LETTER
The rapid rise and dissemination of multidrug-resistant (MDR) bacteria are a major threat to public health worldwide and have narrowed the treatment options for infections caused by these bacteria (1). Colistin is one of the last-resort drugs for the treatment of infections caused by MDR Gram-negative bacteria. However, within the past 2 years, shortly after the report of the first plasmid-mediated colistin resistance gene, mcr-1, in 2015, four other mobile colistin resistance genes (mcr-2, mcr-3, mcr-4, and mcr-5) and multiple variants have been reported (2–6). Compared to mcr-2, mcr-4, and mcr-5, which have been detected solely in Europe so far, both mcr-1 and mcr-3 have been identified globally. Here, we report a novel mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from a single domestic duck.
A total of 15 cloacal samples were obtained from free-range domestic ducks near a river in a suburban area of Qingdao City, Shandong Province, China, in March 2017. Direct sample testing was then performed on all 15 samples to detect the mcr-3 gene, and only one sample was identified as mcr-3 positive. The sample positive for mcr-3 was further isolated on CHROMagar orientation agar plates (bioMérieux, Lyon, France) containing 2 mg/liter colistin. Three mcr-3-positive strains, A. caviae 17AC, P. mirabilis 17PM, and E. coli 17EC, were obtained, and their sequences were confirmed by using 16S rRNA gene sequencing and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry analysis.
A. caviae 17AC, P. mirabilis 17PM, and E. coli 17EC were then subjected to 150-bp paired-end whole-genome sequencing (WGS) using the Illumina HiSeq 2500 platform (Annoroad, Beijing, China). The draft genomes were assembled using CLC Genomics Workbench 9.0 (CLC Bio, Aarhus, Denmark), and all contigs were searched for mcr-3 by standalone BLAST analysis. WGS analysis identified mcr-3-carrying fragments in each genome, including a 26.2-kb contig from 17AC, a 17.8-kb contig from 17PM, and a 13.0-kb contig from 17EC. An mcr-3 variant gene in three contigs showed 98.83% nucleotide sequence identity to mcr-3 from porcine E. coli. The deduced protein sequence differed from MCR-3.1 by seven amino acid substitutions, one of which (V122G) was located in a putative transmembrane region, while the remaining six (R297L, I313V, E337K, H341Y, D358E, and Q468K) were in the catalytic domain. This novel mcr-3 variant was designated mcr-3.10. A. caviae 17AC and E. coli 17EC belong to novel sequence types (STs), ST513 and ST457, respectively. E. coli of ST457 carrying mcr-1 has been reported from humans in the United States and Vietnam (7, 8). Plasmid replicon typing revealed that the mcr-3.10-carrying plasmid in both 17EC and its transconjugant T-17EC belongs to the IncI2 replicon.
A conjugation experiment showed that mcr-3.10 was able to be transferred from donor strain E. coli 17EC to recipient strain E. coli J53, and transconjugant T-17EC was obtained and its sequence confirmed by PCR targeting of mcr-3.10 with Sanger sequencing. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting indicated that mcr-3.10 is located on a chromosome in A. caviae 17AC and P. mirabilis 17PM and on an ∼45-kb plasmid, named p17EC, in E. coli 17EC (see Fig. S1 in the supplemental material).
An antimicrobial susceptibility test was performed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The breakpoints for each antimicrobial agent were interpreted according to CLSI standards (9) and those of the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org) (10). All three mcr-3.10-carrying isolates showed resistance to most of the tested antimicrobial agents, including colistin (8 to 32 μg/ml), polymyxin B (8 to 32 μg/ml), gentamicin (16 to 512 μg/ml), chloramphenicol (128 to 512 μg/ml), ciprofloxacin (32 to 128 μg/ml), and tetracycline (16 to 128 μg/ml), and they remained sensitive to imipenem (0.13 μg/ml) and meropenem (0.06 μg/ml). Like the original mcr-3 gene, the mcr-3.10 gene resulted in an 8-fold increase in the MIC of colistin (2 mg/liter) for transconjugant T-17EC compared with that for the recipient E. coli J53 (0.25 mg/liter). It also exhibited resistance to gentamicin and tetracycline, which can be conferred by ant(6)-Ia and the tet(D) gene identified in p17EC, respectively (Table S1).
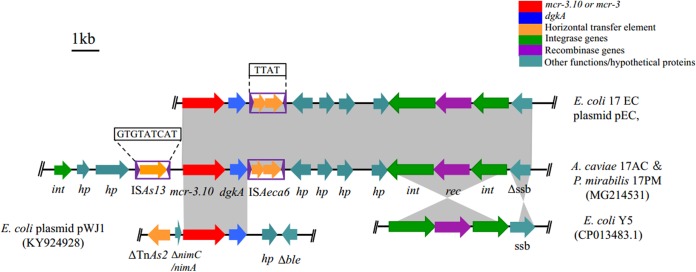
Genetic-environment analysis revealed that the 13.0-kb mcr-3.10-carrying contig in 17EC showed 100% nucleotide sequence identity to the corresponding regions of the 26.2-kb contig from 17AC and the 17.8-kb contig from 17PM. A complete insertion sequence (IS), ISAs13 (https://www-is.biotoul.fr/scripts/ficheIS.php?name=ISAs13), which originated from Aeromonas salmonicida, was located upstream of mcr-3.10 in both 17AC and 17PM. In addition, an IS-like structure with 87% nucleotide identity to ISAs22 was located downstream of mcr-3.10. This IS-like element was 1,198 bp in length and contained two open reading frames flanked by imperfect putative inverted repeats (IRs) (right IR, 12 bp; left IR, 11 bp). This IS-like element was submitted to the ISfinder database (https://www-is.biotoul.fr/scripts/ficheIS.php?name=ISAeca6) and was assigned the designation ISAeca6, which belongs to the IS3 family (Fig. 1). Identical 13.0-kb mcr-3.10-carrying structures shared by three different bacterial species within one duck further confirm the dissemination of mcr-3 between Aeromonas and Enterobacteriaceae in animal guts. Interestingly, the presence of mcr-3.10 in Proteus mirabilis indicated that even the naturally colistin-resistant bacterial species may serve as a reservoir of the mcr-3 variant gene.
FIG 1.
Genetic organizations of scaffolds containing mcr-3.10 obtained from A. caviae 17AC, P. mirabilis 17PM, and E. coli 17EC in this study and structural comparison with plasmid pWJ1 from E. coli WJ1 (GenBank accession no. KY924928) and with the complete genome sequence of E. coli Y5 (GenBank accession no. CP013483.1). The positions and orientations of the genes are indicated by arrows, with the direction of transcription shown by the arrowhead. Gray shading indicates >90% nucleotide sequence identity. The 9-bp direct repeat (5′-GTGTATCAT-3′) of ISAs13 and the 4-bp direct repeats (5′-TTAT-3′) of ISAeca6 are boxed.
Accession number(s).
The nucleotide sequence of the 26.2-kb contig carrying mcr-3.10 in A. caviae 17AC determined in this study has been deposited in GenBank under accession number MG214531.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (31422055 and 81661138002) and the National Basic Research Program of China (2013CB127200).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02106-17.
REFERENCES
- 1.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Wang Y, Walsh TR, Yi L, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 4.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 7.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tada T, Nhung PH, Shimada K, Tsuchiya M, Phuong DM, Anh NQ, Ohmagari N, Kirikae T. 2017. Emergence of colistin-resistant Escherichia coli clinical isolates harboring mcr-1 in Vietnam. Int J Infect Dis 63:72–73. doi: 10.1016/j.ijid.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2013. VET01-A4. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 4th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Bakthavatchalam YD, Veeraraghavan B. 2017. Challenges, issues and warnings from CLSI and EUCAST Working Group on Polymyxin Susceptibility Testing. J Clin Diagn Res 11:DL03–DL04. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.