ABSTRACT
Detection of heteroresistance of Mycobacterium tuberculosis remains challenging using current genotypic drug susceptibility testing methods. Here, we described a melting curve analysis-based approach, termed DeepMelt, that can detect less-abundant mutants through selective clamping of the wild type in mixed populations. The singleplex DeepMelt assay detected 0.01% katG S315T in 105 M. tuberculosis genomes/μl. The multiplex DeepMelt TB/INH detected 1% of mutant species in the four loci associated with isoniazid resistance in 104 M. tuberculosis genomes/μl. The DeepMelt TB/INH assay was tested on a panel of DNA extracted from 602 precharacterized clinical isolates. Using the 1% proportion method as the gold standard, the sensitivity was found to be increased from 93.6% (176/188, 95% confidence interval [CI] = 89.2 to 96.3%) to 95.7% (180/188, 95% CI = 91.8 to 97.8%) compared to the MeltPro TB/INH assay. Further evaluation of 109 smear-positive sputum specimens increased the sensitivity from 83.3% (20/24, 95% CI = 64.2 to 93.3%) to 91.7% (22/24, 95% CI = 74.2 to 97.7%). In both cases, the specificity remained nearly unchanged. All heteroresistant samples newly identified by the DeepMelt TB/INH assay were confirmed by DNA sequencing and even partially by digital PCR. The DeepMelt assay may fill the gap between current genotypic and phenotypic drug susceptibility testing for detecting drug-resistant tuberculosis patients.
KEYWORDS: Mycobacterium tuberculosis, isoniazid-resistant tuberculosis, heteroresistance, melting curve analysis, clamping
INTRODUCTION
Heteroresistance in Mycobacterium tuberculosis is defined as mixed populations of drug-susceptible and -resistant organisms in a single clinical specimen from tuberculosis (TB) patients (1). Patients infected with heteroresistant clones have been increasingly described as having multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB (2–7). As a precursor to full resistance, sensitive detection of heteroresistance is critical for making and tailoring patient treatment plans.
However, detection of heteroresistance is challenging using current molecular assays. The global frontline molecular diagnostic tool for drug-resistant M. tuberculosis, Xpert MTB/RIF (8), identifies patients harboring rifampin-resistant strains for the initiation of MDR-TB treatment, but this assay may detect the presence of resistance when the mixture contains at least 65.6% mutant DNA. Folkvardsen et al. (9), after a thoughtful study on mixtures of isoniazid (INH)-resistant and -susceptible M. tuberculosis strains, concluded that culture-based phenotypic drug susceptibility testing (DST) was the most sensitive method, whereas the line probe assay and Sanger sequencing could not detect the clinically relevant 1% proportion of resistant bacteria. In a recent multiplex melting curve analysis (MCA)-based assay developed for the detection of XDR-TB plus resistance to INH for point-of-care use (10), heteroresistance detectability was demonstrated to be equivalent to that of Sanger sequencing, i.e., 20 to 30%. Although mass array-based iPLEX gold assay (11) and deep sequencing (5, 6) were reported to detect 1% of heteroresistance, both require special and costly instruments, precluding their widespread use in clinical settings.
We previously reported an evaluation study on a Chinese Food and Drug Administration-approved MCA-based assay for detecting mutations associated with INH resistance, the MeltPro TB/INH assay (12), which features wide coverage of mutations, high-throughput analysis, and ease-of-use. However, its heteroresistance detectability is only 20 to 40%. To address the low sensitivity of the MCA-based assay for detecting heteroresistance, we developed a new-generation MCA assay, DeepMelt, that detected as low as 1% heteroresistance by using a probe-clamping strategy.
MATERIALS AND METHODS
Samples.
The archived bacterial samples consisted of DNA isolated from 602 M. tuberculosis isolates. Of these, 320 isolates were from the 309th Hospital of Chinese PLA (Beijing, China) and 282 isolates were from Henan Provincial Center for Disease Control and Prevention (Zhengzhou, Henan, China). All isolates had been characterized by DST on Löwenstein-Jensen (L-J) solid medium using standard proportion protocol (13). All DNA samples were detected by MeltPro TB/INH as described previously (12).
Patient samples consisted of 109 smear-positive sputa collected from Guangzhou Chest Hospital (Guangzhou, Guangdong, China). Each sample was decontaminated by using the N-acetyl-l-cysteine/NaOH method for 15 min, followed by neutralization with sterile phosphate-buffered saline (PBS; pH 6.8). After centrifugation at 3,000 × g for 15 min, the pellet was resuspended in 2 ml of PBS buffer. A 0.5-ml portion of the decontaminated specimens was cultured on mycobacterial growth indicator tubes (Becton Dickinson, USA). Drug susceptibility of the culture-positive isolates was detected by L-J solid medium using standard proportion protocol (13). INH resistance was defined as the growth of more than 1% colonies on L-J medium containing the critical concentration of 0.2 μg/ml of INH compared to growth on drug-free control medium. The remaining 1.5-ml proportion of the decontaminated specimens was subjected to DNA extraction using Lab-Aid 824 MTB DNA extraction kit (Zeesan Biotech, Xiamen, China).
Wild-type M. tuberculosis genomic DNA (gDNA) and mutant M. tuberculosis gDNA were purified from an H37Rv reference strain (ATCC 27294) and a phenotypically INH-resistant M. tuberculosis clinical isolate harboring katG S315T (AGC→ACC), respectively, by using the AxyPrep bacterial genomic DNA miniprep kit (Axygen Scientific, Union City, CA) and quantified using a NanoDrop microvolume spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The concentrations of both gDNA were adjusted to 5 ng/μl (∼106 genomes/μl) with TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.5).
DeepMelt assay for katG S315T.
A singleplex DeepMelt assay was established to detect the most common mutation katG S315T (AGC→ACC). The primers and probe were listed in Table S1 in the supplemental material. PCR mixture contained 1× PCR buffer [75 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, and 0.01% (vol/vol) Tween 20], 3.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.06 μM katG-F1, 1.2 μM katG-R, 0.4 μM katG-P, 1.5 U of KlenTaq-S DNA polymerase (Jem Biotech, Chesterfield, MO), and 5 μl of template DNA. The reaction started with a denaturation step at 95°C for 3 min, followed by 55 cycles of 95°C for 10 s, 61°C for 15 s, and 67.5°C for 15 s. Melting curve analysis was started with a denaturation step for 2 min at 95°C, hybridization step for 2 min at 45°C, and continuous temperature increase from 45°C to 90°C at a ramp rate of 0.04°C/s. Fluorescence was recorded at FAM channel. Amplification and melting curve analysis was conducted using a SLAN-96S real-time PCR machine (Hongshi Tech, Shanghai, China).
DeepMelt TB/INH assay.
A multiplex DeepMelt TB/INH assay was established to detect all commonly found mutations associated with INH resistance. Three sets of primers and four dually labeled probes were designed to detect mutations in three drug resistance-associated genes in a single reaction (see Table S1 in the supplemental material). The reaction mixture contained 20 μl of PCR mixture and 5 μl of template. The PCR mixture solution consisted of 1× PCR buffer [75 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, and 0.01% (vol/vol) Tween 20], 4.0 mM MgCl2, 0.2 mM dNTPs, 0.12 μM katG-F2, 1.2 μM katG-R, 0.06 μM inhA-F, 0.6 μM inhA-R, 0.08 μM ahpC-F, 1.0 μM ahpC-R, 0.16 μM concentrations of each probe, and 2 U of KlenTaq-S DNA polymerase. PCR started with a denaturation step at 95°C for 3 min, followed by 50 cycles of 95°C for 10 s, 61°C for 15 s, and 71°C for 30 s. Melting curve analysis started with a denaturation step for 2 min at 95°C, hybridization step for 2 min at 45°C, and continuous temperature increase from 45 to 90°C at a ramp rate of 0.04°C/s. Fluorescence was recorded at FAM, HEX, ROX, and Cy5 channels. Amplification and melting curve analysis was conducted on a SLAN-96S real-time PCR machine.
DNA sequencing.
For Sanger sequencing, three fragments of the katG gene, inhA promoter, and ahpC promoter were amplified as previously described (12). For mutation-enriched Sanger sequencing, mutations were enriched during the PCR stage according to the DeepMelt principle protocol prior to Sanger sequencing as described in the supplemental material.
Clarity digital PCR.
A digital PCR assay was performed using a Clarity digital PCR system (JN Medsys, Singapore) (14) to detect two common mutations, katG S315T (AGC→ACC) and katG S315N (AGC→AAC). Each reaction mixture consisted of 0.5 μM concentrations of primers, 0.16 μM concentrations of probes, 1× GoTaq colorless Mix (Promega, Madison, WI), 1× Clarity JN solution, 3 μl of DNA sample, and water to 15 μl. The PCR started with a denaturation step at 95°C for 10 min, followed by 50 cycles at 95°C for 50 s and 62°C for 90 s. After PCR amplification, the fluorescent signals were detected on a Clarity Reader, and the data were analyzed using its attached Clarity software. The forward primer was 5′-CGTCGGCGGTCACACTTTCGGTAAGA-3′, the reverse primer was 5′-TCGTCAGCTCCCACTCGTAGCCGTA-3′, the wild-type probe (katG 315AGC) was 5′-FAM-ATCACCAGCGGCATCG-BHQ1-3′, the mutant probe (katG 315ACC) was 5′-HEX-ATCACCACCGGCATCG-BHQ1-3′, and the mutant probe (katG 315AAC) was 5′-HEX-CGATCACCAACGGCATCGA-BHQ1-3′, where underlined bases in the probe sequences represent LNA modification.
Statistical analysis.
Results were analyzed by OpenEpi (v3.01; A. G. Dean, K. M. Sullivan, and M. M. Soe) at a 95% confidence interval (CI).
RESULTS
Working principle of DeepMelt assay.
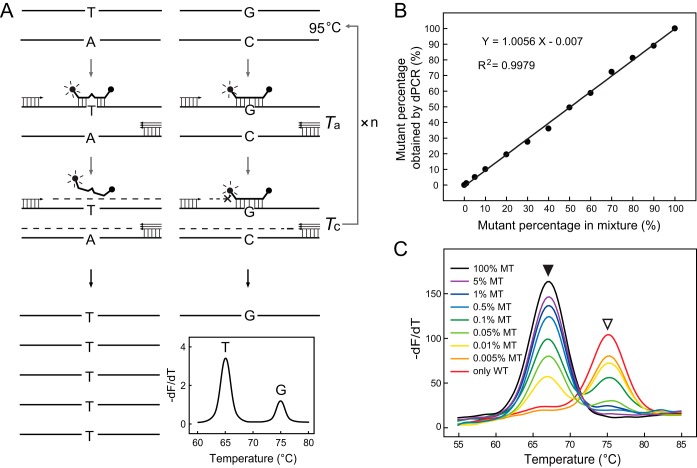
The working principle of the DeepMelt assay is illustrated in Fig. 1A. During PCR amplification of both wild-type and mutant templates, a dually labeled probe was used to specifically block chain elongation of the wild-type template, but it did not interfere with chain elongation of the mutant template. The dually labeled probe also functioned as the detection probe generating melting curves for both wild-type and mutant templates. Therefore, the mutant template was enriched after PCR as demonstrated by the increased melting peak. For this purpose, the probe was designed to perfectly match the wild-type template. A critical extension temperature (Tc) was chosen to fall between the melting temperature (Tm) of the wild type and the mutant so that the wild-type template was tightly bound by the probe and its elongation was hindered. To prevent probe hydrolysis during primer extension, KlenTaq-S DNA polymerase lacking 5′-to-3′ exonuclease activity was used.
FIG 1.
DeepMelt assay for detecting low levels of mutation in mixed templates. (A) Working principle of the DeepMelt assay. G/C, wild-type template; T/A, mutant template; Ta, annealing temperature; Tc, critical extension temperature. (B) Prepared mutant (katG S315T) percentages detected by dPCR. (C) Melting curve analysis of DeepMelt assay with various percentages of mutant katG S315T gDNA in 105 M. tuberculosis genomes/μl. WT, wild type; MT, mutant. Open and filled arrowheads point to wild-type and mutant melting peaks, respectively.
To investigate the ability of the DeepMelt assay to detect less-abundant mutants in mixed templates, we prepared a series of mixtures containing different percentages of mutant katG S315T M. tuberculosis gDNA in the presence of wild-type gDNA. We first evaluated the accuracy of the mutant percentages of these artificial mixtures by digital PCR (dPCR). The results showed that the mutant percentages obtained by dPCR were nearly identical to the prepared percentages (Fig. 1B). The ability of the DeepMelt assay to detect low percentages of mutant gDNA was studied using mixtures containing 0.005, 0.01, 0.05, 0.1, 0.5, 1, 5, and 100% mutant in 105 M. tuberculosis genomes/μl. The results showed that as low as 0.01% mutant gDNA could be detected (Fig. 1C).
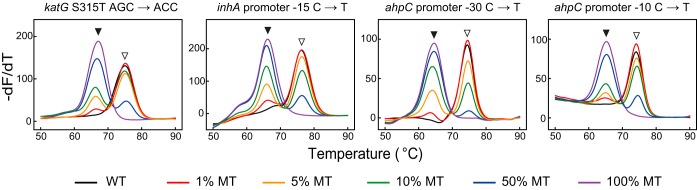
Analytical performance of multiplex DeepMelt TB/INH assay.
A multiplex assay for detecting INH resistance-associated mutations was established based on the DeepMelt principle. The DeepMelt TB/INH assay detects mutations in the katG 315 codon, the inhA promoter (−17∼−8), and the ahpC promoter (−44∼−30, −15∼4) using four probes labeled with FAM, ROX, HEX, and Cy5 fluorophores, respectively. Under the working conditions, the multiplex DeepMelt TB/INH assay detected 1% mutant in the mixture (Fig. 2).
FIG 2.
DeepMelt TB/INH assay for detecting four common mutant types of various mutant percentages in 104 M. tuberculosis genomes/μl. Open and filled arrowheads point to wild-type and mutant melting peaks, respectively.
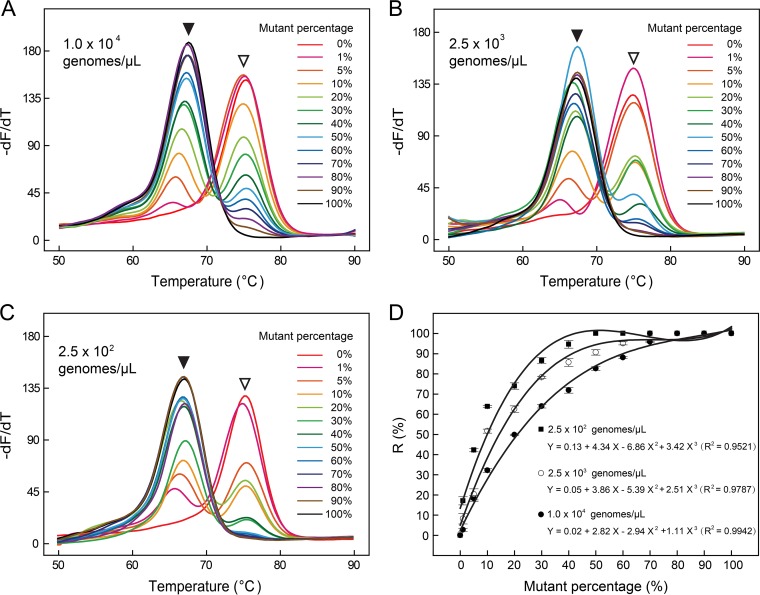
Interestingly, there seemed to be a quantitative relationship between the peak ratio and mutant percentage. To verify this possibility, we prepared three series of mixtures containing 1, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100% katG S315T mutant DNA at three different total DNA concentrations, i.e., 104, 2.5 × 103, and 2.5 × 102 genomes/μl. The ratio (R) of the mutant peak height to the sum of the wild-type peak height and mutant peak height was calculated and plotted with respect to the mutant percentage. The results confirmed the quantitative relationship between the R value and mutant percentage (Fig. 3). Notably, this relationship was dependent on the amount of total DNA, i.e., less total DNA led to a larger R value. Thus, the less-abundant mutant was detected more easily as total DNA levels decreased.
FIG 3.
Melting curve analysis for detecting katG S315T of three different total DNA concentrations, i.e., 104 (A), 2.5 × 103 (B) and 2.5 × 102 (C) genomes/μl. Open and filled arrowheads point to wild-type and mutant melting peaks, respectively. The calculated R values were plotted with respect to the mutant percentage (D), where solid circles, hollow circles, and solid squares represented 104, 2.5 × 103 and 2.5 × 102 genomes/μl, respectively.
We then studied whether 1% mutant was detectable when the total DNA concentration was higher than 104 genomes/μl. The results showed that when the total DNA concentration was increased from 104 to 105 and 106 genomes/μl, 1% mutant was less clearly detected at 105 genomes/μl and nearly undetectable at 106 genomes/μl. The same trend was observed in the detection of 5% mutant, which was detected at all concentrations but became less clearly detected with increased total amounts of gDNA.
Clinical performance of multiplex DeepMelt TB/INH assay.
We first analyzed 602 archived DNA samples that had been evaluated using MeltPro TB/INH assay and phenotypic DST. Of the 188 DST-resistant isolates, 180 isolates were found to contain mutations by the DeepMelt assay. Compared to the MeltPro results, four additional mutation-containing samples were detected. As expected, these four samples were heteroresistant. Of the 414 DST-susceptible samples, one additional heteroresistant sample was detected in addition to the 10 mutation-containing samples detected by the MeltPro assay. Subsequent DNA sequencing analysis confirmed the heteroresistant status of these samples, three of which were verified by mutation-enriched Sanger sequencing due to low levels of mutations (Table 1).
TABLE 1.
Detection results for 20 heteroresistant samples found in this study
| Sample | Type | DSTa | Test resultb |
Sanger sequencingc | dPCR (%)d | |
|---|---|---|---|---|---|---|
| MeltPro | DeepMelt | |||||
| BJ047 | Culture | R | HET | HET | ahpC promoter −10C→Y | – |
| BJ149 | Culture | R | HET | HET | katG 315 AGC→ASC | 70 |
| BJ155 | Culture | R | HET | HET | inhA promoter −15C→T; ahpC promoter −9G→R; −1ins C | – |
| BJ246 | Culture | R | HET | HET | ahpC promoter −39C→Y; ahpC promoter −12C→Y | – |
| HN012 | Culture | S | HET | HET | ahpC promoter −10C→Y | – |
| HN013 | Culture | S | HET | HET | katG 315 AGC→ARC | 51 |
| HN014 | Culture | S | HET | HET | katG 315 AGC→ARC | 46 |
| HN020 | Culture | R | HET | HET | inhA promoter −15C→Y | – |
| HN053 | Culture | R | HET | HET | ahpC promoter −30C→Y; −10C→Y;-6G→R | – |
| HN190 | Culture | R | HET | HET | katG 315 AGC→ASC; inhA promoter −15C→Y | 54 |
| HN349 | Culture | R | HET | HET | katG 315 AGC→ACC; inhA promoter −15C→Y | – |
| HN403 | Culture | R | HET | HET | ahpC promoter −32G→R; −10C→Y | – |
| GZ065 | Sputum | R | HET | HET | katG 315 AGC→ARC; ahpC promoter −30C→Y | 87 |
| BJ030 | Culture | R | WT | HET | ahpC promoter −12C→Y* | – |
| HN018 | Culture | S | WT | HET | katG 315 AGC→ARC; ahpC promoter-10C→Y* | 6 |
| HN084 | Culture | R | WT | HET | ahpC promoter −30C→Y* | – |
| HN104 | Culture | R | WT | HET | inhA promoter −15C→Y | – |
| HN105 | Culture | R | WT | HET | katG 315 AGC→ASC | 20 |
| GZ036 | Sputum | R | WT | HET | katG 315 AGC→ARC* | 7 |
| GZ071 | Sputum | R | WT | HET | ahpC promoter −32G→R* | – |
DST, drug susceptibility testing; S, susceptible; R, resistant.
HET, heteroresistant; WT, wild type.
Sanger sequencing was used except that data labeled with an asterisk (*) were obtained using mutation-enriched Sanger sequencing.
katG 315 mutation percentages obtained by dPCR. –, Not applicable.
We then analyzed 109 smear-positive sputum samples. Among the 24 phenotypically resistant samples, the DeepMelt assay revealed 19 mutant samples and 3 heteroresistant samples. In comparison, the MeltPro assay identified the 19 mutant samples but only one of the three heteroresistant samples. Of the 85 INH-susceptible specimens, the DeepMelt assay and MeltPro assay showed identical results, i.e., 82 samples were wild type, and 3 samples were fully mutant. Three samples detected as fully mutant by both assays were further confirmed by DNA Sanger sequencing, of which two samples harbored katG 315 AGC→ACC, and one sample harbored ahpC −9 G→A. Moreover, the two heteroresistant samples newly identified by the DeepMelt assay were verified by mutation-enriched Sanger sequencing (Table 1).
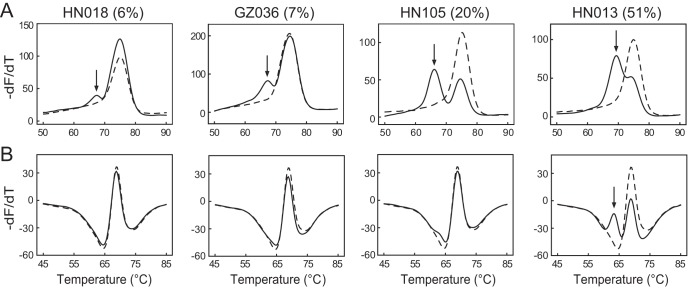
To precisely determine the mutant percentages of heteroresistant samples detected by the DeepMelt assay but missed by the MeltPro assay, all eight samples harboring the mutation katG 315 AGC→ASC or katG 315 AGC→ARC were subjected to a digital PCR assay, which reveals the exact percentage of mutant DNA by counting the numbers of both wild-type and mutant templates. The results showed that five samples detectable by both assays contained more than 40% mutated DNA, while three samples detectable by DeepMelt but undetectable by MeltPro contained less than 20% mutated DNA (Table 1). Representative results are shown in Fig. 4.
FIG 4.
Representative results of four heteroresistant samples harboring katG 315 mutations by DeepMelt TB/INH assay (A) and MeltPro TB/INH assay (B). Solid lines and dashed lines represent the melting curves of the sample and the wild-type control, respectively. The numerical values in parentheses represent the mutant percentages obtained by dPCR. Black arrows point to detectable mutant melting peaks.
DISCUSSION
Heteroresistance has been widely reported in M. tuberculosis. However, it is challenging to detect less-abundant mutants in heteroresistant samples using existing molecular assays. In the present study, through the combined use of a probe for clamping and detection and DNA polymerase lacking 5′-to-3′ exonuclease activity, proportions of mutant genomes as low as 0.01% were detected in samples containing 105 genomes/μl in a singleplex DeepMelt assay. Moreover, using the same strategy, 1% mutants in samples containing 104 genomes/μl could be detected in a multiplex DeepMelt TB/INH assay designed to detect all common mutations associated with INH resistance. Analysis of clinical samples confirmed that the DeepMelt TB/INH assay detected more heteroresistant samples than the previous MeltPro TB/INH assay in both bacterial isolates and sputum samples.
The DeepMelt strategy is a refined version of the traditional MCA assay. MCA-based assays have been increasingly used to detect drug-resistant mutations in TB (12, 15–18). One advantage is the detectability of heteroresistant mutations at a level similar to that detected by Sanger sequencing, i.e., 20 to 40%, which is otherwise impossible using real-time PCR-based assays (8, 19). However, these levels are far from the 1% achieved by the traditional culture-based phenotypic DST method for primary drugs (20–22). To increase the detectability of MCA for the less-abundant mutant, we used the detection probe as a clamp to inhibit wild-type amplification and KlenTaq-S DNA polymerase lacking 5′-to-3′ exonuclease activity to keep the probe intact during the entire amplification procedure. A similar principle was reported for the detection of rare K-ras mutations using a peptide nucleic acid in a LightCycler system (23). We chose locked nucleic acid (LNA) because of its flexible design and predicable Tm value (24, 25). This is important because a number of LNA bases can be easily introduced into the mutation-targeting portion of the probe to obtain a wide and adjustable Tm window between the mutant and the wild type. Within this window, a common Tc value can be easily determined among different mutation sites. As shown by the results of the multiplex DeepMelt TB/INH assay using a common Tc value of 71°C, all four mutations were detected at a 1% level. Thus, the DeepMelt strategy can be extended to other mutations associated with drug resistance in TB.
Interestingly, mutation enrichment efficiency reduced with increasing total DNA levels. This may be because the clamping effect of the probe became less efficient with increasing amounts of target DNA. This may compromise the performance of the DeepMelt TB/INH assay for detecting heteroresistance. However, because the DeepMelt TB/INH assay could detect 1% heteroresistance in DNA samples containing 104 M. tuberculosis genomes/μl, which corresponds to a very strong positive sample, it can certainly do so in most other clinical samples containing fewer than 104 M. tuberculosis genomes/μl.
The clinical value of the DeepMelt assay was demonstrated in the detection of two batches of clinical samples. In the first batch of 602 bacterial isolates, five more heteroresistance results were identified by the DeepMelt TB/INH assay than by the MeltPro assay. This increased the sensitivity from 93.6% (176/188, 95% CI = 89.2 to 96.3%) to 95.7% (180/188, 95% CI = 91.8 to 97.8%), despite the slightly decreased specificity from 97.6% (404/414, 95% CI = 95.6 to 98.7%) to 97.3% (403/414, 95% CI = 95.3 to 98.5%). In the second batch of 109 sputum specimens, two additional heteroresistant specimens were detected among 24 resistant specimens, increasing the sensitivity from 83.3% (20/24, 95% CI = 64.2 to 93.3%) to 91.7% (22/24, 95% CI = 74.2 to 97.7%), whereas the specificity (96.5%) was unchanged (82/85, 95% CI = 90.1 to 98.8%). The increased sensitivity of the DeepMelt assay demonstrated that heteroresistant samples containing low levels of mutant DNA that cannot be detected by MeltPro assay can be correctly identified in agreement with the phenotypic DST method.
Finally, we used digital PCR to determine the mutant percentages of eight heteroresistant samples harboring katG 315 mutations, further confirming that three samples newly identified by DeepMelt contained low levels of mutated DNA. Digital PCR is an absolutely quantitative method for counting copy number and has been widely used in the quantification of low percentage of mutation (26–28). Nevertheless, digital PCR needs independent reaction for each mutation to be detected, precluding its use for simultaneous detection of multiple mutant sites in a single assay.
In summary, the DeepMelt assay efficiently enriched low levels of mutant M. tuberculosis DNA in heteroresistant samples. The assay detected 1% isoniazid resistance and may be applicable for other drug-resistant mutations. This method could potentially fill the gap between current genotypic and phenotypic methods for detecting drug-resistant M. tuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant 81572068) and Important National Science and Technology Specific Project 2017ZX10302301.
We have applied for a Chinese patent for the DeepMelt assay.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01239-17.
REFERENCES
- 1.Rinder H. 2001. Hetero-resistance: an under-recognised confounder in diagnosis and therapy? J Med Microbiol 50:1018–1020. doi: 10.1099/0022-1317-50-12-1018. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann-Thiel S, van Ingen J, Feldmann K, Turaev L, Uzakova GT, Murmusaeva G, van Soolingen D, Hoffmann H. 2009. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur Respir J 33:368–374. doi: 10.1183/09031936.00089808. [DOI] [PubMed] [Google Scholar]
- 3.Rinder H, Mieskes KT, Loscher T. 2001. Heteroresistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 5:339–345. [PubMed] [Google Scholar]
- 4.Zhang X, Zhao B, Liu L, Zhu Y, Zhao Y, Jin Q. 2012. Subpopulation analysis of heteroresistance to fluoroquinolone in Mycobacterium tuberculosis isolates from Beijing, China. J Clin Microbiol 50:1471–1474. doi: 10.1128/JCM.05793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eilertson B, Maruri F, Blackman A, Herrera M, Samuels DC, Sterling TR. 2014. High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 58:3270–3275. doi: 10.1128/AAC.02066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Operario DJ, Koeppel AF, Turner SD, Bao Y, Pholwat S, Banu S, Foongladda S, Mpagama S, Gratz J, Ogarkov O, Zhadova S, Heysell SK, Houpt ER. 2017. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLoS One 12:e0176522. doi: 10.1371/journal.pone.0176522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brossier F, Pham A, Bernard C, Aubry A, Jarlier V, Veziris N, Sougakoff W. 2017. Molecular investigation of resistance to second-line injectable drugs in multidrug-resistant clinical isolates of Mycobacterium tuberculosis in France. Antimicrob Agents Chemother 61:e01299-16. doi: 10.1128/AAC.01299-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol 48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folkvardsen DB, Svensson E, Thomsen VO, Rasmussen EM, Bang D, Werngren J, Hoffner S, Hillemann D, Rigouts L. 2013. Can molecular methods detect 1% isoniazid resistance in Mycobacterium tuberculosis? J Clin Microbiol 51:1596–1599. doi: 10.1128/JCM.00472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravorty S, Roh SS, Glass J, Smith LE, Simmons AM, Lund K, Lokhov S, Liu X, Xu P, Zhang G, Via LE, Shen Q, Ruan X, Yuan X, Zhu HZ, Viazovkina E, Shenai S, Rowneki M, Lee JS, Barry CE III, Gao Q, Persing D, Kwiatkawoski R, Jones M, Gall A, Alland D. 2017. Detection of isoniazid-, fluoroquinolone-, amikacin-, and kanamycin-resistant tuberculosis in an automated, multiplexed 10-color assay suitable for point-of-care use. J Clin Microbiol 55:183–198. doi: 10.1128/JCM.01771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Zhao B, Huang H, Zhu Y, Peng J, Dai G, Jiang G, Liu L, Zhao Y, Jin Q. 2013. Co-occurrence of amikacin-resistant and -susceptible Mycobacterium tuberculosis isolates in clinical samples from Beijing, China. J Antimicrob Chemother 68:1537–1542. doi: 10.1093/jac/dkt082. [DOI] [PubMed] [Google Scholar]
- 12.Hu S, Li G, Li H, Liu X, Niu J, Quan S, Wang F, Wen H, Xu Y, Li Q. 2014. Rapid detection of isoniazid resistance in Mycobacterium tuberculosis isolates by use of real-time-PCR-based melting curve analysis. J Clin Microbiol 52:1644–1652. doi: 10.1128/JCM.03395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinese Antituberculosis Association. 2006. The laboratory science procedure of diagnostic bacteriology in tuberculosis, p 46–51. China Education and Culture Press, Beijing, China. [Google Scholar]
- 14.Low H, Chan SJ, Soo GH, Ling B, Tan EL. 2017. Clarity digital PCR system: a novel platform for absolute quantification of nucleic acids. Anal Bioanal Chem 409:1869–1875. doi: 10.1007/s00216-016-0131-7. [DOI] [PubMed] [Google Scholar]
- 15.Huang Q, Liu Z, Liao Y, Chen X, Zhang Y, Li Q. 2011. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One 6:e19206. doi: 10.1371/journal.pone.0019206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Hu S, Li G, Li H, Liu X, Niu J, Wang F, Wen H, Xu Y, Li Q. 2015. Evaluation of the MeltPro TB/STR assay for rapid detection of streptomycin resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb) 95:162–169. doi: 10.1016/j.tube.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Pang Y, Dong H, Tan Y, Deng Y, Cai X, Jing H, Xia H, Li Q, Ou X, Su B, Li X, Zhang Z, Li J, Zhang J, Huan S, Zhao Y. 2016. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci Rep 6:25330. doi: 10.1038/srep25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho SN, Barry CE III, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J Clin Microbiol 50:2194–2202. doi: 10.1128/JCM.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng J, Yu X, Cui Z, Xue W, Luo Z, Wen Z, Liu M, Jiang D, Zheng H, Wu H, Zhang S, Li Y. 2016. Multi-fluorescence real-time PCR assay for detection of RIF and INH resistance of Mycobacterium tuberculosis. Front Microbiol 7:618. doi: 10.3389/fmicb.2016.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canetti G, Froman S, Grosset J, Hauduroy P, Langerova M, Mahler HT, Meissner G, Mitchison DA, Sula L. 1963. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ 29:565–578. [PMC free article] [PubMed] [Google Scholar]
- 21.Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, Rist N, Smelev NA. 1969. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ 41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 22.American Thoracic Society. 2000. Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med 161:1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 23.Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, Chiou CC. 2006. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res 34:e12. doi: 10.1093/nar/gnj008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You Y, Moreira BG, Behlke MA, Owczarzy R. 2006. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res 34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owczarzy R, You Y, Groth CL, Tataurov AV. 2011. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry 50:9352–9367. doi: 10.1021/bi200904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huggett JF, Cowen S, Foy CA. 2015. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 61:79–88. doi: 10.1373/clinchem.2014.221366. [DOI] [PubMed] [Google Scholar]
- 27.Kuypers J, Jerome KR. 2017. Applications of digital PCR for clinical microbiology. J Clin Microbiol 55:1621–1628. doi: 10.1128/JCM.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pholwat S, Stroup S, Foongladda S, Houpt E. 2013. Digital PCR to detect and quantify heteroresistance in drug-resistant Mycobacterium tuberculosis. PLoS One 8:e57238. doi: 10.1371/journal.pone.0057238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.