are currently in development.
FIG 1.
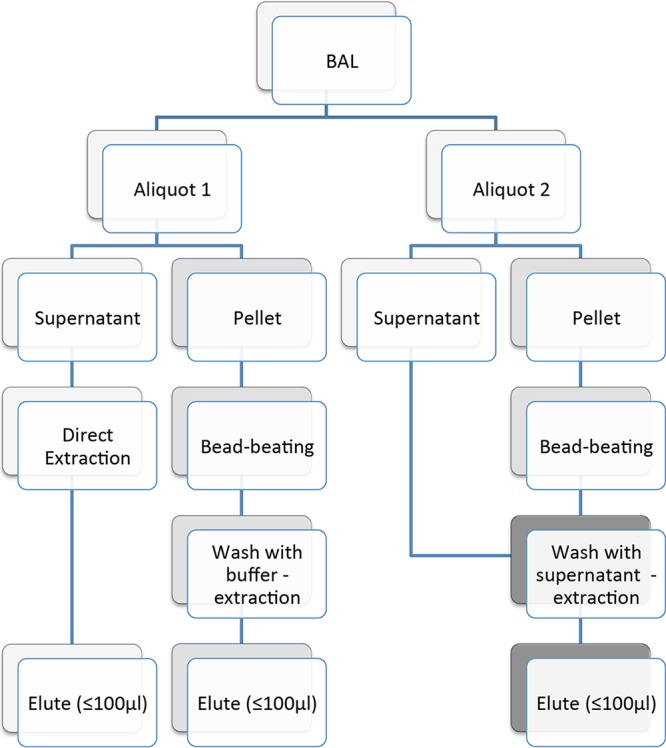
Overview of process used to test BALF specimens.
Clinical signs and symptoms of IMD and microbiological data were recorded using the ClinicalSurveys.net online platform of the University Hospital of Cologne (with software under license from QuestBack GmbH, Cologne, Germany) for each patient with IMD, defined according to revised European Organization for the Research and Treatment of Cancer (EORTC)/Mycoses Study Group (MSG) criteria (11). Bronchoscopy and antifungal treatment were performed in accordance with institutional protocols. The study was approved by the local ethics committees of the UKW (Ethikommission des Universitätsklinikums Würzburg, Würzburg, Germany, approval 270/15) and the committees at the centers in Frankfurt, Mainz, Essen, Cologne, Berlin, and Erlangen. In CAR, samples were sent for routine investigation of IMD, including fungal PCR, and after initial testing, surplus clinical material was retained for performance assessment and quality control purposes.
DNA extraction from BALF.
All steps for extracting DNA from BALF were performed in a class II laminar-flow cabinet. Briefly, samples were vortexed and 0.5 ml of BALF was centrifuged at 5,000 × g for 7 min (UKW) or at 10,000 × g for 5 min (CAR). The supernatant fraction (S) was transferred to a new tube. The remaining cell pellet (P) was bead beaten for 90 s using ceramic beads (MagNA Lyser green beads; Roche), and 50 μl liquid was added to the pellet for bead beating (UKW). Between 50 and 500 μl of liquid was added to the pellet to allow the transfer of material for downstream extraction with commercial products. For combination S/P testing, S was used, whereas when testing the P, DNA-free water was applied.
In Cardiff, the pellet with residual S was bead beaten for 30 s using ceramic beads (MagNA Lyser green beads; Roche). After the bead beating, the samples were pulse centrifuged and washed with 500 μl of molecular-grade water if only the P was being processed or 500 μl of S if P and S in combination were being processed. If the S was being processed, 500 μl was extracted directly with no bead beating. All samples were mixed with 2 ml of NucliSens lysis buffer (bioMérieux), and the mixture was incubated at room temperature for 10 min before being extracted using a bioMérieux EasyMag automated extractor and the generic total nucleic acid (version 2.0) protocol. All nucleic acid was eluted in 60 μl. In each sample, the amount of genomic DNA from Neisseria meningitidis, which acted as a positive extraction control for each specimen, was quantified to monitor for inhibition and DNA extraction efficiency. A negative control in the form of molecular-grade water was included to monitor for contamination borne by the procedure.
DNA was extracted using local procedures: in UKW, DNA obtained from the P after bead beating was extracted using a High Pure template preparation kit (Roche) protocol for the isolation of nucleic acids from mammalian whole blood (low volume). For P/S and S alone, to cope with the increased sample volume (500 μl), a modified protocol was applied by utilizing increased reagent volumes where applicable (400 μl binding buffer, 80 μl proteinase K, 200 μl isopropanol). As an extraction control, 200 μl water with 5,000 copies of Bacillus DNA was used (6). For the combined analysis of the S/P fraction, the bead-beaten pellet fraction was supplemented with the remaining 450 μl BALF supernatant. The proteinase K incubation time was increased to 15 min at 70°C. The elution volume was adjusted to 70 μl.
DNA amplification and detection.
All PCR methods were validated by testing different EAPCRI panels and showed comparable performance in detecting the thresholds set by EAPCRI (12, 14). In Würzburg, an Aspergillus-specific real-time PCR assay (4, 8) was used to detect fungal DNA. Briefly, 20-μl reaction mixtures contained 0.3 μM primers, 0.15 μM hydrolysis probe, 10 μl TaqMan gene expression master mix (Applied Biosystems), and 5 μl template DNA. Amplification was carried out in a StepOnePlus machine (Applied Biosystems) with the following steps: 50°C for 2 min, 95°C for 10 min, and 60 cycles of 95°C for 5 s, 54°C for 15 s (detection step), and 72°C for 1 s. Negative and positive PCR controls were included in each run. Samples were analyzed in duplicate.
In each DNA extraction run, one negative control (DNA-free water spiked with 5,000 copies of a plasmid carrying Bacillus DNA) was included as a quality control. Besides the detection of fungal DNA, Bacillus DNA was detected. This internal control was tested independently of the fungal target (monoplex) but within the same PCR run. The reaction mixtures (total volume, 20 μl) contained the primers and probe cited before (6), 10 μl of TaqMan gene expression master mix (Applied Biosystems), and 5 μl of template DNA. Besides the use as an extraction control, Bacillus DNA was also used to monitor PCR inhibition. For this, 1,000 copies of a plasmid carrying Bacillus DNA were spiked into every PCR mixture. PCR inhibition was claimed if the sample quantification cycle (Cq) value differed by a value of more than 3.3 from the value for the controls.
In Cardiff, the Aspergillus real-time PCR test was performed using a Corbett Rotor-Gene Q HRM instrument targeting the 28S rRNA gene and 5 μl of template in a final reaction volume of 25 μl (15). PCR controls in the form of cloned PCR products (300, 30, and 3 input copies) and no-template molecular-grade water were included to monitor PCR performance. An internal control PCR targeting the ctrA gene of Neisseria meningitidis was performed. The remaining DNA eluates, which had already been tested routinely for the presence of Aspergillus DNA, were stored at −20°C and shipped on dry ice to UKW for further testing.
The detection of Mucorales and human DNA was performed only in Würzburg (13, 16), and the following modifications were used for human DNA. The PCR volume was reduced to 20 μl using 10 μl of TaqMan gene expression master mix (Applied Biosystems) and 2 μl of template DNA. The amounts of primers and probe were reduced to 400 nM and 90 nM, respectively. The elongation temperature was decreased to 60°C.
At both centers, duplicate PCR testing allowed different interpretations of test positivity. In this study, one positive PCR replicate was considered significant.
Statistical analysis.
Patients with EORTC/MSG-defined proven and probable IFD and with microbiological evidence of infection with Aspergillus or Mucorales spp. were classified as cases, and patients with no evidence of fungal disease were categorized as controls. The sensitivities, specificities, positive predictive values (PPV), negative predictive values (NPV), and odds ratios (OR), along with their associated 95% confidence intervals (CI), were calculated according to their definitions using GraphPad Prism software (version 5.04) (see Table 2).
TABLE 2.
Clinical performance of Aspergillus and Mucorales PCR assays when testing different BALF fractionsa
| BALF fraction | Sensitivityb | Specificityb | PPVc | NPVc | OR |
|---|---|---|---|---|---|
| S/P | 19/28 (67.9, 47.7–84.1) | 1/14 (92.9, 66.1–99.8) | 95.0 (75.1–99.9) | 59.1 (36.4–79.3) | 27.4 |
| S | 16/28 (57.1, 37.2–75.5) | 0/14 (100.0, 76.8–100.0) | 100.0 (79.4–100.0) | 53.9 (33.4–73.4) | 38.3 |
| P | 14/28 (50.0, 30.7–69.4) | 0/14 (100.0, 76.8–100.0) | 100.0 (76.8–100.0) | 50.0 (30.7–69.4) | 29 |
Patients with possible IFD were excluded from the analysis. PPV, positive predictive value; NPV, negative predictive value; OR, odds ratio. The OR was calculated by adding 0.5 to the value in each cell to avoid dividing by zero.
Values are the number of positive patients/total number of patients (percent, 95% CI) observed when the indicated number was positive.
Values are percent (95% CI).
A paired t test was used to compare the different amounts of human DNA in BALF specimens between patient subgroups. P values of <0.05 were considered significant.
RESULTS
Positivity rates and comparison of available DNA concentrations.
In total, 99 samples were collected from 96 patients. Twenty-eight samples (28%) showed Aspergillus DNA in 4 different combinations (Table 1). All 28 samples were positive by the combined approach (S/P), whereas the single fractions alone showed reduced positivity (for S, 23/28 samples [82%] were positive [P = 0.0515]; for P, 12/28 samples [43%] were positive [P < 0.0001]). No sample was positive by the testing of only S or P. In contrast, 4 samples showed positive results for Aspergillus DNA when only the combination of S and P was tested.
TABLE 1.
Distribution of positivity across the various BALF fractions tested by Aspergillus and Mucorales PCR assays
| PCR target | No. (%) of the following samples that were PCR positivea: |
|||
|---|---|---|---|---|
| S/P, S, and P | S/P and S only | S/P and P only | S/P only | |
| Aspergillus | 11 (39) | 12 (43) | 1 (4) | 4 (14) |
| Mucorales | 3 (50) | 1 (17) | 0 | 2 (33) |
S, supernatant fraction; P, cell pellet; S/P, combination of both fractions.
For most positive samples (57%, 16/28), testing of the combination of S and P generated an earlier (lower) Cq value than testing of S or P alone, indicating the presence of a larger amount of available DNA in the combination of S and P, which thereby improved the chance of successful detection by PCR. If all three combinations tested positive, the mean Cq values for S/P, P, and S were 39.2, 40.1, and 41.3, respectively. The same trend was seen for the amount of human DNA present in the BALF extracts. The combination of S and P significantly showed the highest mean amount of human DNA compared to the amount for S or P alone, 488, 323, and 235 ng/μl, respectively (P = 0.0003). Two samples (2%) containing large amounts of human DNA (9,151 and 1,566 ng/μl) inhibited the PCR.
For the detection of Mucorales DNA, 6 samples (6%) were positive by use of the three different combinations (Table 1). All 6 samples were positive by use of combination of S and P, whereas the single fractions alone showed reduced positivity (for S, 4/6 samples [67%] were positive; for P, 3/6 samples [50%] were positive). No sample was positive by the testing of S or P only. In contrast, 2 samples showed positive results when only the combination of S and P was tested. For all positive samples, testing of the combination of S and P generated an earlier (lower) Cq value than testing of S or P alone, indicating the presence of a larger amount of available DNA. When the PCR result was positive, the mean Cq values for S/P, P, and S were 27.0, 28.9, and 29.8, respectively. Interestingly, in three patients, the DNA of Mucorales as well as Aspergillus spp. was detected in parallel in S/P alone.
Correlation of PCR assay results to the EORTC/MSG classification for IFD.
All study patients were categorized according to the current EORTC/MSG classification for IFD (11), resulting in 35 patients with proven/probable IFD (36%), 47 patients with possible IFD (49%), and 14 patients with undetermined IFD (15%). As our PCR assays allowed the detection of Mucorales and Aspergillus DNA only, patients with EORTC/MSG-defined proven and probable IFD and with microbiological evidence of Aspergillus or Mucorales spp. were classified as cases and patients with no evidence of fungal disease were categorized as controls. There were 2 proven cases, 1 of which was proven to be positive for Aspergillus and 1 of which was proven to be positive for Mucorales, and 26 probable cases. Among the probable cases, 4 cases were proven to be positive for galactomannan (GM) in serum, 17 cases were proven to be positive for GM in BALF, Aspergillus was cultured from BALF in 11 cases, and Mucorales was cultured from BALF in 1 case. The combination approach with S and P showed the highest sensitivity and negative predictive value (Table 2), whereas use of the S or P fractions alone showed higher specificities and positive predictive values (PPV) than the use of the combination of S and P. As no Mucorales or Aspergillus DNA was detected in any of the patients in whom IFD was undetermined by use of S or P only, the specificity and PPV reached 100% (Table 2). Of the six patients positive only by the testing of S/P (Table 1), three were classified as having probable IFD, two were classified as having possible IFD, and one was classified as having undetermined IFD.
DISCUSSION
Aspergillus, Pneumocystis, and Cryptococcus spp. are the most common fungi causing lung infections in humans (17). However, not only fungi but also other pathogens of the lower respiratory tract are major causes of morbidity and mortality in patients with hematological malignancies (18). As a consequence, sensitive, accurate, and early diagnosis is essential to identify and confirm the presence of infectious agents in the lung. While the detection of galactomannan or fungal DNA in blood, computed tomography scans of the lung, or analysis of samples from the upper respiratory tract allow only an indirect diagnosis, bronchoscopy markedly improves the ability to identify an infectious agent.
The early and specific diagnosis of IA leads to the early initiation of specific antifungal therapy, which is directly associated with improved clinical outcomes (1). However, this demand might be hard to achieve in the daily clinical routine. In a study by Harris and Geyer, the time from the time of detection of lung infiltrates to the time of bronchoscopy was a median of 3 days (range, 1 to 13 days), despite the fact that positive yields from BALF are the greatest when bronchoscopy is performed within the first 24 h of presentation (19).
Our multicenter study showed the necessity to obtain high-quality BALF for the PCR-based detection of fungal pathogens. We observed strong center-specific effects revealing substantially different human DNA concentrations in all BALF fractions (from 0.2 ng DNA/μl to up to 9 μg DNA/μl). As a consequence, the reproducibility of the results may vary or PCR amplification might be inhibited for some specimens. Hence, to reveal the optimal sensitivity, specificity, and reproducibility, the quality control measures and the precautions against contamination that are taken must be stringent.
In a systematic review by Avni et al. evaluating the diagnostic accuracy of PCR detection of fungi in BALF compared to that of the detection of GM, PCR demonstrated a statistically higher sensitivity with no loss of specificity when a GM optical density threshold of ≥1.0 was considered a positive result (20). In addition, the Aspergillus-specific PCR is the only indirect mycological test for which independently available control material and international quality control schemes to calibrate and allow the impartial comparison of different assays are available (10). Furthermore, the European Aspergillus PCR Initiative (www.eapcri.eu), which has defined standards for the comparison of Aspergillus PCRs between laboratories (14), is currently working on standardized recommendations for BALF as well.
The question whether the cell-free supernatant or the cell pellet of a BALF specimen should be used for the PCR-based diagnosis of a mold infection is currently still under debate. While the supernatant potentially contains free-floating DNA molecules, the pellet fraction contains a large amount of DNA from phagocytosed fungi, e.g., within alveolar macrophages. Interestingly, free-floating DNA proved to be stable within BALF for at least 5 days at room temperature (data not shown). This observation is of special relevance for future multicenter studies or centralized diagnostic laboratory services.
Our current study demonstrates that the use of both BALF fractions (instead of a single fraction alone) for DNA extraction reduces the risk of fungal DNA loss. The PCR result was positive for 28% (28/99) of the BALF samples when the approach with the combination of S and P was used. When samples from HM/hematopoietic stem cell transplantation patients were tested, the PCR result showed reduced rates of positivity of 23% and 12%, respectively, when either single fraction (S or P only) was used. This observation is consistent with the findings of our former study combining plasma with a whole-blood fraction, which yielded a sensitivity and a reproducibility superior to those obtained when a single fraction (plasma or the leukocyte pellet) was used for patients at high risk for IA (8).
According to our knowledge, only one additional study (16) has so far compared the use of the BALF pellet and supernatant. These authors detected Aspergillus DNA in 7 of 10 episodes of IA for the pellet fraction and in only 4 of 10 episodes for the supernatant fraction. Analysis of the results obtained with the BALF pellet and supernatant together conferred a sensitivity and a specificity identical to those obtained with the BALF pellet alone (16). These results are in contrast to those of our current study, where the approach with the combination of S and P yielded a higher sensitivity. However, while Khot et al. (16) combined the analytical values for the pellet and supernatant, our approach covers the combination of DNA eluates prior to PCR amplification. Furthermore, in contrast to the findings of Khot et al. (16), our study demonstrated a slightly higher sensitivity for the BALF supernatant. However, the DNA extraction protocols, primers, and PCR assay designs that were used varied broadly between the two studies.
It is remarkable that the sensitivity of the PCR-based diagnosis of proven/probable IFI by the use of BALF varies broadly. Two recent systematic reviews and meta-analyses demonstrated that the sensitivity obtained by the testing of BALF ranges from 30% to 100% (9, 20). Accordingly, the sensitivity values in our study ranged from 68% (for S/P) to 50% (for P), which is in the above-mentioned range.
In conclusion, this multicenter study demonstrates that the use of BALF allows the detection of Aspergillus and Mucorales DNA in patients with hematological malignancies suffering from proven and probable IFD. The appropriate protocol for DNA extraction is key to the successful detection of fungal DNA in BALF. By testing both BALF fractions (pellet, supernatant), a sensitivity superior to that achieved by testing a single fraction was obtained.
ACKNOWLEDGMENTS
This work was supported by the Wilhelm Sander Stiftung (grant number 2015-083.1).
D.T. has received honoraria and travel grants from Gilead and Merck Sharp & Dohme (MSD) and travel grants from Astellas and Jazz Pharmaceuticals and is a consultant on the advisory boards for Gilead, MSD, and Pfizer. S.S. has received personal fees and nonfinancial support from Amgen, Basilea, Gilead, MSD, Jazz Pharmaceuticals, and Pfizer. L.M. has received travel grants from Amgen, Astellas, Medac, Merck, MSD, Novartis, and Pfizer. S.W.K., T.E., J.S., H.E., I.W., D.K., R.B.P., T.L., S.F., and V.R. state that they do not have any conflicts of interest. J.L. is founding member of EAPCRI and received project funding from Bruker and Renishaw Diagnostics Limited, a travel grant from Astellas, and financial support from MSD. J.K. has received travel grants from Gilead, Basilea, and Astellas and speaker's fees from MSD. P.L.W. is a founding member of EAPCRI; received project funding from Bruker, Myconostica, Luminex, and Renishaw Diagnostics Limited; was sponsored by Myconostica, MSD, Launch Diagnostics, Bruker, and Gilead Sciences to attend international meetings; is on the speaker's bureau for Gilead Sciences; and was provided consultancy fees from Renishaw Diagnostics Limited and Gilead Sciences. J.J.V. has served on the speaker's bureaus of Pfizer, Merck, Gilead, Basilea, and Astellas; received research funding from Astellas, Gilead, Infectopharm, Merck/MSD, Basilea, and Pfizer; has received travel assistance from Astellas, Gilead, Merck/MSD, and Basilea; and is a consultant to Astellas, Gilead, Basilea, and Merck/MSD. O.A.C. is supported by the German Federal Ministry of Research and Education; has received research grants from Actelion, Amplyx, Arsanis, Astellas, AstraZeneca, Basilea, Bayer, Cidara, Duke University (NIH grant UM1AI104681), F2G, Gilead, GSK, Leeds University, Matinas, MedPace, Melinta, Merck/MSD, Miltenyi, Pfizer, Rempex, Roche, Sanofi Pasteur, Scynexis, Seres, and The Medicines Company; is a consultant to Achaogen, Anacor, Amplyx, Actelion, Astellas, Basilea, Cidara, Da Volterra, F2G, Gilead, Janssen, Matinas, Menarini, Merck/MSD, Paratek, PSI, Scynexis, Seres, Summit, Tetraphase, and Vical; and has received lecture honoraria from Astellas, Basilea, Gilead, Merck/MSD, and Pfizer. W.J.H. received research grants from MSD/Merck and Pfizer; serves on the speaker's bureaus of Alexion, Astellas, Basilea, Bristol-Myers Squibb, Gilead, Janssen, MSD, and Pfizer; and received travel grants from Alexion, Astellas, Lilly, MSD, Novartis, and Pfizer.
REFERENCES
- 1.Drgona L, Khachatryan A, Stephens J, Charbonneau C, Kantecki M, Haider S, Barnes R. 2014. Clinical and economic burden of invasive fungal diseases in Europe: focus on pre-emptive and empirical treatment of Aspergillus and Candida species. Eur J Clin Microbiol Infect Dis 33:7–21. doi: 10.1007/s10096-013-1944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruhnke M, Groll AH, Mayser P, Ullmann AJ, Mendling W, Hof H, Denning DW, University of Manchester in association with the LIFE program. 2015. Estimated burden of fungal infections in Germany. Mycoses 58(Suppl 5):S22–S28. doi: 10.1111/myc.12392. [DOI] [PubMed] [Google Scholar]
- 3.Pegorie M, Denning DW, Welfare W. 2017. Estimating the burden of invasive and serious fungal disease in the United Kingdom. J Infect 74:60–71. doi: 10.1016/j.jinf.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Springer J, Loeffler J, Heinz W, Schlossnagel H, Lehmann M, Morton O, Rogers TR, Schmitt C, Frosch M, Einsele H, Kurzai O. 2011. Pathogen-specific DNA enrichment does not increase sensitivity of PCR for diagnosis of invasive aspergillosis in neutropenic patients. J Clin Microbiol 49:1267–1273. doi: 10.1128/JCM.01679-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers TR, Morton CO, Springer J, Conneally E, Heinz W, Kenny C, Frost S, Einsele H, Loeffler J. 2013. Combined real-time PCR and galactomannan surveillance improves diagnosis of invasive aspergillosis in high risk patients with haematological malignancies. Br J Haematol 161:517–524. doi: 10.1111/bjh.12285. [DOI] [PubMed] [Google Scholar]
- 6.Springer J, Lackner M, Nachbaur D, Girschikofsky M, Risslegger B, Mutschlechner W, Fritz J, Heinz WJ, Einsele H, Ullmann AJ, Loffler J, Lass-Florl C. 2016. Prospective multicentre PCR-based Aspergillus DNA screening in high-risk patients with and without primary antifungal mould prophylaxis. Clin Microbiol Infect 22:80–86. doi: 10.1016/j.cmi.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 7.White PL, Barnes RA, Springer J, Klingspor L, Cuenca-Estrella M, Morton CO, Lagrou K, Bretagne S, Melchers WJG, Mengoli C, Donnelly JP, Heinz WJ, Loeffler J, Eapcri. 2015. Clinical performance of Aspergillus PCR for testing serum and plasma: a study by the European Aspergillus PCR Initiative. J Clin Microbiol 53:2832–2837. doi: 10.1128/JCM.00905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer J, Schlossnagel H, Heinz W, Doedt T, Soeller R, Einsele H, Loeffler J. 2012. A novel extraction method combining plasma with a whole-blood fraction shows excellent sensitivity and reproducibility for patients at high risk for invasive aspergillosis. J Clin Microbiol 50:2585–2591. doi: 10.1128/JCM.00523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heng SC, Morrissey O, Chen SC, Thursky K, Manser RL, Nation RL, Kong DC, Slavin M. 2015. Utility of bronchoalveolar lavage fluid galactomannan alone or in combination with PCR for the diagnosis of invasive aspergillosis in adult hematology patients: a systematic review and meta-analysis. Crit Rev Microbiol 41:124–134. doi: 10.3109/1040841X.2013.804033. [DOI] [PubMed] [Google Scholar]
- 10.Smibert OC, Slavin MA. 2017. Cart before the horse: use of Aspergillus PCR to increase the diagnostic yield from BAL in hematological patients at risk of invasive aspergillosis. Leuk Lymphoma 58:2773–2776. doi: 10.1080/10428194.2017.1330479. [DOI] [PubMed] [Google Scholar]
- 11.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White PL, Mengoli C, Bretagne S, Cuenca-Estrella M, Finnstrom N, Klingspor L, Melchers WJ, McCulloch E, Barnes RA, Donnelly JP, Loeffler J. 2011. Evaluation of Aspergillus PCR protocols for testing serum specimens. J Clin Microbiol 49:3842–3848. doi: 10.1128/JCM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer J, Goldenberger D, Schmidt F, Weisser M, Wehrle-Wieland E, Einsele H, Frei R, Loffler J. 2016. Development and application of two independent real-time PCR assays to detect clinically relevant Mucorales species. J Med Microbiol 65:227–234. doi: 10.1099/jmm.0.000218. [DOI] [PubMed] [Google Scholar]
- 14.White PL, Bretagne S, Klingspor L, Melchers WJ, McCulloch E, Schulz B, Finnstrom N, Mengoli C, Barnes RA, Donnelly JP, Loeffler J. 2010. Aspergillus PCR: one step closer to standardization. J Clin Microbiol 48:1231–1240. doi: 10.1128/JCM.01767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White PL, Linton CJ, Perry MD, Johnson EM, Barnes RA. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin Infect Dis 42:479–486. doi: 10.1086/499949. [DOI] [PubMed] [Google Scholar]
- 16.Khot PD, Ko DL, Hackman RC, Fredricks DN. 2008. Development and optimization of quantitative PCR for the diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. BMC Infect Dis 8:73. doi: 10.1186/1471-2334-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maschmeyer G, Donnelly JP. 2016. How to manage lung infiltrates in adults suffering from haematological malignancies outside allogeneic haematopoietic stem cell transplantation. Br J Haematol 173:179–189. doi: 10.1111/bjh.13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols WG, Guthrie KA, Corey L, Boeckh M. 2004. Influenza infections after hematopoietic stem cell transplantation: risk factors, mortality, and the effect of antiviral therapy. Clin Infect Dis 39:1300–1306. doi: 10.1086/425004. [DOI] [PubMed] [Google Scholar]
- 19.Harris B, Geyer AI. 2017. Diagnostic evaluation of pulmonary abnormalities in patients with hematologic malignancies and hematopoietic cell transplantation. Clin Chest Med 38:317–331. doi: 10.1016/j.ccm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avni T, Levy I, Sprecher H, Yahav D, Leibovici L, Paul M. 2012. Diagnostic accuracy of PCR alone compared to galactomannan in bronchoalveolar lavage fluid for diagnosis of invasive pulmonary aspergillosis: a systematic review. J Clin Microbiol 50:3652–3658. doi: 10.1128/JCM.00942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


