ABSTRACT
The search for antiprion compounds has been encouraged by the fact that transmissible spongiform encephalopathies (TSEs) share molecular mechanisms with more prevalent neurodegenerative pathologies, such as Parkinson's and Alzheimer's diseases. Cellular prion protein (PrPC) conversion into protease-resistant forms (protease-resistant PrP [PrPRes] or the scrapie form of PrP [PrPSc]) is a critical step in the development of TSEs and is thus one of the main targets in the screening for antiprion compounds. In this work, three trimethoxychalcones (compounds J1, J8, and J20) and one oxadiazole (compound Y17), previously identified in vitro to be potential antiprion compounds, were evaluated through different approaches in order to gain inferences about their mechanisms of action. None of them changed PrPC mRNA levels in N2a cells, as shown by reverse transcription-quantitative real-time PCR. Among them, J8 and Y17 were effective in real-time quaking-induced conversion reactions using rodent recombinant PrP (rPrP) from residues 23 to 231 (rPrP23–231) as the substrate and PrPSc seeds from hamster and human brain. However, when rPrP from residues 90 to 231 (rPrP90–231), which lacks the N-terminal domain, was used as the substrate, only J8 remained effective, indicating that this region is important for Y17 activity, while J8 seems to interact with the PrPC globular domain. J8 also reduced the fibrillation of mouse rPrP23–231 seeded with in vitro-produced fibrils. Furthermore, most of the compounds decreased the amount of PrPC on the N2a cell surface by trapping this protein in the endoplasmic reticulum. On the basis of these results, we hypothesize that J8, a nontoxic compound previously shown to be a promising antiprion agent, may act by different mechanisms, since its efficacy is attributable not only to PrP conversion inhibition but also to a reduction of the PrPC content on the cell surface.
KEYWORDS: prion protein, therapy, antiscrapie compounds, RT-QuIC, prion strains, antiprion, antiscrapie, prions, PrP
INTRODUCTION
The prion protein (PrP) is to date the only agent attributed to be responsible for the transmissible spongiform encephalopathies (TSEs). These diseases comprise a group of fatal neurodegenerative disorders which arise after the conversion of the cellular prion protein (PrPC), normally present on the surface of many cell types, into a form called scrapie (PrPSc), which is disease associated (1). This conversion requires contact between both PrP forms, wherein PrPSc catalyzes the misfolding of PrPC, giving rise to new PrPSc molecules (2–4). Since the encounter between PrPC and PrPSc is essential, blocking this contact seems to be a promising strategy for the therapy of TSEs (5–7).
Another interesting feature of these diseases is that PrPSc may adopt different structures, which are associated with different prion strains (8–11). This is particularly important for the treatment of TSEs, since some antiprion compounds appear to have a strain-dependent effect (12–16). For this reason, it seems reasonable to look for molecules that target PrPC rather than PrPSc, since the substrate is the same in all conversion reactions. Albeit PrPC possesses physiological functions (17), research groups showed that PrP knockout animals, PrP(0/0), did not have significant clinical or physiological impairments (18, 19). Thus, compounds that dynamically bind to PrPC may be of interest.
Most cases of prion diseases occur spontaneously and predominantly affect people who are about 60 years old. The aging of the population, added to the fact that other neurodegenerative diseases share the same characteristics of prion diseases, has driven several research groups to search for new compounds with antiprion activity since these diseases are still incurable. Many strategies have been exploited to treat TSEs, such as PrPC stabilization, PrPSc clearance, and prevention of PrPC-PrPSc binding (20–22). However, no treatment available to date has been able to slow or interrupt the progression of the disease in humans. Besides strain selectivity, high toxicity and an unfavorable pharmacokinetic profile (especially, low levels of permeation through the blood-brain barrier) are factors that may hamper the attainment of therapeutic compounds (21). Our group has previously reported four trimethoxychalcones (compounds J1, J8, J20, and J35) and two oxadiazoles (compounds Y13 and Y17) that reduced protease-resistant PrP (PrPRes) levels in prion-infected murine neuroblastoma (ScN2a) cells, with 50% inhibitory concentrations ranging from 1 to 10 μM (23). Furthermore, at a concentration of 25 μM, these compounds were not toxic to noninfected murine neuroblastoma (N2a) cells. In a real-time quaking-induced conversion (RT-QuIC) assay (24), compounds Y13 and Y17 delayed and partially blocked the conversion of PrPC to PrPRes at a concentration of 50 μM (23). Using in silico approaches, these molecules were predicted to be sufficiently brain permeant, satisfactorily bioavailable after oral administration, and capable of binding to the same hydrophobic pocket of PrPC (23). Moreover, none of these compounds elicited acute toxicity in Swiss mice (25). Hence, the aromatic compounds J1, J8, J20, J35, Y13, and Y17 have been shown to be promising candidates for prion disease therapy.
Herein, we investigated the mechanisms underlying the antiprion role played by the trimethoxychalcones J1, J8, and J20 and the oxadiazole Y17 using in vitro strategies. As assessed by the RT-QuIC reaction, compounds J8 and Y17 inhibited PrPC-to-PrPRes conversion when the reaction mixture was seeded with hamster PrPSc and even with Creutzfeldt-Jakob disease (CJD) agent-infected human brain-derived PrPSc. Regarding their mechanisms of action, we showed that the PrPC N terminus is crucial to the activity of Y17, while J8 seems to bind to the PrPC globular domain. Furthermore, J1, J8, and Y17 decreased the PrPC content on the N2a cell surface by trapping this protein in the endoplasmic reticulum, which may be related to their antiprion effect in infected cells.
RESULTS
Effects of the compounds on PrPRes accumulation in cells infected with different prion strains.
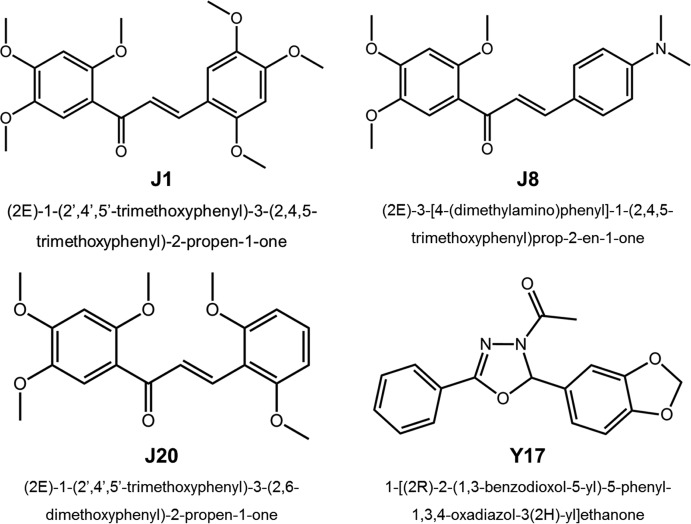
In a previous study, we analyzed a panel consisting of over 200 aromatic compounds and found four chalcones (compounds J1, J8, J20, and J35) and two oxadiazoles (compounds Y13 and Y17) able to decrease PrPRes levels by at least 50% in RML strain scrapie-infected N2a (ScN2a-RML) cells with 50% effective concentrations up to 10 μM. Additionally, these compounds were devoid of toxicity for ScN2a cells and were predicted to have an adequate drug-like profile (23). The acute toxicity of these substances was evaluated in vivo, and we found that single oral administration (300 mg/kg of body weight) or repeated intraperitoneal administration (10 mg/kg 3 times a week for 4 weeks) did not cause toxic effects in mice (25). Based on these findings, in the study described here we analyzed by the RT-QuIC assay the efficacies of four of these compounds (compounds J1, J8, J20, and Y17, the structures of which are shown in Fig. 1) against other prion strains using different infected cell cultures and also in a cell-free PrPC-PrPSc system.
FIG 1.
Chemical structures of the antiscrapie compounds evaluated.
The existence of distinct prion strains is an intriguing characteristic of TSEs. Each prion strain confers a different conformation to PrP, and the strains are distinguished by differences in incubation times (26, 27), the times of onset of clinical symptoms (28), neuropathological patterns and the extent of cerebral damage (29), glycosylation profiles, and resistance to digestion by proteinase K (PK) (30). For these reasons, testing the antiprion activity of the antiscrapie drug candidates against different strains is crucial.
The initial screening was performed by dot blotting using ScN2a-RML cells (23). Here, in addition to the RML strain, we also performed our analyses on scrapie agent strain 22L-infected N2a (ScN2a-22L) cells. The latter strain is naturally more resistant, since it originated from the reinfection of ScN2a-RML cells that had been cured after seven passages in medium containing 1 μg/ml pentosan polysulfate (PPS) (31). We also used chronic wasting disease agent-infected mule deer brain (MDB-CWD) cells (32) in order to test the effects of the compounds on a nonmurine cell line.
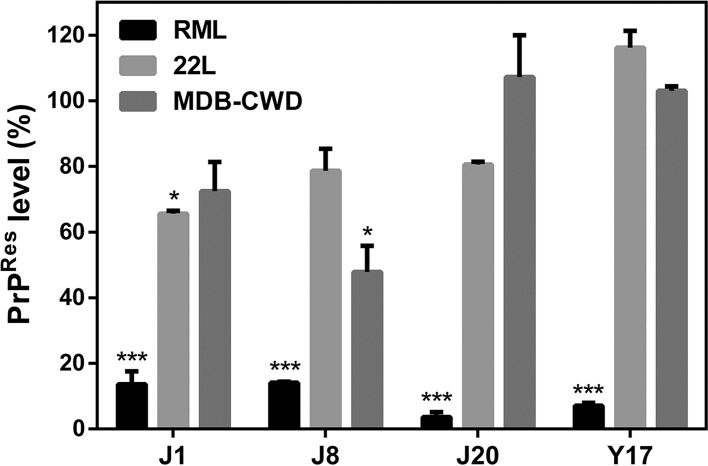
As previously demonstrated (23), all compounds decreased the PrPSc content in ScN2a-RML cells by ∼80% (Fig. 2). However, the same effect was not observed in ScN2a-22L cells and MDB-CWD cells, in which only compounds J1 and J8 had a significant effect. J1 reduced the PrPSc levels in ScN2a-22L cells by ∼40%, while J8 decreased the level of PrPSc accumulation in MDB-CWD cells by ∼50%. These results are in agreement with published data showing that the effect against other prions strains is less intense than that observed against the RML strain (31, 33, 34). Since there is no human cell line persistently infected with PrPSc up to the present moment, we evaluated the efficacy of the compounds by the RT-QuIC assay, which allowed us to use infected human brain homogenate as a seed in this cell-free conversion assay.
FIG 2.
Effects of the compounds on PrPSc accumulation in ScN2a-RML, ScN2a-22L, and MDB-CWD cells. The cells were incubated in the presence of the compounds at 10 μM for 4 days in 96-well plates. All conditions were tested in quadruplicate in each experiment. Cell lysates were treated with PK and submitted to dot blotting. PrPRes was detected with the anti-PrP antibody 6D11. Quantification of the assay was done by integrating the density of each dot using ImageJ software. One-way ANOVA was performed. Error bars indicate standard errors of the means (SEM) for at least two replicates. *, P < 0.05 relative to the control; ***, P < 0.001 relative to the control. The data shown are representative of those from two independent experiments.
Evaluation of the effects of the compounds by RT-QuIC assay.
One of the most important features of prions is the ability of the infectious prion molecule, PrPSc, to recruit PrPC and convert it into the abnormal form. This is the central event in prion diseases (1). Therefore, inhibition of this conversion has been exploited as a therapeutic strategy, and investigators have searched for compounds able to inhibit this conversion (5–7, 23).
The RT-QuIC assay is an in vitro amplification method used to detect very small quantities of PrPSc particles in biological fluids, such as cerebrospinal fluid, olfactory mucosa (35), and saliva (36). In RT-QuIC reactions, recombinant prion protein (rPrP) is used as the substrate and infected brain homogenate is used as a source of the PrPSc seed, which promotes the conversion (37, 38). The in vivo conversion of PrPC into PrPSc is characterized by a conformational change wherein there is a reduction in the α-helical content accompanied by an increase in the β-sheet structure (1, 39). This gain in β-sheets allows PrPSc molecules to assemble and give rise to structured aggregates (40), such as amyloid fibrils. These fibers interact with thioflavin T (ThT), an established dye for amyloid fibrils (41), leading to an increase in its fluorescence intensity. Since the PrPRes formed in vitro keeps the same properties, ThT is used to monitor PrPRes formation. Using this assay, it is also possible to identify molecules able to decrease or inhibit PrPRes amplification. For this reason, this approach has been used to screen antiprion compounds and also to characterize their mechanisms of action (7, 23, 38, 42, 43). Recently, Hughson and colleagues successfully employed this assay to test the ability of hypochlorous acid to inactivate prions (44). Thus, we applied this technique to check the efficacy of the compounds investigated here.
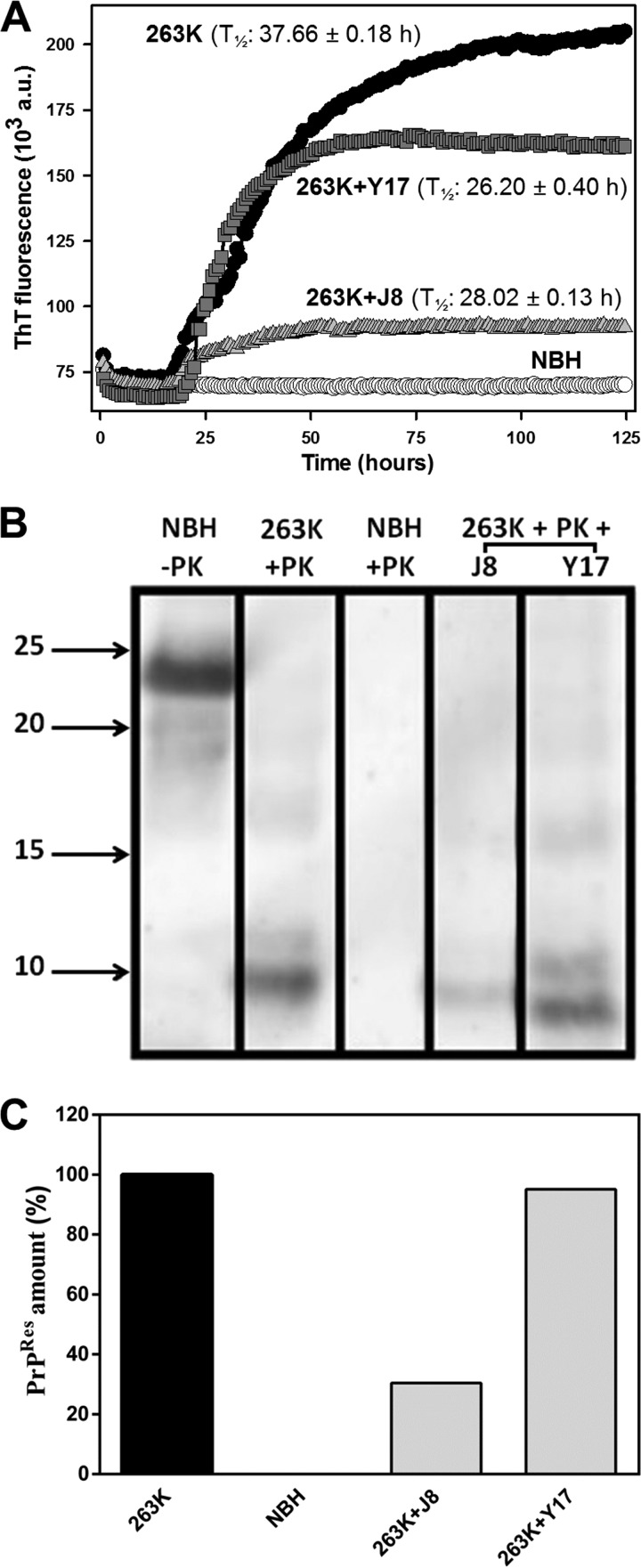
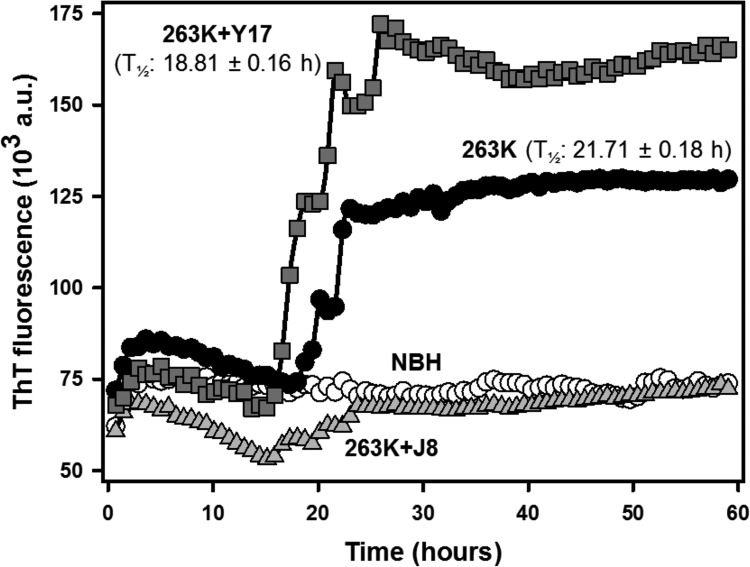
Using 263K scrapie agent-infected hamster brain homogenate as the seed, we verified that compounds J8 and Y17 decreased the amount of PrPRes formed (Fig. 3A). J8 was the most effective one, sharply inhibiting this reaction. These compounds did not delay the beginning of the ThT-probed fibrillation process (lag phase), as indicated by comparing the times to the half-maximal ThT fluorescence (T1/2) of the reactions (Fig. 3A). Compounds J1 and J20 were not inhibitory in this assay (data not shown).
FIG 3.
Effects of the test compounds on rPrP23–231 fibrillation in reaction mixtures seeded with 263K scrapie agent-infected hamster brain homogenate. (A) RT-QuIC reactions with full-length hamster rPrPSen at 0.1 mg/ml (4.3 μM) in the absence (positive control) or in the presence of the test compounds at 40 μM (1:10 molar ratio). Normal brain homogenate (NBH) was used as a negative control. T1/2 values are shown. All conditions were tested in quadruplicate in each experiment. a.u., absorbance units. (B) Western immunoblots of the reaction end products. The contents from each experiment (4 wells) were mixed, treated with 10 μg/ml (0.35 μM) PK at 37°C for 1 h, and subjected to Western blotting. PrPRes was labeled with the antibody R20. Molecular size markers (in kilodaltons) are shown on the left. (C) PrPRes quantities normalized to the values for the positive control (100%) and the negative control (0%). The experiment was performed three times, and the data shown here are representative of those from one experiment. The lanes were cut from the same blot, with no differential adjustments being made to the image contrast or intensity. The results for compounds J1 and J20 were removed, since they were not effective in this assay. The original blot is shown as Fig. S1 in the supplemental material.
To confirm that the decreases in the ThT fluorescence intensity could be attributed to interference with rPrP conversion rather than a direct artifactual quenching of ThT fluorescence, we performed Western blotting to quantify the amount of rPrPRes after the end of the reactions. There was less intense antibody staining of the rPrPRes bands in the lanes corresponding to the samples with J8 and Y17 (Fig. 3B; see also Fig. S1 in the supplemental material). Normal brain homogenate (NBH) treated with PK revealed no labeling, as expected, while a fragment of about 23 kDa corresponding to full-length rPrP was identified in the untreated lane. The quantification of the gel (Fig. 3C) showed that J8 decreased the PrPRes content by ∼70% in the RT-QuIC reaction. However, Y17 had only a minor effect, reducing the PrPRes content by less than 10%.
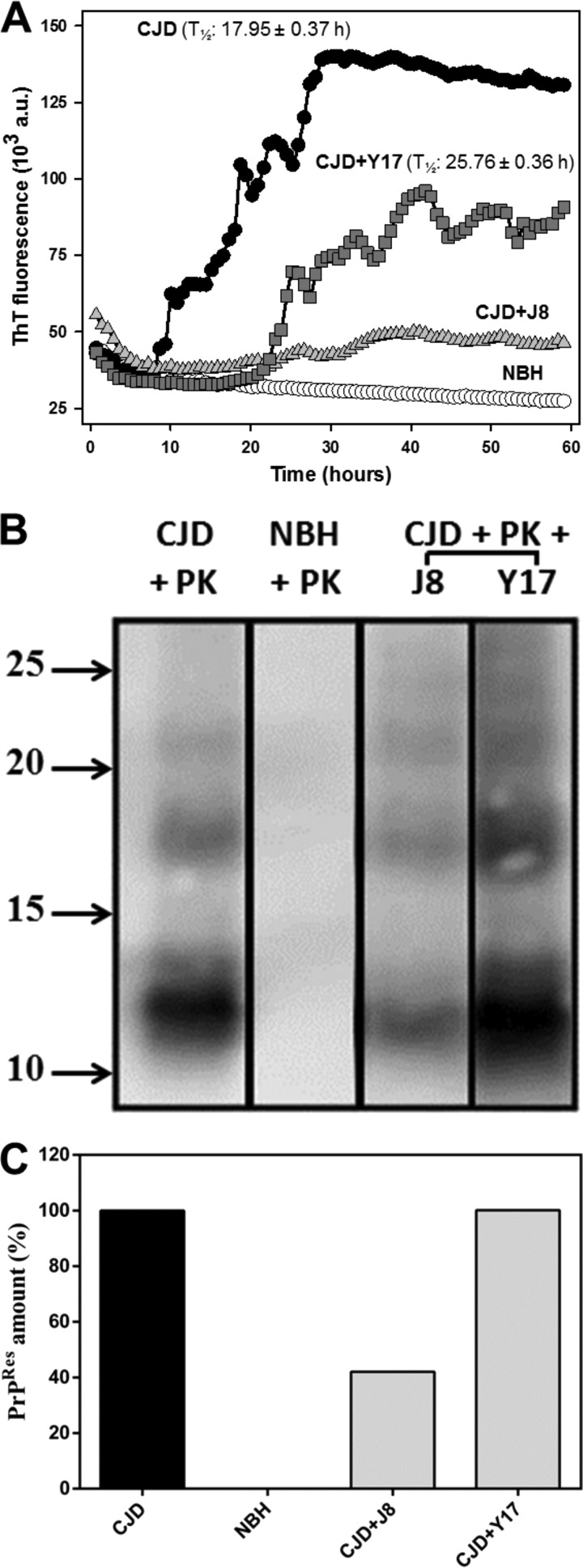
We then performed the RT-QuIC reaction using CJD agent-infected human brain homogenate as the seed. As with hamster scrapie agent-seeded reactions, compounds J8 and Y17 decreased the level of fibril amplification seeded by human PrPSc (Fig. 4A). This time, these compounds not only decreased the amount of rPrPRes formed but also affected the reaction kinetics, retarding the lag phase and increasing the T1/2 values. A similar effect was observed by Vieira and colleagues (42), wherein low-molecular-weight heparin affected both the extent and the kinetics of PrP fibrillation. For the same reason described above, we subjected the RT-QuIC reaction end products to Western blotting (Fig. 4B and S2), wherein a reduction in the amount of rPrPRes was evoked by J8 (Fig. 4C).
FIG 4.
Effects of the test compounds on rPrP23–231 fibrillation in reaction mixtures seeded with CJD agent-infected human brain homogenate. (A) RT-QuIC reactions with full-length hamster rPrPSen at 0.1 mg/ml (4.3 μM) in the absence (positive control) or in the presence of the test compounds at 40 μM (1:10 molar ratio). Normal brain homogenate (NBH) was used as a negative control. T1/2 values are shown for all experiments except the experiment with the CJD agent and J8, in which the T1/2 values could not be correctly determined. All conditions were tested in quadruplicate. (B) Western immunoblots of the reaction end products. Molecular size markers (in kilodaltons) are shown on the left. (C) Quantities of PrPRes relative to the values for samples containing the CJD agent seed (positive control) and NHB (negative control). Lanes were cut from the same blot, with no differential adjustments being made to the image contrast or intensity. The results for compounds J1 and J20 were removed, since they were not effective in this assay. The original blot is shown as Fig. S2 in the supplemental material.
Among the mechanisms of action already identified for bioactive compounds with antiprion activity are (i) direct binding to PrPC or PrPSc, which can stabilize either one or both forms and prevent further interaction between them; (ii) the ability to decrease the level of PrPC expression on the cell surface, which reduces the availability of substrate for the conversion; and (iii) PrPSc clearance (discussed in references 5 and 43).
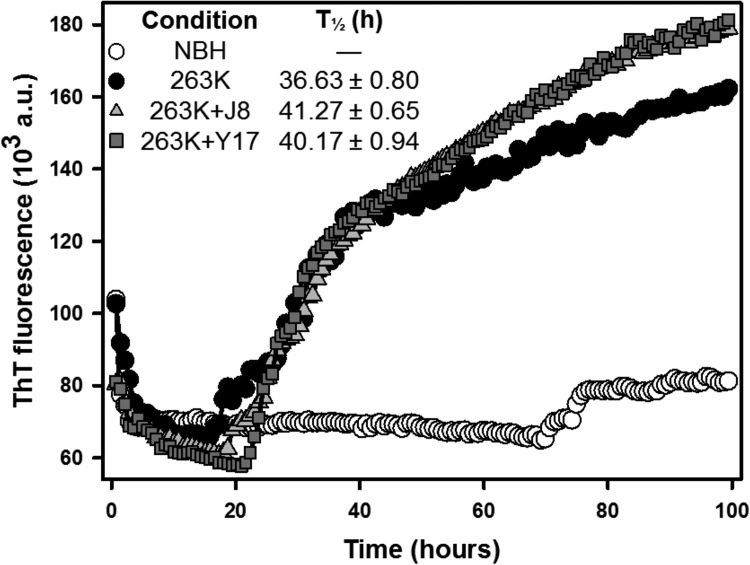
Some of our compounds were effective in the RT-QuIC assay, leading us to believe that they act by direct binding to PrP molecules (either PrPC or PrPSc). To assess what region of PrP is crucial for the effects of the compounds in this assay, we performed an RT-QuIC reaction with truncated PrP (rPrP from residues 90 to 231 [rPrP90–231]). This construction lacks the N-terminal region, which comprises a critical binding site for PrPSc and is therefore crucial for prion propagation. Mice expressing PrPC with a deletion in this region (a deletion from residues 23 to 31 [Δ23–31]) exhibit resistance to prion infection, as evidenced by elongated incubation times and delayed accumulation of PrPSc (45). PrP interaction with ligands putatively associated with its physiological function can also be mediated by this region (reviewed in references 17 and 46). ThT fluorescence was less intense when we used the rPrP90–231 construct (Fig. 5) than when we used the rPrP23–231 construct (Fig. 3) as the substrate. J8 remained effective against the truncated construct, indicating that this compound probably interacts with the PrPC C-terminal, globular domain (Fig. 5). On the other hand, Y17 was ineffective in this assay, showing that the N-terminal region is essential to its activity.
FIG 5.
Effects of test compounds on RT-QuIC kinetics with rPrP90–231 as the substrate and strain 263K as the seed. The reactions were performed with truncated hamster rPrPSen at 0.1 mg/ml (6.3 μM) in the absence (positive control) or in the presence of the test compounds at 40 μM. Normal brain homogenate (NBH) was used as a negative control. T1/2 values are shown for all experiments except the experiment with 263K and J8, in which the T1/2 values could not be correctly determined.
RT-QuIC reactions were also performed after preincubation of the compounds with the seed only (in the absence of the substrate), which resulted in the loss of their efficacy (Fig. 6). However, the screening in cell lines infected with different prion strains showed a strong strain-dependent effect for all compounds (Fig. 2), indicating that an alternative mechanism of action may exist. Based on our results, we cannot rule out the possibility that the compounds bind to both PrP forms (PrPC and PrPSc) or that another substance in the brain homogenate might be quenching the compounds, thus making them unavailable to bind to PrPC (or even PrPSc). Additionally, the interaction of the compound with PrPSc from the brain homogenate might somehow allow the infectious form to convert PrPC in vitro.
FIG 6.
Effect of preincubation of the test compounds with the strain 263K seed on rPrP23–231 fibrillation. The reactions were performed with full-length hamster rPrPSen at 0.1 mg/ml (4.3 μM) in the absence (positive control) or in the presence of the test compounds at 40 μM. Normal brain homogenate (NBH) was used as a negative control. T1/2 values for the RT-QuIC reaction kinetics are shown.
Effect on fibrillation kinetics seeded with in vitro-produced fibrils.
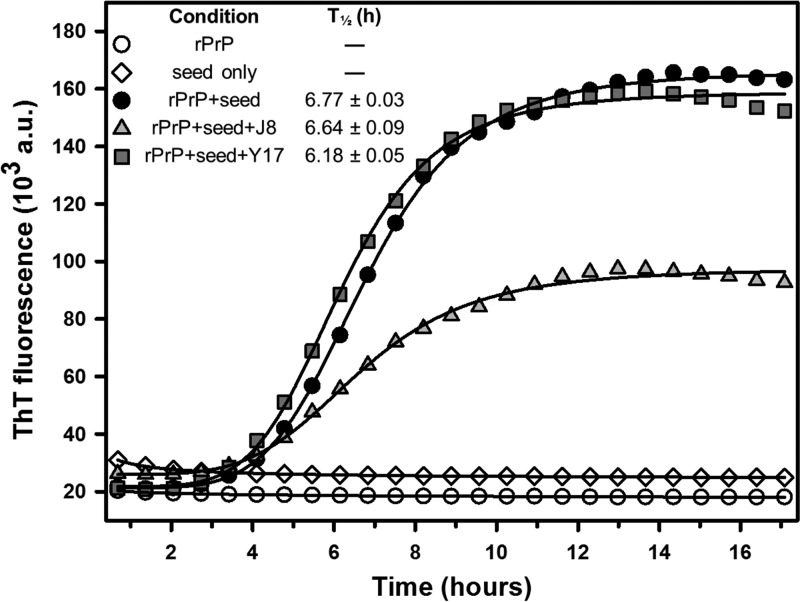
Baskakov and colleagues developed a technique to induce rPrP fibrillation in vitro under partially destabilizing conditions (1 M guanidinium HCl and 3 M urea) without the need for a PrPSc seed (47, 48). Herein, we used this method as a tool to generate fibrils and employ them to seed the in vitro conversion of murine rPrP23–231 under nondenaturing conditions, as in the RT-QuIC assay. This technique provided a simplified and economical approach to investigate the ability of the compounds to influence rPrP fibrillation in vitro. Although none of the compounds delayed the lag phase, as indicated by comparing the T1/2 values for the reactions, the total conversion was significantly reduced in the presence of J8 in relation to the level of conversion of the control (Fig. 7). This finding corroborates the results from the RT-QuIC assays, reinforcing our hypothesis that J8 interacts with PrPC and prevents its conformational transition into misfolded, amyloid forms.
FIG 7.
Kinetics of murine rPrP23–231 fibrillation in reaction mixtures seeded with in vitro-produced fibrils. The assay was performed with 2 μM rPrP, 1% (mol/mol) seed, and the test compounds at 20 μM (1:10 molar ratio). T1/2 values are shown. The data shown are representative of those from five independent experiments.
Molecular docking simulation.
Previous works have indicated that some residues in PrPC α-helices 2 and 3 have a reduced conformational stability compared to others. Conformational fluctuations in these regions could account for the transition from PrPC to high-energy intermediate states and PrPSc (49, 50). Kuwata et al. (51) reported an effective antiprion compound, named GN8, which reduced the PrPRes content in cells infected with four prion strains, prolonged diseased mouse survival after intraventricular and subcutaneous administration, stabilized the PrPC structure against thermal denaturation, and directly bound to PrPC globular domain. As evidenced by nuclear magnetic resonance (NMR) and computer simulation analyses, this group showed that GN8 interacted with the Asn159 (located in the α1-β2 loop) and Glu196 (located in the α2-α3 loop) residues by hydrogen bonding and proposed that the intercalation between these two hot-spot regions stabilizes the PrPC conformation. Subsequent computational studies reinforced the chemical chaperone activity exhibited by GN8 (52, 53).
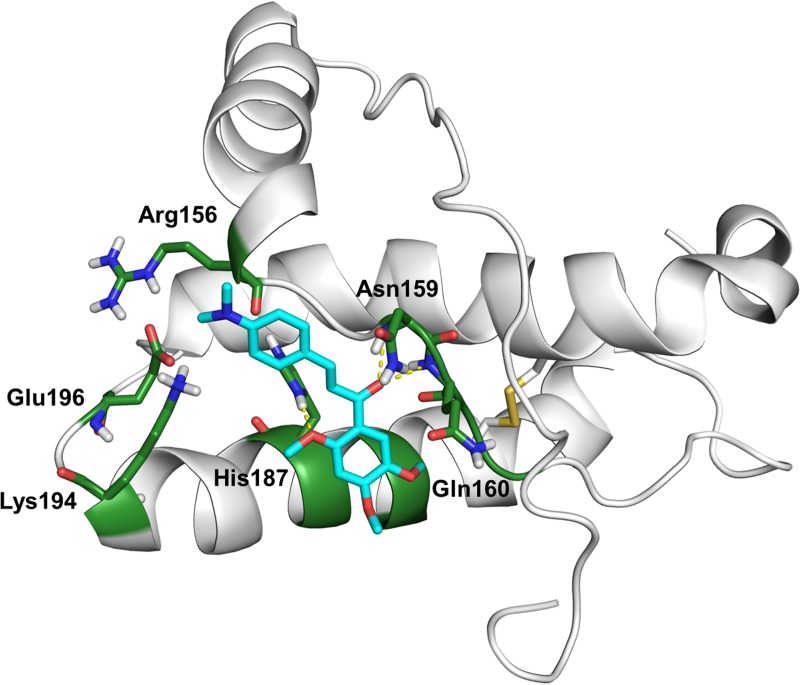
In order to obtain further information about the mode of interaction of J8 with the PrPC globular domain, we performed a molecular docking simulation between PrP121–231 (PDB ID 1AG2) and J8 using AutoDock (version 4.2.6) software. To validate our method, we first executed the docking analysis with GN8 and found a molecular orientation and interactions very similar to those evidenced in previous work (Fig. S3) (51–53). J8 was predicted to form hydrogen bonds with α1-β2 loop residues Asn159 and Gln160 and with α2 residue His187 (Fig. 8). We thus hypothesize that the connection between these two sites made by J8 may restrict conformational fluctuations in the PrPC globular domain and hamper the conversion into β-sheet-rich structures, like those in PrPSc.
FIG 8.
Molecular docking simulation of J8 with PrP121–231 (PDB ID 1AG2). The carbon atoms in the main residues of the PrPC hot-spot regions are colored in green, and the carbon atoms in J8 are colored in cyan. Hydrogen bonds are represented as yellow dashed lines. Hydrogen bonds were formed between the J8 2-methoxy group and the His187 side chain (3.1 Å) and between the J8 carbonyl group and both the Asn159 main chain (2.7 Å) and the Gln160 main chain (2.7 Å). The binding energy was −7.35 kcal/mol.
Effects on cellular PrP gene expression.
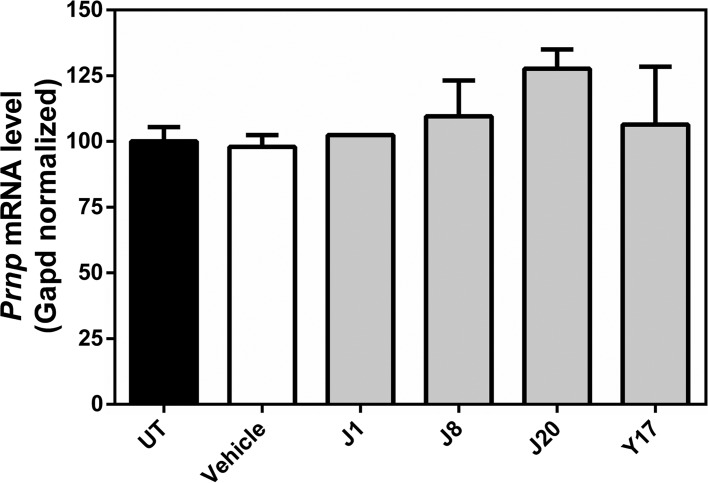
Our next approach was to investigate whether changes in PrPC expression could be a key event in the antiprion activity displayed by the compounds. We treated N2a cells with the compounds and analyzed PrPC gene expression by reverse transcription-quantitative real-time PCR (RT-qPCR). Treatment with J1, J8, J20, or Y17 did not alter the Prnp mRNA content in relation to that in untreated cells (Fig. 9). This finding strongly suggests that the mechanisms of action of these compounds are independent of the regulation of Prnp mRNA levels.
FIG 9.
RT-qPCR analysis of Prnp mRNA expression in N2a cells. Cells were treated with the test compounds (J1, J8, J20, and Y17) at 10 μM or the vehicle (DMSO 0.1%). Prnp mRNA levels were normalized to the Gapd mRNA content and were expressed relative to the value for untreated cells (UT). One-way ANOVA was performed using GraphPad Prism software. Error bars indicate standard errors of the means (SEM). The number of independent experiments for each condition was 4 for untreated cells, 1 for vehicle-treated cells, 1 for J1-treated cells, 3 for J8-treated cells, 2 for J20-treated cells, and 1 for Y17-treated cells.
Effects of the compounds on PrPC localization.
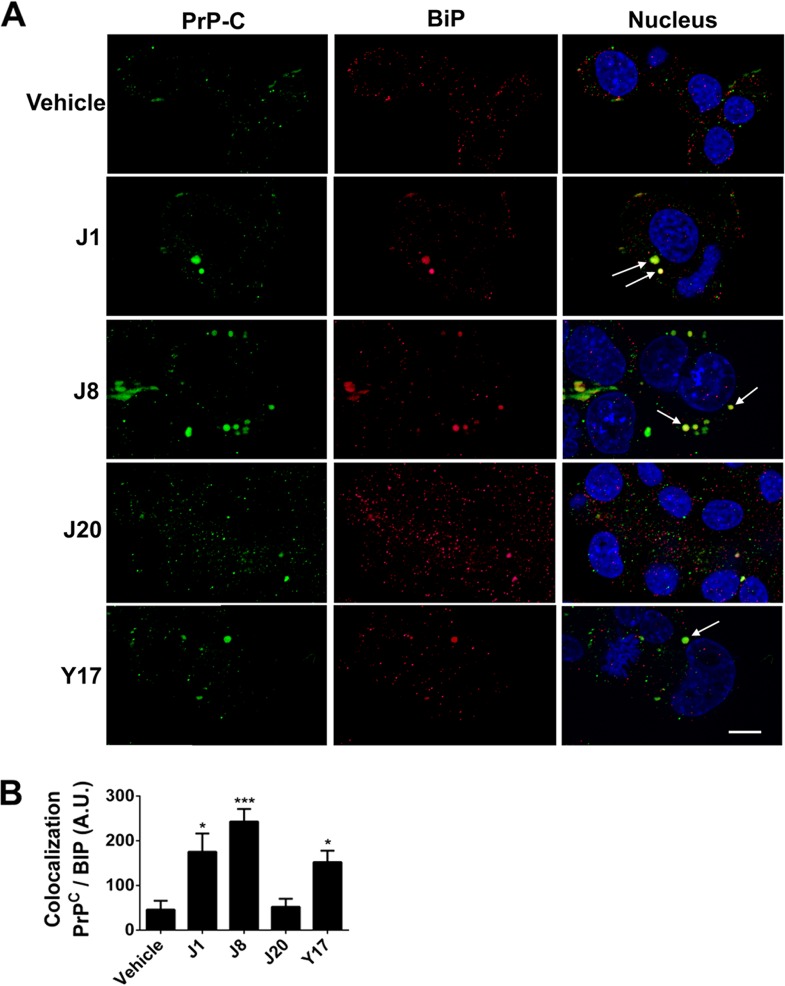
As the compounds did not alter the PrPC mRNA content, we evaluated if the antiprion effect could be due to changes in PrP cellular location by analyzing the N2a cells treated with the compounds by immunofluorescence. One of the ways to prevent PrPSc formation is to promote the internalization of PrPC, thus reducing the amount of substrate available on the cell surface to be converted into PrPSc. Our results showed that J1, J8, and Y17 were able to drastically reduce the level of PrPC exposure on the cell surface (Fig. S4). In order to determine if the test compounds were able to restrain the PrPc at the endoplasmic reticulum, we measured by structured confocal microscopy the amount of PrPc colocalized with GRP78/BiP (endoplasmic reticulum-resident chaperone GRP78/binding immunoglobulin protein) (Fig. 10A). We verified that cells treated with the same compounds able to reduce PrPC membrane location (J1, J8, and Y17) demonstrated high levels of GRP78/BiP-PrPC colocalization (Fig. 10B). The same analysis was performed using as the target microtubule-associated protein 1A/1B light chain 3B (LC3), a marker of autophagosomes, but no association between PrPC and LC3 was demonstrated (Fig. S5), indicating that the mechanism of action of compounds J1, J8, and Y17 is related to PrPC arrest in the endoplasmic reticulum.
FIG 10.
PrPC neuronal location after test compound treatment. (A) Representative fields of images from structured illumination confocal immunofluorescence analysis of PrPC (green) and BiP (red) cellular colocalization in mouse neuroblastoma N2a cells treated with 10 μM test compounds (J1, J8, J20, and Y17) or 0.1% DMSO (vehicle). Arrows, areas of PrPC and BiP colocalization, indicating PrPC arrest at the endoplasmic reticulum. The nuclei were stained with DAPI (blue). Bar, 10 μm. (B) Colocalization analysis was performed with ImageJ software. Twenty fields with more than 30 cells each were analyzed per condition. One-way ANOVA was performed. Error bars indicate standard errors of the means (SEM). *, P < 0.05 relative to the control; ***, P < 0.001 relative to the control.
Effects of the compounds on rPrP stability in vitro.
We then investigated whether the antiprion activity exhibited by the compounds could be attributable to an ability to stabilize the PrPC structure. This was performed by subjecting mouse rPrP23–231 to temperature-induced denaturation in the presence of the compounds. Circular dichroism was used to monitor the changes in the PrP secondary structure along the process. The ellipticity values at 222 nm, which are distinctive for the α-helical content (54), were recorded during temperature scanning from 25 to 80°C. None of the compounds altered the stability of rPrPC at molar ratios of 1:1 and 1:10 (Fig. S6), as they did not elicit any significant difference in T1/2 values in relation to the value for the vehicle. This finding may be explained by transient binding to the protein.
DISCUSSION
The unceasing search for antiprion drugs has been stimulated not only because TSEs comprise a group of untreatable diseases but also because prion diseases share some pathological characteristics with other neurodegenerative diseases, such as Alzheimer's and Parkinson's diseases (6, 55). The most common strategy exploited to discover new antiprion drug candidates is based on screening for molecules that are able to decrease the amount of PrPSc in prion-infected cells or that are effective in inhibiting PrPC-to-PrPSc conversion. In this sense, several antiscrapie compounds have previously been identified through in silico and in vitro methodologies, including 2-aminothiazoles, tetrapyrroles, methoxychalcones, oxadiazoles, pyrazolones, and polyanionic or polycationic molecules (23, 56–61). Members of several classes of compounds have extended the survival time of prion-infected animals when they were administered prophylactically or well in advance of the clinical phase of disease, but no treatments are known to be effective in humans at any stage of prion infection. Thus, it remains important to continue the search for more effective compounds that may directly interact with PrPC or PrPSc, affect PrPSc clearance, or act via additional mechanisms of action. Regarding the direct interaction with PrPC, Hosokawa-Muto and coworkers (62) investigated a library of 60,000 compounds and identified, through in silico screening using AutoDock software, 590 potential ligands able to interact with PrP. From these, 205 were selected for evaluation in prion-infected cells, and only 24 significantly inhibited the accumulation of PrPSc. Besides that, some of the compounds decreased PrPSc levels but had a low affinity of binding to PrP, as evidenced by surface plasmon resonance. Subsequently, the same group classified antiprion substances into four categories according to their effect: (i) those that specifically stabilize the conformation of PrP, (ii) those that nonspecifically stabilize the conformation of PrP, (iii) those that aggregate and precipitate PrPC, and (iv) those that interact with molecules other than PrPC. Considering the multifactorial nature of prion diseases, a molecule with two motifs, one for prion recognition and another for metal chelation, was synthesized (63). Beyond its antiprion activity in ScN2a cells, this compound (multitarget-directed ligand 1) extended the lag phase in an in vitro amyloid seeding assay, inhibited metal-induced prion aggregation, and decreased the amounts of reactive oxygen species (63), indicating that multitarget-directed ligands may be of value for the treatment of TSEs. It is crucial to consider that beyond PrP aggregation and accumulation, these diseases also involve brain inflammation, synaptic alterations, and neuronal death (64), and therefore, it is reasonable to identify compounds that act against other targets, besides PrP. In this work, we investigated the mechanisms of action of four compounds previously identified to have antiprion activity in ScN2a cells (23). It is worth mentioning that one of the main reasons for the failure of various antiprion compounds as drugs is in vivo toxicity. In this sense, the selected compounds, which were previously tested in healthy mice (25), did not elicit acute toxicity, thus encouraging us to further investigate their mechanisms of action. Since two of the most common targets of antiprion drugs are PrPC and PrPSc, we performed RT-QuIC reactions to check this possibility. We found that J8 was effective when both seeds (263K and the CJD agent) were used, while Y17 was active only against the CJD agent seed. Furthermore, J8 and Y17 also delayed the lag phase, increasing the T1/2 of the reaction from ∼18 h to 32 and 26 h, respectively. We performed docking assays with J8 using murine PrP (PDB ID 1AG2) to support our proposal that this compound interacts with the globular domain of PrPC (Fig. 9), as it was effective both in RT-QuIC reactions and also when rPrP90–231 was used as the substrate. In the absence of the PrP N-terminal region, the inhibitory effect was intensified, probably because the unstructured region could hamper the interaction between this molecule and the protein. Performance of the RT-QuIC assay with preincubation of the compounds with the brain homogenate showed a loss of compound efficacy (Fig. 6), indicating that these compounds may interact with other molecules in the brain homogenate (or even with PrPSc). As discussed above, other mechanisms apart from direct PrPC/PrPSc binding can contribute to antiprion activity. None of the compounds altered significantly the PrPC mRNA content, as verified by RT-qPCR (Fig. 9). To explore that, we analyzed the effects of compounds in the cellular environment by structured illumination confocal microscopy. We observed that J1, J8, and Y17 decreased the amount of PrPC on the cell surface, causing PrP arrest in the endoplasmic reticulum (Fig. 10). The association of PrPC with GRP78/BiP could significantly impact the outcome of the disease, due to the involvement of GRP78/BiP in the reduction of the level of PrPSc replication and the disease phenotype associated with the inhibitory activity on PrP misfolding. Park and collaborators demonstrated that reducing the amount of GRP78/BiP in vitro and in vivo led to a significant increase in the level of PrPSc replication and disease progression (65). More interestingly, overexpression of GRP78/BiP reduced the level of PrPSc formation in cells. In summary, we suggest that compound J8 is an interesting candidate for further in vivo analysis in diseased mice, as it was effective in the in vitro conversion and aggregation assays, and this effect might be due to a direct interaction with PrPC. Nevertheless, we propose that compounds with mechanisms other than direct binding to PrPC/PrPSc and also clearance of PrPSc be investigated in the search for antiprion compounds. This proposal is supported by the fact that several inhibitors of PrPC-to-PrPSc conversion are identified annually but in fact there is no proportional progress in preclinical and clinical studies of these compounds.
MATERIALS AND METHODS
Compounds.
The chalcones (compounds J1, J8, and J20) and oxadiazole (compound Y17) (Fig. 1) analyzed in this work were synthesized as previously described (66–68). Structural characterization was performed by 1H and 13C nuclear magnetic resonance (NMR) in CDCl3 or dimethyl sulfoxide-d6 (DMSOd6) solution and by electrospray ionization mass spectrometry (ESI-MS). The NMR and MS-ESI spectra obtained for the chalcones (compounds J1, J8, and J20) and oxadiazole (compound Y17) were according to the assigned structures and the structural data available in work previously published in the literature (66–68).The purity of the tested compounds was assessed by analytical high-performance liquid chromatography (HPLC) using a Shimadzu LC-20AD chromatograph with a Kromasil 100-5C18 column (4.6 mm by 250 mm) and a Shimadzu SPD-M20A detector. The isocratic HPLC solvent system was acetonitrile-water at 60:40 or 80:20, and the flow rate was 1.0 ml/min. All compounds presented purities higher than 95%. These substances were solubilized in dimethyl sulfoxide (DMSO) and then diluted in phosphate-buffered saline, glucose, and phenol red (PBBS) at pH 7.3 for scrapie agent-infected cell assays or in phosphate-buffered saline (PBS) at pH 7.0 for spectroscopy measurements.
rPrP expression and purification.
rPrP was expressed in E. coli and purified by affinity chromatography as previously described (24).
Cell lines.
Murine neuroblastoma (N2a) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 10 μg/ml gentamicin (complete medium). We also used a murine neuroblastoma cell line, ScN2a, infected with the prion strains RML (69) and 22L (70) and cells of a transformed mule deer cell line persistently infected with the chronic wasting disease agent (MDB-CWD cells) (32). Scrapie agent-infected cells were grown in Opti-MEM medium (Gibco Life Technologies) supplemented with 2 mM glutamine, 10% FBS, penicillin, and streptomycin. All cells were incubated at 37°C in a 5% CO2 atmosphere.
Dot blot assays.
Dot blot assays were performed as previously described (23, 31). Cells that had previously been cultured in bottles until they reached confluence were seeded into a 96-well plate and incubated for 4 h to enable adhesion to the plate surface. After this period, the compounds were applied into the medium to a final concentration of 10 μM, and the culture was incubated for 4 days. Each treatment was tested in quadruplicate. The vehicle corresponded to DMSO at 0.1%. Before proceeding to subsequent steps, the cells were inspected by optical microscopy, and no change in cellular appearance relative to that of the control cells was observed. After removal of the culture medium, the cells were lysed and treated with 25 μg/ml (0.88 μM) proteinase K (PK; Calbiochem) for 1 h at 37°C; then, 200 μl of Pefabloc at 1 mM (Boehringer Manheim) was added to each well to inhibit PK activity. The lysates were applied to the dot blot apparatus (Minifold 1; Schleicher & Schuell, BioScience GmbH) and transferred to a polyvinyl difluoride membrane. The Prnp(0/0) mouse-derived monoclonal IgG2a anti-mouse PrP antibody 6D11 (1:7,500; code sc-58581), which recognizes PrP residues 97 to 100 (71, 72), was diluted 1:7,500 in a 5% nonfat milk plus Tris-buffered saline–Tween 20 (TBS-T) solution for 1 h. After 3 washings in TBS-T, the alkaline phosphatase-conjugated secondary antibody was added and the mixture was incubated for at least 30 min. The membrane was then washed with TBS-T, and the AttoPhosH reagent (Promega) was applied to the center of the membrane, which was left to dry prior to being read in a Typhoon scanner (GE Healthcare). The intensity density for each well was quantified using ImageJ software (http://imageJ.nih.gov/ij).
RT-QuIC assays.
Real-time quaking-induced conversion (RT-QuIC) reactions were performed as previously described (23, 24). Each well of a black-walled, clear-bottom 96-well plate (Nunc, Rockford, IL, USA) was loaded with a reaction mixture containing the following components at the indicated final concentrations: 10 mM phosphate buffer (pH 7.4), 300 mM NaCl, 10 μM thioflavin T (ThT), 1 mM EDTA tetrasodium salt, test compounds at 40 μM, substrate (rPrPSen) at 0.1 mg/ml, and 2 μl of a 1 × 10−7 dilution of 263K scrapie agent-infected hamster brain homogenate and CJD agent-infected human brain homogenate containing ∼10 fg of PrPRes, as quantified by Western blotting. Two different substrates were used: full-length hamster PrP (PrP23–231) and truncated hamster PrP (PrP90–231), which does not have the N-terminal region. Each treatment was tested in quadruplicate. The vehicle corresponded to 0.4% DMSO. The plates were sealed with a sealer (Nalgene Nunc International) and placed in a FLUOstar Omega microplate reader (BMG Labtech, Germany). Experiments with continuous cycles of 1 min of double-orbital shaking at 700 rpm and 1 min of rest were performed. Recordings of the ThT fluorescence (excitation wavelength, 450 ± 10 nm; emission wavelength, 480 ± 10 nm) at the bottoms of the wells were taken every 45 min. The final curves represent the means for 4 wells. The results were fitted to four-parameter logistic regression curves described by the following equation on SigmaPlot software (Systat Software, Inc.):
| (1) |
where y is the fluorescence intensity, y0 is the minimum value of the fluorescence intensity, x is time, x0 is the time required for 50% of the maximum fluorescence intensity (T1/2), a is the maximum value of the fluorescence intensity, and b is the curve slope. The RT-QuIC reaction products were submitted to Western blotting. The contents from 4 wells were treated with 10 μg/ml (0.35 μM) PK and then submitted to Western blotting. PrP was labeled with anti-PrP antibody R20, which recognizes residues 218 to 232 (73). Band intensity density was quantified using ImageJ software (http://imageJ.nih.gov/ij).
Fibril amplification seeded with in vitro-produced fibrils.
Initially, rPrP fibrils were produced in vitro following the protocol described by Baskakov and colleagues (47, 48). Briefly, a stock solution of murine rPrP23–231 in 6 M guanidinium HCl was diluted to a final concentration of 10 μM in 10 mM phosphate buffer (pH 6.8) containing 1 M guanidinium HCl, 3 M urea, and 150 μM NaCl. The subsequent steps were based on the RT-QuIC assay and were performed as described above, with the following differences: (i) the reaction mixtures contained 10 mM phosphate buffer (pH 7.4), 300 mM NaCl, 10 μM ThT, test compounds at 20 μM, substrate (murine rPrP23–231) at 2 μM, and in vitro-produced fibrils at 1% (mol/mol); (ii) the vehicle corresponded to 0.2% DMSO; and (iii) recording of the ThT fluorescence signals at the bottoms of the wells was performed every 41 min. Results were fitted to four-parameter logistic regression curves described by equation 1 in SigmaPlot software (Systat Software, Inc.).
Molecular docking simulation.
The method used for molecular docking simulation was based on that described by Hyeon et al. (74). Initially, the energy-minimized three-dimensional structures of the tested compounds were generated in Avogadro (version 1.2.0) software (75). The compounds were then docked into the mouse PrPC globular domain (PDB ID 1AG2) using AutoDock (version 4.2.6) software (76).The search space was delimited in a grid of 64 by 58 by 54 points with a default spacing of 0.375 Å and centered on x, y, and z coordinates of 4.913, 7.305, and 2.540, respectively. A total of 50 docking simulation runs were executed with the Lamarckian genetic algorithm (LGA), using the default parameters. The results were subjected to the clustering of conformations, and the conformer selected for further analysis was the one which presented the lowest Gibbs free energy of binding in the most populated cluster.
RT-qPCR.
The reverse transcription quantitative real-time PCR (RT-qPCR) protocol was executed as previously described (77, 78). N2a cells were seeded in complete medium at a density of ∼1 × 104 cells/cm2 into 6-well plates and were incubated for 3 h at 37°C in a 5% CO2 atmosphere. The compounds were then applied to the cultures at a final concentration of 10 μM. The vehicle corresponds to 0.1% DMSO. After 4 days of incubation, the medium was removed and the cultures were washed with PBS. Total RNA extraction was performed using the TRIzol reagent (Thermo) in accordance with the manufacturer's instructions. Afterwards, genomic DNA contamination was eliminated by digestion with DNase I from a DNA-free kit (Ambion). The RNA concentration was measured by determination of its absorbance at 260 nm, and its integrity was verified by electrophoresis on 1% agarose and ethidium bromide staining. In order to ensure that the samples were devoid of DNA, conventional PCR was carried out using three primers for the proto-oncogene myc. RNA from noncontaminated samples was then reverse transcribed using a first-strand cDNA synthesis kit (GE Healthcare). Subsequently, RT-qPCR was performed using an Applied Biosystems 7500 system. Amplification mixtures (20 μl) included TaqMan master mix buffer, template cDNA, primers, and TaqMan probes with a 6-carboxyfluorescein (FAM) reporter at the 5′ end and a black hole quencher (BHQ) at the 3′ end. The sequences of PrP (Prnp) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Gapd) primers and probes are shown in Table S1 in the supplemental material. Prnp mRNA levels were normalized to the Gapd mRNA content.
Fluorescence microscopy.
N2a cells were seeded at a density of ∼1,500 cells/cm2 into 24-well plates containing coverslips. The cells were exposed for 48 h to the different test compounds at 10 μM, using a 0.1% solution of DMSO as the vehicle. Following the incubation, the medium was removed and the cells were fixed for 2 h with 4% paraformaldehyde, permeabilized for 1 h with 50 mM glycine, and blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). After that, the primary antibodies anti-mouse PrP 6D11 (1:500), rabbit-derived polyclonal IgG anti-mouse GRP78/BiP (1:500; code sc-13968), and rabbit-derived polyclonal IgG anti-mouse LC3 (1:3,000; code ab51520) were added and the mixture was incubated overnight. The cells were then incubated with Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin antibody (Invitrogen, CA, USA) and Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin antibody (Invitrogen, CA, USA) for 2 h, and the nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich, St. Louis, MO, USA). The coverslips were then removed from the wells, mounted on slides with Vectashield mounting medium (Vector Laboratories), and analyzed on a Zeiss Axio Observer Z1 fluorescence microscope, operated by AxioVision (release 4.8.2.0) software (Carl Zeiss, Heidenheim, Germany). Confocal images were generated with a structured illumination system (Apotome 2; Carl Zeiss, Heidenheim, Germany). Samples were excited by XBO illumination using a Zeiss Plan-Neofluar 63× objective (numerical aperture, 1.40). Images were acquired with an HMR Axiocan monochrome camera controlled by AxioVision (version 3.2) software (Carl Zeiss, Heidenheim, Germany). Colocalization analysis was performed with ImageJ software. Briefly, the red (BiP) and green (PrPC) signals were extracted, the background pixilation was removed from the analysis, and then merged channels were analyzed using the built-in colocalization threshold function of the software. A minimum of 100 cells per sample were scored in each experiment.
Statistical analysis.
Data from the dot blot and RT-qPCR assays were analyzed using one-way analysis of variance (ANOVA) with Tukey's multiple comparisons. Microscopy data were analyzed using a nonparametric one-way ANOVA, with a Kruskal-Wallis test conducted as a posttest to compare relevant groups. All statistical evaluations were performed with GraphPad Prism software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Research Program of the NIAID and also by grants from the National Council of Technological and Scientific Development (CNPq) and the Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ) (to Y.C.). We thank Gregory J. Raymond (NIH, RML) for excellent scientific support and Tuane Vieira (IFRJ) for assistance with the fibrillation assay. We also thank Luiz Carlos Ribeiro for technical assistance with the Leica (TCS-SPE) confocal microscope at CENABIO-UFRJ.
N.C.F., L.M.A., R.A.P.M., B.C., and Y.C. designed the research; N.C.F., L.M.A., A.G.H., G.R.C., C.F.G., R.A.D.C., J.R.H., D.S.S., and F.A.L. performed the research; P.N.F. purified the recombinant PrP; Y.C., R.A.P.M., M.L.B., B.C., and F.A.L. contributed new reagents/analytic tools; A.M.T.S. assisted with the docking analysis. N.C.F., L.M.A., R.A.P.M., G.R.C., B.C., F.A.L., and Y.C. analyzed the data; M.L.C.B. and R.A.D.C. performed the synthesis and structural characterization of the organic compounds; and N.C.F., L.M.A., and Y.C. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01441-17.
REFERENCES
- 1.Prusiner SB. 1998. Prions. Proc Natl Acad Sci U S A 95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, Torchia M, Yang SL, Serban D, Carlson GA, Hoppe PC, Westaway D, DeArmond SJ. 1990. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63:673–686. doi: 10.1016/0092-8674(90)90134-Z. [DOI] [PubMed] [Google Scholar]
- 3.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B. 1994. Cell-free formation of protease-resistant prion protein. Nature 370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 4.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sim V. 2012. Prion disease: chemotherapeutic strategies. Infect Disord Drug Targets 12:144–160. doi: 10.2174/187152612800100161. [DOI] [PubMed] [Google Scholar]
- 6.Bolognesi ML, Legname G. 2015. Approaches for discovering anti-prion compounds: lessons learned and challenges ahead. Expert Opin Drug Discov 10:389–397. doi: 10.1517/17460441.2015.1016498. [DOI] [PubMed] [Google Scholar]
- 7.Hyeon JW, Kim SY, Lee SM, Lee J, An SSA, Lee MK, Lee YS. 2017. Anti-prion screening for acridine, dextran, and tannic acid using real time-quaking induced conversion: a comparison with PrPSc-infected cell screening. PLoS One 12:e0170266. doi: 10.1371/journal.pone.0170266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. 1995. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 9.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat Med 4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 10.Chien P, Weissman JS, DePace AH. 2004. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem 73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 11.Castilla J, Morales R, Saá P, Barria M, Gambetti P, Soto C. 2008. Cell-free propagation of prion strains. EMBO J 27:2557–2566. doi: 10.1038/emboj.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronier S, Beringue V, Bellon A, Peyrin J-M, Laude H. 2007. Prion strain- and species-dependent effects of antiprion molecules in primary neuronal cultures. J Virol 81:13794–13800. doi: 10.1128/JVI.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawasaki Y, Kawagoe K, Chen C-J, Teruya K, Sakasegawa Y, Doh-ura K. 2007. Orally administered amyloidophilic compound is effective in prolonging the incubation periods of animals cerebrally infected with prion diseases in a prion strain-dependent manner. J Virol 81:12889–12898. doi: 10.1128/JVI.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller-Vedam L, Ghaemmaghami S. 2013. Strain specificity and drug resistance in anti-prion therapy. Curr Top Med Chem 13:2397–2406. doi: 10.2174/15680266113136660168. [DOI] [PubMed] [Google Scholar]
- 15.Bian J, Kang H-E, Telling GC. 2014. Quinacrine promotes replication and conformational mutation of chronic wasting disease prions. Proc Natl Acad Sci U S A 111:6028–6033. doi: 10.1073/pnas.1322377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giles K, Olson SH, Prusiner SB. 2017. Developing therapeutics for PrP prion diseases. Cold Spring Harb Perspect Med 7:a023747. doi: 10.1101/cshperspect.a023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linden R. 2017. The biological function of the prion protein: a cell surface scaffold of signaling modules. Front Mol Neurosci 10:77. doi: 10.3389/fnmol.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp H-P, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. 1992. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 19.Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, Sathiyaseelan J, Wu H, Matsushita H, Koster J, Kato S, Ishida I, Soto C, Robl JM, Kuroiwa Y. 2007. Production of cattle lacking prion protein. Nat Biotechnol 25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cashman NR, Caughey B. 2004. Prion diseases—close to effective therapy? Nat Rev Drug Discov 3:874–884. doi: 10.1038/nrd1525. [DOI] [PubMed] [Google Scholar]
- 21.Cordeiro Y, Ferreira NC. 2015. New approaches for the selection and evaluation of anti-prion organic compounds. Mini Rev Med Chem 15:84–92. doi: 10.2174/1389557515666150227111629. [DOI] [PubMed] [Google Scholar]
- 22.Sim VL, Caughey B. 2009. Recent advances in prion chemotherapeutics. Infect Disord Drug Targets 9:81–91. doi: 10.2174/1871526510909010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira NC, Marques IA, Conceição WA, Macedo B, Machado CS, Mascarello A, Chiaradia-Delatorre LD, Yunes RA, Nunes RJ, Hughson AG, Raymond LD, Pascutti PG, Caughey B, Cordeiro Y. 2014. Anti-prion activity of a panel of aromatic chemical compounds: in vitro and in silico approaches. PLoS One 9:e84531. doi: 10.1371/journal.pone.0084531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B. 2010. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog 6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueiredo CP, Ferreira NC, Passos GF, da Costa R, Neves FS, Machado CSC, Mascarello A, Chiaradia-Delatorre LD, Neuenfeldt PD, Nunes RJ, Cordeiro Y. 2015. Toxicological evaluation of anti-scrapie trimethoxychalcones and oxadiazoles. An Acad Bras Cienc 87:1421–1434. doi: 10.1590/0001-3765201520140712. [DOI] [PubMed] [Google Scholar]
- 26.Fraser H, Dickinson AG. 1973. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol 83:29–40. [DOI] [PubMed] [Google Scholar]
- 27.Bruce ME, McConnell I, Fraser H, Dickinson AG. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol 72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 28.Collinge J, Clarke AR. 2007. A general model of prion strains and their pathogenicity. Science 318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 29.Poggiolini I, Saverioni D, Parchi P. 2013. Prion protein misfolding, strains, and neurotoxicity: an update from studies on mammalian prions. Int J Cell Biol 2013:910314. doi: 10.1155/2013/910314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bessen RA, Marsh RF. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 68:7859–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. 2003. New inhibitors of scrapie-associated prion protein formation in a library of 2,000 drugs and natural products. J Virol 77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond GJ, Olsen EA, Lee KS, Raymond LD, Bryant PK, Baron GS, Caughey WS, Kocisko DA, McHolland LE, Favara C, Langeveld JPM, van Zijderveld FG, Mayer RT, Miller MW, Williams ES, Caughey B. 2006. Inhibition of protease-resistant prion protein formation in a transformed deer cell line infected with chronic wasting disease. J Virol 80:596–604. doi: 10.1128/JVI.80.2.596-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M, Appelhans D, Schwarz S, Klajnert B, Bryszewska M, Voit B, Rogers M. 2010. Influence of surface functionality of poly(propylene imine) dendrimers on protease resistance and propagation of the scrapie prion protein. Biomacromolecules 11:1314–1325. doi: 10.1021/bm100101s. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy JM, Franke M, Resenberger UK, Waldron S, Simpson JC, Tatzelt J, Appelhans D, Rogers MS. 2013. Anti-prion drug mPPIg5 inhibits PrPC conversion to PrPSc. PLoS One 8:e55282. doi: 10.1371/journal.pone.0055282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bongianni M, Orrù C, Groveman BR, Sacchetto L, Fiorini M, Tonoli G, Triva G, Capaldi S, Testi S, Ferrari S, Cagnin A, Ladogana A, Poleggi A, Colaizzo E, Tiple D, Vaianella L, Castriciano S, Marchioni D, Hughson AG, Imperiale D, Cattaruzza T, Fabrizi GM, Pocchiari M, Monaco S, Caughey B, Zanusso G. 2017. Diagnosis of human prion disease using real-time quaking-induced conversion testing of olfactory mucosa and cerebrospinal fluid samples. JAMA Neurol 74:155. doi: 10.1001/jamaneurol.2016.4614. [DOI] [PubMed] [Google Scholar]
- 36.Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B, Hoover EA. 2013. Rapid antemortem detection of CWD prions in deer saliva. PLoS One 8:e74377. doi: 10.1371/journal.pone.0074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groveman BR, Orrú CD, Hughson AG, Bongianni M, Fiorini M, Imperiale D, Ladogana A, Pocchiari M, Zanusso G, Caughey B. 2017. Extended and direct evaluation of RT-QuIC assays for Creutzfeldt-Jakob disease diagnosis. Ann Clin Transl Neurol 4:139–144. doi: 10.1002/acn3.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz M, Cramm M, Llorens F, Müller-Cramm D, Collins S, Atarashi R, Satoh K, Orrù CD, Groveman BR, Zafar S, Schulz-Schaeffer WJ, Caughey B, Zerr I. 2016. The real-time quaking-induced conversion assay for detection of human prion disease and study of other protein misfolding diseases. Nat Protoc 11:2233–2242. doi: 10.1038/nprot.2016.120. [DOI] [PubMed] [Google Scholar]
- 39.Caughey B, Baron GS, Chesebro B, Jeffrey M. 2009. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem 78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groveman BR, Kraus A, Raymond LD, Dolan MA, Anson KJ, Dorward DW, Caughey B. 2015. Charge neutralization of the central lysine cluster in prion protein (PrP) promotes PrPSc-like folding of recombinant PrP amyloids. J Biol Chem 290:1119–1128. doi: 10.1074/jbc.M114.619627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khurana R, Coleman C, Ionescu-Zanetti C, Carter SA, Krishna V, Grover RK, Roy R, Singh S. 2005. Mechanism of thioflavin T binding to amyloid fibrils. J Struct Biol 151:229–238. doi: 10.1016/j.jsb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Vieira TCRG, Cordeiro Y, Caughey B, Silva JL. 2014. Heparin binding confers prion stability and impairs its aggregation. FASEB J 28:2667–2676. doi: 10.1096/fj.13-246777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imberdis T, Heeres JT, Yueh H, Fang C, Zhen J, Rich CB, Glicksman M, Beeler AB, Harris DA. 2016. Identification of anti-prion compounds using a novel cellular assay. J Biol Chem 291:26164–26176. doi: 10.1074/jbc.M116.745612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughson AG, Race B, Kraus A, Sangaré LR, Robins L, Groveman BR, Saijo E, Phillips K, Contreras L, Dhaliwal V, Manca M, Zanusso G, Terry D, Williams JF, Caughey B. 2016. Inactivation of prions and amyloid seeds with hypochlorous acid. PLoS Pathog 12:e1005914. doi: 10.1371/journal.ppat.1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turnbaugh JA, Unterberger U, Saa P, Massignan T, Fluharty BR, Bowman FP, Miller MB, Supattapone S, Biasini E, Harris DA. 2012. The N-terminal, polybasic region of PrPC dictates the efficiency of prion propagation by binding to PrPSc. J Neurosci 32:8817–8830. doi: 10.1523/JNEUROSCI.1103-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linden R, Martins VR, Prado MAM, Cammarota M, Izquierdo I, Brentani RR. 2008. Physiology of the prion protein. Physiol Rev 88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 47.Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. 2005. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrPSc. J Mol Biol 346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 48.Baskakov IV, Bocharova OV. 2005. In vitro conversion of mammalian prion protein into amyloid fibrils displays unusual features. Biochemistry 44:2339–2348. doi: 10.1021/bi048322t. [DOI] [PubMed] [Google Scholar]
- 49.Kuwata K, Li H, Yamada H, Legname G, Prusiner SB, Akasaka K, James TL. 2002. Locally disordered conformer of the hamster prion protein: a crucial intermediate to PrPSc? Biochemistry 41:12277–12283. doi: 10.1021/bi026129y. [DOI] [PubMed] [Google Scholar]
- 50.Kuwata K, Kamatari YO, Akasaka K, James TL. 2004. Slow conformational dynamics in the hamster prion protein. Biochemistry 43:4439–4446. doi: 10.1021/bi036123o. [DOI] [PubMed] [Google Scholar]
- 51.Kuwata K, Nishida N, Matsumoto T, Kamatari YO, Hosokawa-Muto J, Kodama K, Nakamura HK, Kimura K, Kawasaki M, Takakura Y, Shirabe S, Takata J, Kataoka Y, Katamine S. 2007. Hot spots in prion protein for pathogenic conversion. Proc Natl Acad Sci U S A 104:11921–11926. doi: 10.1073/pnas.0702671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikawa T, Ishikura T, Kuwata K. 2009. Theoretical study of the prion protein based on the fragment molecular orbital method. J Comput Chem 30:2594–2601. doi: 10.1002/jcc.21265. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto N, Kuwata K. 2009. Regulating the conformation of prion protein through ligand binding. J Phys Chem B 113:12853–12856. doi: 10.1021/jp905572w. [DOI] [PubMed] [Google Scholar]
- 54.Johnson WC. 1988. Secondary structure of proteins through circular dichroism spectroscopy. Annu Rev Biophys Biophys Chem 17:145–166. doi: 10.1146/annurev.bb.17.060188.001045. [DOI] [PubMed] [Google Scholar]
- 55.Brettschneider J, Del Tredici K, Lee VM-Y, Trojanowski JQ. 2015. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat Rev Neurosci 16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caughey B, Caughey WS, Kocisko DA, Lee KS, Silveira JR, Morrey JD. 2006. Prions and transmissible spongiform encephalopathy (TSE) chemotherapeutics: a common mechanism for anti-TSE compounds? Acc Chem Res 39:646–653. doi: 10.1021/ar050068p. [DOI] [PubMed] [Google Scholar]
- 57.Supattapone S, Wille H, Uyechi L, Safar J, Tremblay P, Szoka FC, Cohen FE, Prusiner SB, Scott MR. 2001. Branched polyamines cure prion-infected neuroblastoma cells. J Virol 75:3453–3461. doi: 10.1128/JVI.75.7.3453-3461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yudovin-Farber I, Azzam T, Metzer E, Taraboulos A, Domb AJ. 2005. Cationic polysaccharides as antiprion agents. J Med Chem 48:1414–1420. doi: 10.1021/jm049378o. [DOI] [PubMed] [Google Scholar]
- 59.Kimata A, Nakagawa H, Ohyama R, Fukuuchi T, Ohta S, Doh-ura K, Suzuki T, Miyata N. 2007. New series of antiprion compounds: pyrazolone derivatives have the potent activity of inhibiting protease-resistant prion protein accumulation. J Med Chem 50:5053–5056. doi: 10.1021/jm070688r. [DOI] [PubMed] [Google Scholar]
- 60.Gallardo-Godoy A, Gever J, Fife KL, Silber BM, Prusiner SB, Renslo AR. 2011. 2-Aminothiazoles as therapeutic leads for prion diseases. J Med Chem 54:1010–1021. doi: 10.1021/jm101250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massignan T, Cimini S, Stincardini C, Cerovic M, Vanni I, Elezgarai SR, Moreno J, Stravalaci M, Negro A, Sangiovanni V, Restelli E, Riccardi G, Gobbi M, Castilla J, Borsello T, Nonno R, Biasini E. 2016. A cationic tetrapyrrole inhibits toxic activities of the cellular prion protein. Sci Rep 6:23180. doi: 10.1038/srep23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hosokawa-Muto J, Kamatari YO, Nakamura HK, Kuwata K. 2009. Variety of antiprion compounds discovered through an in silico screen based on cellular-form prion protein structure: correlation between antiprion activity and binding affinity. Antimicrob Agents Chemother 53:765–771. doi: 10.1128/AAC.01112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolognesi ML, Bongarzone S, Aulic S, Ai Tran HN, Prati F, Carloni P, Legname G. 2015. Rational approach to an antiprion compound with a multiple mechanism of action. Future Med Chem 7:2113–2120. doi: 10.4155/fmc.15.79. [DOI] [PubMed] [Google Scholar]
- 64.Soto C, Satani N. 2011. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol Med 17:14–24. doi: 10.1016/j.molmed.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park K-W, Eun Kim G, Morales R, Moda F, Moreno-Gonzalez I, Concha-Marambio L, Lee AS, Hetz C, Soto C. 2017. The endoplasmic reticulum chaperone GRP78/BiP modulates prion propagation in vitro and in vivo. Sci Rep 7:44723. doi: 10.1038/srep44723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borchhardt DM, Mascarello A, Chiaradia LD, Nunes RJ, Oliva G, Yunes RA, Andricopulo AD. 2010. Biochemical evaluation of a series of synthetic chalcone and hydrazide derivatives as novel inhibitors of cruzain from Trypanosoma cruzi. J Braz Chem Soc 21:142–150. doi: 10.1590/S0103-50532010000100021. [DOI] [Google Scholar]
- 67.Nunes RJ, Mascarello A, Yunes RA, Stumpf TR, Leal PC, Yunes JA, Pereira de Souza Melo C, Canevarolo RR, Domeneghini Chiaradia L, Bortolini Silveira A, Laranjeira ABA. 2013. Acyl-hydrazone and oxadiazole compounds, pharmaceutical compositions containing the same and uses thereof. Patent WO 2013075199 A1.
- 68.Costa A, Chiaradia-Delatorre LD, dos Santos Bubniak L, Mascarello A, Marzarotto MAL, de Moraes ACR, Stumpf TR, Cordeiro MNS, Yunes RA, Nunes RJ, Santos-Silva MC. 2014. Apoptotic effect of synthetic 2′,4′,5′-trimethoxychalcones in human K562 and Jurkat leukemia cells. Med Chem Res 23:4301–4319. doi: 10.1007/s00044-014-1002-4. [DOI] [Google Scholar]
- 69.Caughey B, Raymond GJ. 1993. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J Virol 67:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Birkett CR, Hennion RM, Bembridge DA, Clarke MC, Chree A, Bruce ME, Bostock CJ. 2001. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J 20:3351–3358. doi: 10.1093/emboj/20.13.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pankiewicz J, Prelli F, Sy MS, Kascsak RJ, Kascsak RB, Spinner DS, Carp RI, Meeker HC, Sadowski M, Wisniewski T. 2006. Clearance and prevention of prion infection in cell culture by anti-PrP antibodies. Eur J Neurosci 23:2635–2647. doi: 10.1111/j.1460-9568.2006.04805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spinner DS, Kascsak RB, LaFauci G, Meeker HC, Ye X, Flory MJ, Kim JI, Schuller-Levis GB, Levis WR, Wisniewski T, Carp RI, Kascsak RJ. 2007. CpG oligodeoxynucleotide-enhanced humoral immune response and production of antibodies to prion protein PrPSc in mice immunized with 139A scrapie-associated fibrils. J Leukoc Biol 81:1374–1385. doi: 10.1189/jlb.1106665. [DOI] [PubMed] [Google Scholar]
- 73.Caughey B, Raymond GJ, Ernst D, Race RE. 1991. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol 65:6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hyeon JW, Choi J, Kim SY, Govindaraj RG, Jam Hwang K, Lee YS, An SSA, Lee MK, Joung JY, No KT, Lee J. 2015. Discovery of novel anti-prion compounds using in silico and in vitro approaches. Sci Rep 5:14944. doi: 10.1038/srep14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. 2012. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cavalheiro GR, Matos-Rodrigues GE, Gomes AL, Rodrigues PMG, Martins RAP. 2014. c-Myc regulates cell proliferation during lens development. PLoS One 9:e87812. doi: 10.1371/journal.pone.0087812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martins RAP, Linden R, Dyer MA. 2006. Glutamate regulates retinal progenitors cells proliferation during development. Eur J Neurosci 24:969–980. doi: 10.1111/j.1460-9568.2006.04966.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.