ABSTRACT
The increasing incidence of multidrug-resistant Acinetobacter baumannii (MDRAb) infections worldwide has necessitated the development of novel antibiotics. Human defensin 5 (HD5) is an endogenous peptide with a complex architecture and antibacterial activity against MDRAb. In the present study, we attempted to simplify the structure of HD5 by removing disulfide bonds. We found that the Cys2-4 bond was most indispensable for HD5 to inactivate MDRAb, although the antibacterial activity of the derivative was significantly attenuated. We then replaced the noncationic and nonhydrophobic residues with electropositive Arg to increase the antibacterial activity of HD5 derivative that contains a Cys2-4 bond, obtaining another derivative termed HD5d5. The in vitro antibacterial assay and irradiation-wound-infection animal experiment both showed that HD5d5 was much more effective than HD5 at eliminating MDRAb. Further investigations revealed that HD5d5 efficiently bound to outer membrane lipid A and penetrated membranes, leading to bacterial collapse and peptide translocation. Compared to HD5, more HD5d5 molecules were located in the cytoplasm of MDRAb, and HD5d5 was more efficient at reducing the activities of superoxide dismutase and catalase, causing the accumulation of reactive oxygen species that are detrimental to microbes. In addition, HD5 failed to suppress the pathogenic outer membrane protein A of Acinetobacter baumannii (AbOmpA) at concentrations up to 50 μg/ml, whereas HD5d5 strongly bound to AbOmpA and exhibited a dramatic toxin-neutralizing ability, thus expanding the repertoire of drugs that is available to treat MDRAb infections.
KEYWORDS: Acinetobacter baumannii, defensin, membrane penetration, reactive oxygen species, outer membrane protein A
INTRODUCTION
Acinetobacter baumannii is one of the leading causes of nosocomial infections. The emergence of multidrug-resistant A. baumannii (MDRAb) is increasing at alarming rates worldwide (1). A recent study characterizing 39,320 A. baumannii isolates revealed that the prevalence of MDRAb had escalated from 21.4% (2003 to 2005) to 35.2% (2009 to 2012) (2). This pathogen primarily invades the human body via the respiratory tract, wounds, and catheters, with 30-day mortality rates reaching 49% (3). Due to the lack of effective antibiotics, the development of novel bactericides to eliminate MDRAb is urgently needed.
Defensin is an evolutionarily ancient component of the mammalian defense system with bactericidal potency (4). The electrostatic interaction between defensin and negatively charged membranes is an important step in killing bacteria, because it enables subsequent membrane penetration (5, 6). Because microbes cannot easily alter their anionic properties, defensin might not encounter rapid resistance (7). Human defensin is divided into α and β subfamilies according to the sequence homology and disulfide connectivity. Human defensin 5 (HD5) is an α-subfamily peptide that enables the inactivation of pathogenic bacteria in vivo and is thus a potential bactericide that could eliminate MDRAb (8). We previously prepared bioactive HD5 in Pichia pastoris and designed potent antibiotics based on the topology of HD5 (9–11).
HD5 is a 32-residue cationic peptide with a specific conformation donated by three disulfide bonds (Fig. 1A). HD5 forms this complex architecture because it mainly functions in the intestine, where disulfide bonds protect peptides from degradation by trypsin, an Arg- and Lys-targeting protease that is abundant in the enteric canal (12). Nevertheless, when the peptide is topically used on the eyes or skin, where the trypsin content is negligible, disulfide removal is theoretically tolerated (13, 14). Because the specific disulfide bonds of HD5 burden its preparation, structural simplification by disulfide reduction definitely promotes its development into a topical bactericide. Notably, simplified peptides may be superior at killing bacteria, since the action of human β-defensin 1 (HBD1) is enhanced upon disulfide reduction (15), and linear LL-37 is more potent against A. baumannii than disulfide-containing HBD3 (16). Researchers have not yet determined whether disulfide reduction can boost the antibacterial activity of HD5 against A. baumannii in addition to simplifying its preparation.
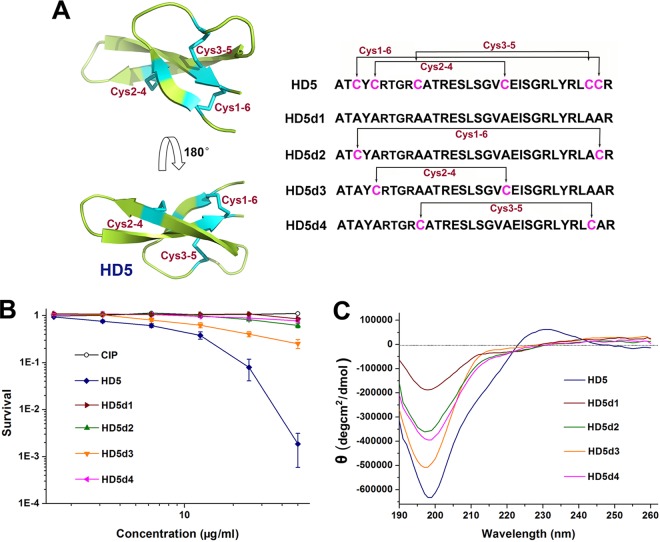
FIG 1.
Structural analysis and antibacterial assessment of HD5 and HD5d1-4. (A) Stereoview of HD5 and alignment of peptide sequences. The conformation of HD5 (PDB 1ZMP) was visualized using PyMOL and is presented as ribbon diagram. Disulfide bonds are highlighted in cyan. (B) Virtual colony count assay. The bacterial survival rate was calculated as the ratio of the number of surviving colonies after treatment with peptides and CIP to the number of surviving colonies after treatment with sterile water. The results are shown as means ± the standard deviations (SD). (C) CD spectra revealing the conformational difference between HD5 and HD5d1-4. The spectral signal was obtained at 190 to 260 nm with an interval of 1 nm.
The outer membrane protein A of A. baumannii (AbOmpA) is a major envelope protein that mediates the invasion of microbes into host cells (17). In addition, it is an important pathogenic factor, as recombinant AbOmpA directly induces the early-onset apoptosis and delayed-onset necrosis of dendritic cells, in which the overload of reactive oxygen species (ROS) is implicated (18). Moreover, AbOmpA can enter epithelial cells and target mitochondria, resulting in the production of proapoptotic molecules (19). Human α-defensin can bind and suppress bacterial toxin in addition to its prominent antibacterial action (20, 21). HD5 is recognized to govern multivalent binding capabilities (22), and its spatial conformation affects toxin neutralization (23). The interaction of HD5 and its simplified derivatives with AbOmpA warrants investigation.
In the present study, we evaluated the effect of disulfide reduction on the antibacterial action of HD5 against A. baumannii. A simplified derivative with remarkable bactericidal potency against MDRAb was designed and termed HD5d5. The underlying mechanism by which HD5d5 inactivated MDRAb was investigated using biolayer interferometry (BLI), scanning electron microscopy (SEM), and laser confocal microscopy (LSCM). The interaction between HD5d5 and AbOmpA was analyzed as well.
RESULTS
Structural simplification by disulfide reduction.
We initially used a modified broth microdilution method established by the Hancock laboratory to analyze the antibacterial action of HD5 against A. baumannii (24). Ciprofloxacin (CIP) was used as an antibiotic reference. HD5 was inferior to CIP at killing A. baumannii ATCC 19606 but was comparable with CIP at inactivating MDRAb isolated from the infected wound of a burn patient (Table 1). The MICs of HD5 and CIP against MDRAb were both 320 μg/ml. However, using a virtual colony count assay, HD5 was more efficient than CIP at eliminating MDRAb (Fig. 1B). The average concentration of HD5 required to inactivate 50% of MDRAb (LD50) was 9.3 μg/ml, which was at least 5.4-fold lower than CIP (Table 2).
TABLE 1.
MICs of antimicrobial agents required to eliminate A. baumannii, calculated using the broth microdilution method
| Antimicrobial agent | MIC (μg/ml) |
|
|---|---|---|
| MDRAb | ATCC 19606 | |
| HD5 | 320 | 320 |
| HD5d1 | 2,560 | 2,560 |
| HD5d2 | 2,560 | 1,280 |
| HD5d3 | 1,280 | 1,280 |
| HD5d4 | 2,560 | 1,280 |
| HD5d5 | 40 | 40 |
| HD5d6 | 80 | 80 |
| CIP | 320 | ≤2.5 |
TABLE 2.
Lethal doses that kill 50 and 90% of MDRAb, calculated using the virtual colony count assay
| Antimicrobial agent | LD (μg/ml)a |
|
|---|---|---|
| LD50 | LD90 | |
| HD5 | 9.3 ± 0.6 | 24.2 ± 1.1 |
| HD5d1 | >50 | >50 |
| HD5d2 | >50 | >50 |
| HD5d3 | 19.7 ± 0.9 | >50 |
| HD5d4 | >50 | >50 |
| HD5d5 | <1.56 | 2.8 ± 0.7 |
| HD5d6 | <1.56 | 4.4 ± 0.9 |
| CIP | >50 | >50 |
Although the bactericidal effect of HD5 on MDRAb was ambiguous, the effect of disulfide reduction was uncovered by both antibacterial experiments. The MICs and LDs of derivatives without (HD5d1) or containing only one disulfide bond (HD5d2-4) were all increased compared to HD5 (Tables 1 and 2). Circular dichroism (CD) spectroscopy revealed that HD5d1-4 did not form rigid β-sheet structures in aqueous solution, as manifested by an elevated signal intensity at 200 nm (Fig. 1C), which indicates an increase in random coil structures. Disulfide reduction conceivably simplifies the structure of HD5 at the expense of bactericidal attenuation, which was attributed to the lack of a rigid conformation.
Functional enhancement by Arg introduction.
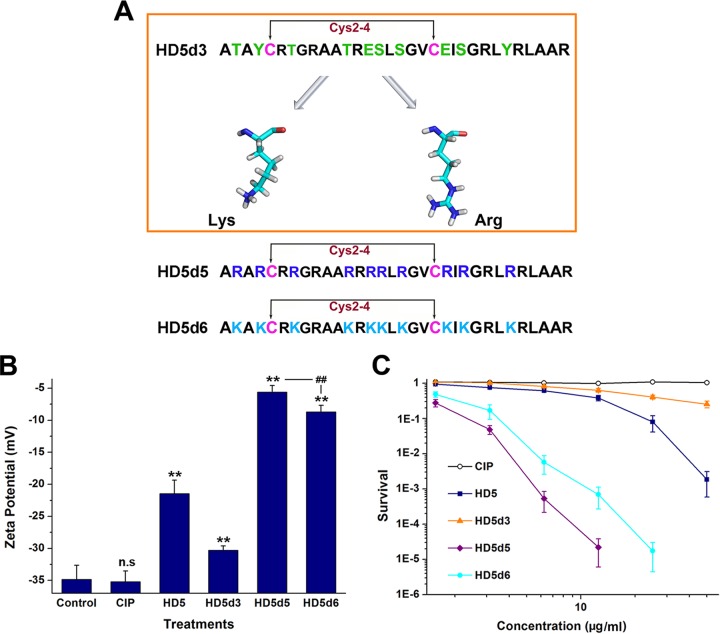
Arg and Lys are electropositive residues that highly prevalent in human defensins (25). Because an increase in the positive charge may improve the bactericidal potency of peptides (11, 26) and HD5d3 was better at inactivating MDRAb than the other derivatives (Tables 1 and 2), we replaced the noncationic and nonhydrophobic residues of HD5d3 with Arg and Lys, respectively, obtaining two additional derivatives termed HD5d5 and HD5d6 (Fig. 2A). Despite the same net charge, HD5d5 bound to MDRAb membranes more tightly than HD5d6, causing a sharp increase in the surface potential (Fig. 2B). The modified broth microdilution method and virtual colony count assay concurrently revealed that HD5d5 was not only more efficient than HD5d6 but also superior to HD5 and CIP at killing MDRAb (Table 1 and Fig. 2C).
FIG 2.
Arg is more instrumental in the ability of HD5d3 to eliminate MDRAb than Lys. (A) Amino acid sequences of peptides. The residues of HD5d3 selected for substitution are colored green. Lys and Arg are presented as sticks (C, cyan; N, blue; O, red; H, white). (B) Zeta potential determination. Peptides were prepared at a concentration of 20 μM in sterile water. **, P < 0.01 (compared to the potential of the control group); ##, P < 0.01. (C) Virtual colony count assay revealing the antibacterial actions of HD5d5 and HD5d6 against MDRAb. Points lower than 10−6 were scored as zero survival and are not plotted. The results derived from three independent experiments conducted in duplicate are presented as means ± the SD.
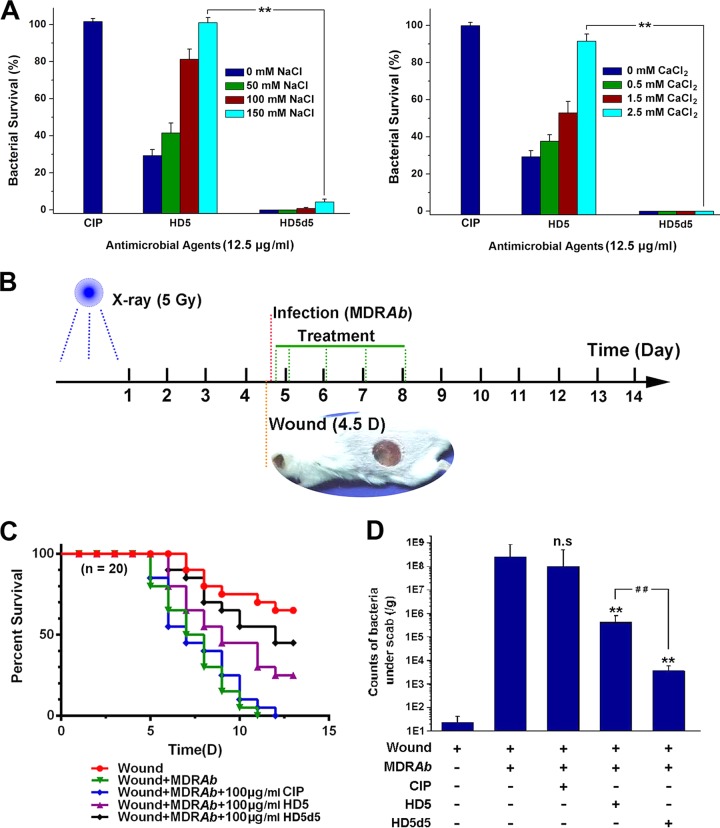
Due to the shielding effects of salt ions, the efficiency of many antibacterial peptides that are endowed with excellent activities against A. baumannii in vitro is attenuated in vivo (27). We further determined the bactericidal potency of HD5d5 in the context of increasing concentrations of monovalent Na+ and divalent Ca2+. Treatment with 12.5 μg/ml HD5d5 efficiently killed MDRAb in solutions containing 150 mM NaCl and 2.5 mM CaCl2, whereas HD5 was nearly inactive (Fig. 3A), highlighting the medicinal value of the newly designed peptide in MDRAb infection therapy.
FIG 3.
Antibacterial actions of HD5 and HD5d5 in saline solution and in vivo. (A) Antibacterial activities of HD5 and HD5d5 in the presence of increasing concentrations of NaCl and CaCl2. Mean values were obtained from three independent experiments. Error bars indicate the SD. **, P < 0.01. (B) Diagram depicting the IWI model. BALB/c mice were irradiated with a single 5-Gy dose. A 1-cm-diameter full-thickness skin wound was prepared 4 days after irradiation, which was then contaminated with MDRAb after 2 h. (C) Survival curve. Mice subjected to irradiation and MDRAb infection were treated with CIP, HD5, or HD5d5 (n = 20) at 100 μg/ml. The results are presented as the survival rate. (D) Counts of microbes under the scabs that were resistant to CIP on day 9. The results are presented as means ± the SD. n.s, not significant; **, P < 0.01 (compared to the value of the infected group); ##, P < 0.01.
In vivo antibacterial assessment.
A. baumannii is an opportunistic pathogen that threatens individuals with compromised immunity. Considering that acute irradiation destroys the human immune system, we established an irradiation-wound-infection (IWI) model to evaluate the bactericidal effects of CIP, HD5, and HD5d5 on MDRAb in vivo (Fig. 3B). These bactericides were all harmless to erythrocytes and human epithelial HaCaT cells at concentrations below 100 μg/ml (see Fig. S1 in the supplemental material). According to the results of the IWI experiment, all infected mice that did not receive treatment died within 11 days, whereas the application of 100 μg/ml HD5 and HD5d5, but not CIP, elevated the survival rate (Fig. 3C). In addition, HD5d5 was more efficient at treating the infection than HD5. This finding was consistent with the counts of CIP-resistant microbes under the wound scabs (Fig. 3D), which convincingly revealed that HD5d5, a simplified derivative of HD5, is a promising antibiotic against MDRAb in vivo.
Insights into the bactericidal mechanism of HD5d5.
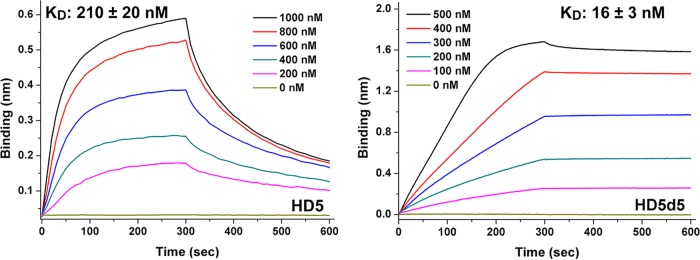
Lipid A is one of the important targets of defensins in the outer membrane of Gram-negative bacteria (11). We conducted BLI to analyze the interaction between HD5d5 and lipid A loaded on amine-reactive second-generation (AR2G) biosensors to examine the mechanism underlying the better bactericidal performance of HD5d5 against MDRAb in vivo than HD5. The equilibrium dissociation constant (KD) of HD5d5 binding to lipid A in the presence of 150 mM NaCl averaged 16 nM, which was 13.1-fold lower than HD5 (Fig. 4), illustrating that HD5d5 exhibits stronger membrane recruitment than HD5.
FIG 4.
Lipid A binding kinetics of HD5 and HD5d5. Lipid A was immobilized at 20 μg/ml. Peptides were prepared in 10 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl at concentrations of 200, 400, 600, 800, and 1,000 nM. The periods allowed for association and disassociation were both 300 s. Representative results from three independent experiments are shown. KD values are reported as means ± the SD.
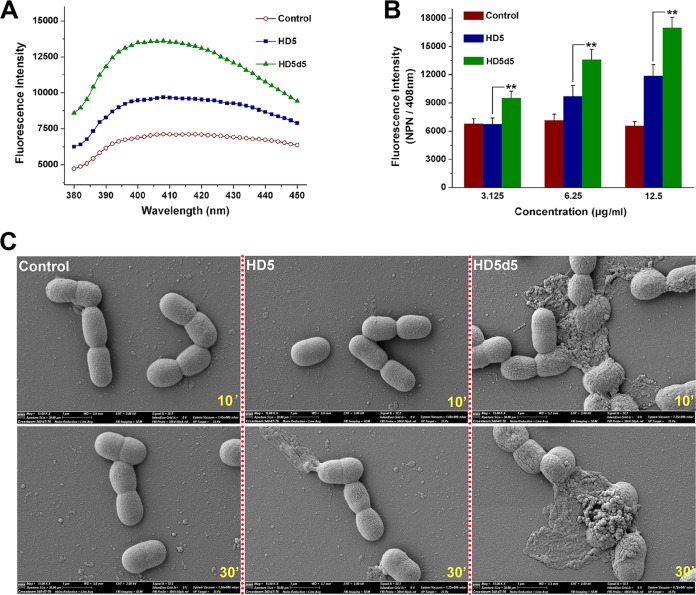
1-N-Phenylnaph-thylamine (NPN) is a hydrophobic dye that incorporates into impaired membranes and fluoresces upon excitation at 350 nm, exhibiting a maximum emission at approximately 408 nm (Fig. 5A). The fluorescence intensity of MDRAb treated with HD5d5 was significantly higher than MDRAb treated with HD5 (Fig. 5B), suggesting that HD5d5 penetrates MDRAb to a greater extent than HD5 after membrane recruitment. SEM images supported the hypothesis that MDRAb cells that are attacked by HD5 generally maintain their integrity after 10 min of incubation, whereas a subset of the cells exposed to HD5d5 were visibly injured, as indicated by the loss of smooth membrane, leakage of cytoplasmic content, and aggregation of microbes (Fig. 5C). Both peptides enabled the disintegration of MDRAb in 30 min, but consistent with the bactericidal discrepancy, HD5d5 was more destructive than HD5.
FIG 5.
HD5d5 penetrates MDRAb membranes to a greater extent than HD5. (A) NPN uptake assay. A representative fluorescence spectrum of NPN equilibrated with MDRAb in the presence of 6.25 μg/ml peptides is shown. NPN was excited at 350 nm. The fluorescent intensity was scanned from 380 to 450 nm at intervals of 2 nm. (B) Histogram revealing the dose-dependent membrane penetration of peptides. HD5 and HD5d5 were prepared at concentrations of 3.125, 6.25, and 12.5 μg/ml in sterile water. The emission wavelength of NPN was fixed at 408 nm. The results are presented as means ± the SD. **, P < 0.01. (C) SEM images of the morphological changes in MDRAb cells exposed to peptides. Bacteria cultured to mid-logarithmic phase were coincubated with peptides at 100 μg/ml and 37°C for 10 and 30 min. The extra high tension of SEM is 2,000 V.
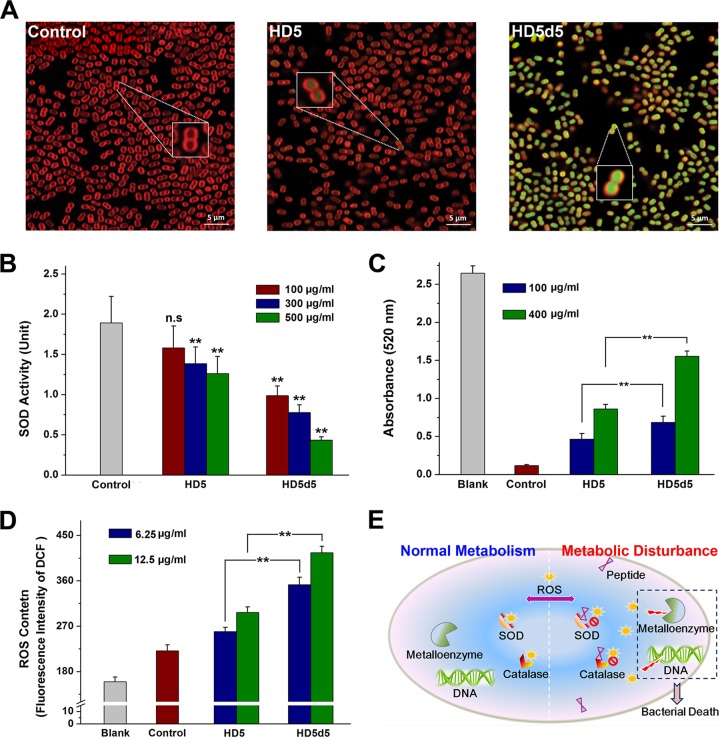
Membrane penetration is not the only way for peptides to inactivate bacteria (28). Because a portion of MDRAb cells exposed to peptides were freed from catastrophic collapse (see Fig. S2 in the supplemental material), we suspected that HD5d5 may have also inactivated MDRAb through other mechanisms. In addition to mechanical injury, antibacterial peptides have been reported to be capable of targeting intracellular elements, disturbing metabolic processes and inducing bacterial death in a nondisruptive manner (29, 30). Based on our LSCM analysis, HD5 and HD5d5 both entered the cytoplasm of MDRAb (Fig. 6A), enabling interactions between the peptides and critical proteins or enzymes inside cells. Notably, HD5d5 was superior to HD5 not only in the number of molecules located in the cytoplasm but also in its ability to elicit protein agglutination once in contact with microbial contents (see Fig. S3 in the supplemental material). The activities of superoxide dismutase (SOD) and catalase, two conspicuous enzymatic scavengers of ROS, were subsequently attenuated (Fig. 6B and C), resulting in the evident accumulation of ROS in the microbes (Fig. 6D). In addition, a significantly greater ROS content was detected in MDRAb exposed to HD5d5 than in cells treated with HD5. We propose that the oxidative stress caused by metabolic inhibition also contributes to HD5d5-mediated MDRAb inactivation (Fig. 6E).
FIG 6.
Intracellular targeting of HD5 and HD5d5 in MDRAb. (A) LSCM images showing the molecular location of peptides in MDRAb. Bacteria cultured to mid-logarithmic phase were coincubated with HD5 and HD5d5 at 100 μg/ml at 37°C for 20 min. Peptides were labeled with FITC (488/525 nm, green fluorescence), and the MDRAb membrane was stained with FM 4-64FX (565/744 nm, red fluorescence). Scale bar, 5 μm. Representative cells are magnified in the inset photographs. (B) SOD activity analysis. The cytoplasmic content of MDRAb was coincubated with 100, 300, and 500 μg/ml peptides at 37°C for 10 min. Results are presented as means ± the SD. n.s, not significant; **, P < 0.01 (compared to the control group). (C) Catalase activity analysis. The cytoplasmic content of MDRAb was coincubated with peptides at 100 and 400 μg/ml and 37°C for 10 min. Blank, hydrogen peroxide only; control, peptide-free. **, P < 0.01. (D) ROS content in MDRAb treated with peptides. HD5 and HD5d5 were prepared at concentrations of 6.25 and 12.5 μg/ml in sterile water. The fluorescent intensity was obtained at 525 nm upon excitation at 488 nm. The results are presented as means ± the SD. Blank, DCFH-DA-free; control, peptide-free. **, P < 0.01. (E) Diagram illustrating the metabolic disturbance induced by peptides to inactivate MDRAb. Normally, intracellular ROS are detrimental to metalloenzymes and DNA and are eliminated by SOD and catalase. The translocation of HD5d5 enables its interaction with the cytoplasmic content, which reduces the enzymatic activity and results in ROS accumulation.
Evaluation of AbOmpA neutralization by HD5d5.
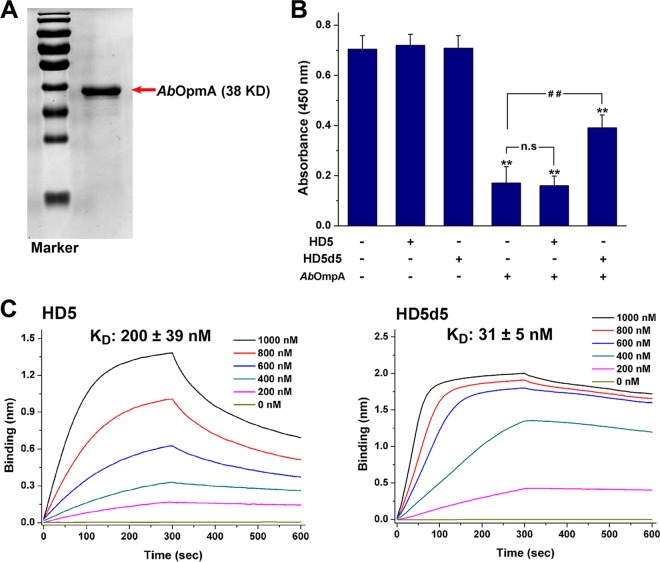
HD5 is noted for its multivalent binding and toxin neutralization activities, in addition to the bactericidal activity (22, 31). Since AbOmpA is toxic to the human epithelium (19), we measured the survival of laryngeal epithelial Hep-2 cells exposed to this virulence factor in the presence or absence of HD5 and HD5d5. Purified AbOmpA prepared by genetic recombination was toxic to Hep-2 at 5 μg/ml (Fig. 7A and B). HD5 failed to alleviate the cytotoxic effect at concentrations up to 50 μg/ml, showing a low affinity of 200 ± 39 nM for AbOmpA (Fig. 7C). In contrast, HD5d5 exerted a marked dose-dependent mitigative effect (see Fig. S4 in the supplemental material), with a KD of 31 ± 39 nM, which was ∼6.5-fold lower than HD5, indicating that the potent toxin-binding ability contributes to the AbOmpA-neutralizing effect of HD5d5.
FIG 7.
HD5d5 suppresses AbOmpA by binding with high affinity. (A) SDS-PAGE and Coomassie blue staining. AbOmpA was prepared by genetic recombination using a pET-30a-OmpA expression plasmid. Portions (2 μg) of AbOmpA were analyzed by SDS–10% PAGE. Protein bands were stained with Coomassie blue, and the image was obtained using a Bio-Rad ChemiDoc XRS+ system. (B) AbOmpA neutralization. HEp-2 cells were coincubated with 5 μg/ml AbOmpA in the absence or presence of peptides at 37°C for 9 h. HD5 and HD5d5 were prepared at a concentration of 50 μg/ml in sterile water. The results are presented as means ± the SD. n.s, not significant; **, P < 0.01 (compared to the absorbance of the AbOmpA-free group); ##, P < 0.01. (C) AbOmpA binding kinetics. AbOmpA was immobilized on AR2G biosensors at 20 μg/ml. HD5 and HD5d5 were prepared in 10 mM sodium phosphate buffer (pH 7.4) at concentrations of 200, 400, 600, 800, and 1,000 nM. The periods allowed for association and disassociation were both 300 s. Representative results from three independent experiments are shown. The KD values are reported as means ± the SD.
DISCUSSION
The increasing incidence of MDRAb infections worldwide is challenging the human antibiotic arsenal. HD5, a cationic peptide primarily detected in intestine Paneth cells, is a potential candidate to antagonize MDRAb, as it has evolved the ability to suppress pathogens (32). We used a modified broth microdilution method and virtual colony count assay to determine the antibacterial action of HD5 against MDRAb in this study. The broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) is widely used to analyze the antibiotic susceptibility of bactericides (33, 34). However, its accuracy is lowered when bactericides, such as cationic peptides, precipitate in Mueller-Hinton broth (MHB) or adsorb to microtiter plates (35). Wu et al. specifically applied the polypropylene plate and prepared peptides in 0.01% acetic acid containing 0.2% bovine serum albumin (BSA), obtaining a modified method that was instrumental to the bactericidal evaluation of numerous peptides (36, 37).
Nevertheless, this method ignores the fact that MHB lowers the antibacterial activity of certain peptides (38). HD5 is a lectin-like molecule with multivalent binding abilities (39), which enables its interaction with MHB. Incubation of HD5 with increasing concentrations of MHB revealed that the bactericidal potency of HD5 was gradually decreased (see Fig. S5 in the supplemental material). This finding may help explain the different susceptibilities of MDRAb to HD5 observed in the modified broth microdilution method (90% MHB) and virtual colony count assay (1% MHB). Based on the agreement between the results of the virtual colony assay and IWI experiment, as well as the greater complexity of the in vivo environment than 1% MHB, we postulate that the antibacterial action of HD5 against MDRAb is underestimated and overestimated by the modified broth microdilution method and virtual colony assay, respectively.
Disulfide bonds play an inconclusive role in the ability of human defensin to kill bacteria, as manifested by the observations that disulfide removal impairs the activity of human neutrophil peptide 1 (HNP1) against Staphylococcus aureus (31), but HBD3 inactivates Escherichia coli independently of disulfide bonds (40). The effects of HBD1 and HD6 on anaerobes are even enhanced upon disulfide reduction (15, 41). Consistent with a recent finding that disulfide bonds are crucial for HD5 to eliminate E. coli and Salmonella enterica serovar Typhimurium (6), we discovered that disulfide bonds were also indispensable for MDRAb killing. Notably, the preservation of only one disulfide bond significantly improved the antibacterial activity, and the Cys2-4 bond was more instrumental than the other pairings, potentially because Cys2-4 is the sole disulfide bond shared by human α- and β-defensins (see Fig. S6 in the supplemental material). Conserved elements are often endowed with more functions, as previously described (42).
Positive charges and hydrophobicity are important factors in defensins that lead to bacterial death. An increase in either the net charge or hydrophobicity theoretically enhances bactericidal activity. However, because hydrophobic residues have been connected to the cytotoxicity of antibacterial peptides (43), an electropositive improvement is widely used in defensin-derived antibiotic design (11, 14). Arg and Lys are natural cationic residues that highly abundant in human defensins. Arg has been shown to be more beneficial for electrostatic interactions than Lys (44), and replacing Arg with Lys weakened the bactericidal potency of HNP1 toward E. coli, which was attributed to the larger solvent-accessible surface of Arg in stereo (45). These findings are likely why HD5d5 exerted stronger abilities than HD5d6 to alter the bacterial surface charge and kill MDRAb.
In addition to the contribution to electrostatic interactions, Arg also benefits peptides by inducing negative Gaussian curvature and pore formation on bacterial membranes (46, 47), which explains why HD5d5, an Arg mutant of HD5, was more efficient at penetrating MDRAb. Membrane penetration does not necessarily lead to cell death. Numerous bactericides, such as TWF, a Phe mutant of cathelicidin, and temporin L, are able to penetrate bacterial membranes without affecting cell survival, which is attributed to the absence of catastrophic collapse (28, 48). Based on our SEM observations, both HD5 and HD5d5 caused visible cellular disintegration, which indicates a determinative role of membrane penetration in MDRAb-killing activity.
Different mechanisms by which antibacterial peptides penetrate membranes have been proposed, including the SMH model and the toroidal pore model (39). Notably, peptides would enter the cytoplasm in both of these models. Intracellular nucleic acids, proteins, and enzymes are consequently vulnerable, which has been experimentally confirmed and considered as an alternative pathway for bacterial death (5). HD5d5 entered the cytoplasm of MDRAb, where it interacted with cellular contents and decreased the activities of SOD and catalase, resulting in an accumulation of ROS in the cells. Due to the correlation with metalloenzyme inactivation and DNA destruction (49), the overload of ROS is responsible for the elimination of E. coli and A. baumannii by many antibiotics (50, 51). Intracellular targeting of HD5d5 plausibly contributes to the devitalization of cells following membrane penetration but does not induce lethal collapse.
Toxin neutralization is another dramatic property of defensin that distinguishes it from traditional antibiotics. HD5 governs potent multivalent binding to glycoproteins, which is attenuated when Arg is replaced with Ala (22). Similarly, replacing Arg with Ala weakens the interaction between HD5 and anthrax LF, a Zn2+-dependent protease that is toxic to macrophages, which leads to a striking attenuation of bacteriotoxin suppression (23). According to our data, HD5d5 was more efficient at binding to and inhibiting AbOmpA than HD5, further indicating that Arg is beneficial for peptides that suppress virulence factors. However, further investigation is required to clearly determine whether AbOmpA neutralization by HD5d5 is due to domain blocking or protein unfolding, as occurs with LL-37 and retrocyclin (16, 52), respectively.
Collectively, HD5d5, a simplified derivative of HD5 constructed by disulfide reduction and Arg introduction, is able to bind to and penetrate MDRAb membranes, causing cell death by catastrophic collapse and metabolic disruption. Unlike traditional antibiotics, HD5d5 captures and suppresses AbOmpA, which expands the approaches available to alleviate MDRAb infection. HD5d5 exerts potent antibacterial activity against MDRAb and is thus a promising candidate to antagonize MDRAb in clinical settings.
MATERIALS AND METHODS
Peptide preparation.
Peptides were chemically synthesized by Chinese Peptide Company (FDA 300451884; Hangzhou, Zhejiang, People's Republic of China). The sequence information is presented in Fig. 1A. The purity and molecular mass of peptides were determined by high-performance liquid chromatography (Waters, Milford, MA) and matrix-assisted laser desorption ionization-time of flight (Shimadzu, Kyoto, Japan), respectively (the results of these analyses are presented in Table S1 in the supplemental material).
Isolation of MDRAb.
The MDRAb strain was isolated from the wound of a burn patient at the First Affiliated Hospital of Third Military Medical University (TMMU). The bacterium was identified by API 20NE (bioMérieux, Marcy l'Etoile, FRA) and 16S rRNA gene sequencing (see Fig. S7 in the supplemental material). The resistance to tetracycline, amikacin, cefotaxime, CIP, aztreonam, and trimethoprim-sulfamethoxazole antibiotics was determined using the Kirby-Bauer method (see Fig. S8 in the supplemental material).
Broth microdilution antibacterial assay.
The MICs of HD5 and its derivatives for MDRAb and A. baumannii (ATCC 19606) were determined using the broth microdilution method recommended by the CLSI with the modifications reported by the Hancock laboratory (24). Peptides were prepared in 0.01% acetic acid containing 0.2% BSA at concentrations of 25, 50, 100, 200, 400, 800, 1,600, 3,200, 6,400, 12,800, and 25,600 μg/ml. Bacteria cultured to mid-logarithmic phase were diluted to 2 × 105 CFU/ml in MHB. Portions (100 μl) of the bacterial suspension were coincubated with 11-μl portions of peptides at 37°C in sterile polypropylene microtiter plates (Corning, catalog no. 3365). The absorbance at 600 nm was obtained after 24 h of incubation using an M2e microplate reader (Molecular Devices, Silicon Valley, CA). The MIC was recorded as the lowest concentration of peptide leading to a 70% reduction in bacterial turbidity. The MICs of CIP were analyzed using the standard broth microdilution method outlined by the CLSI. This experiment was repeated three times on different days.
Virtual colony count antibacterial assay.
MDRAb cultured to mid-logarithmic phase were diluted to 106 CFU/ml in 10 mM sodium phosphate buffer (pH 7.4) containing 1% MHB. Peptides and CIP were prepared in sterile water at concentrations ranging in 2-fold dilutions from 15.6 to 500 μg/ml. Portions (90 μl) of bacteria were coincubated with 10-μl portions of peptides or CIP at 37°C for 1 h. A 100-μl aliquot of 2× MHB was then added, and bacterial growth was monitored using an M2e microplate reader as previously described (38). HD5 and HD5d5 (12.5 μg/ml) were coincubated with MDRAb in the presence of increasing concentrations of NaCl (0, 50, 100, and 150 mM) and CaCl2 (0, 0.5, 1.5, and 2.5 mM) to analyze the salt-resistant antibacterial action. This experiment was conducted in duplicate and repeated three times.
CD spectroscopy.
Peptide conformations were measured using an Applied Photophysics Chirascan instrument (Leatherhead, Surrey, UK) at 27°C. A cell with a 1-mm path length was used. The spectral data were obtained from 190 to 260 nm at 1-nm intervals. The time per point was 0.5 s, and the scanning time was approximately 65 s. Peptides were prepared in sterile water at a concentration of 100 μg/ml. The results obtained from three independent scans were averaged and smoothed using Pro-Data Chirascan 4.1 software.
Zeta potential determination.
Nine hundred microliters of MDRAb (108 CFU/ml) grown to mid-logarithmic phase were coincubated with 100 μl of peptides or sterile water, and the solution was then loaded into a disposable zeta cell. Samples were equilibrated at 25°C for 2 min. The results were obtained using a Zetasizer Nano ZS (Malvern Instruments, Malvern, Worcestershire, UK). This experiment was conducted in duplicate and repeated three times.
Animal experiment.
The bactericidal actions of peptides were evaluated in vivo using an IWI model. One hundred female BALB/c mice, 8 to 10 weeks old and weighing 18 to 22 g, were irradiated with 5 Gy in one treatment using an RS2000 X-ray irradiator (Rad Source, Suwanee, GA) and divided randomly into five groups. A full-thickness skin wound of 1 cm in diameter was prepared 4 days after irradiation, and 15 μl of MDRAb (4 × 108 CFU/ml) was evenly coated on the lesion after 2 h. Peptides (100 μg/ml, 20 μl) were applied 3 h later and daily thereafter for 4 days. Animal survival was monitored for 13 days. The number of microbes under the scabs that were resistant to 100 μg/ml CIP was counted on day 9. Scabs from six mice were sampled and immersed in 1 ml of 10 mM sodium phosphate buffer (pH 7.4). Homogenates were prepared using a gentleMACS dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany) and were further diluted and cultured on MHB plates. Animal experiments were approved by the Animal Experimental Ethics Committee of TMMU.
BLI-based binding assay.
The binding kinetics of peptides to lipid A (catalog no. L5399; Sigma) and AbOmpA were analyzed using a ForteBio “Octet Red 96” BLI System (Menlo Park, CA). The AR2G biosensors were used and activated with 20 mM 1-ethyl-3-(3-dimethylaminopropy)carbodiimide hydrochloride (EDC, E1769; Sigma) and 10 mM sulfo-N-hydroxysulfosuccinimide (s-NHS, catalog no. 56485; Sigma). Peptides were prepared in 10 mM sodium phosphate buffer (pH 7.4) at concentrations of 200, 400, 600, 800, and 1,000 nM. The binding responses were recorded at 30°C and processed as previously described (11). The KD was calculated as the ratio of the dissociation rate constant (Koff) to the association rate constant (Kon). This experiment was repeated three times.
Membrane penetration detection.
One hundred ninety microliters of MDRAb (1 × 108 CFU/ml) cultured to mid-logarithmic phase was coincubated with 10 μl of peptides at 37°C for 1 h. A 10-μl aliquot of NPN (104043, Sigma, 200 μM) was added. The fluorescence intensity was measured at 380 to 450 nm using a Tecan Infinite M1000 Pro microplate reader (Männedorf, Zürich, CHE). This experiment was conducted in duplicate and repeated three times.
SEM.
A 1-ml aliquot of MDRAb (108 CFU/ml) cultured to mid-logarithmic phase was centrifuged and resuspended in 50 μl of 10 mM sodium phosphate (pH 7.4). Bacteria were coincubated with 5 μl of peptides (1 mg/ml) at 37°C for 10 min and 30 min at a shaking speed of 130 rpm. The mixture was used to coat a coverslip and dried at room temperature. Samples were fixed with 2.5% glutaraldehyde at 4°C, dehydrated with gradient concentrations of ethanol, and desiccated with tert-butyl alcohol. A gold coating was used to prevent the accumulation of static electric fields in the specimen. A Zeiss Crossbeam 340 scanning electron microscope (Oberkochen, Germany) was used to observe the morphology of MDRAb.
LSCM.
MDRAb (108 CFU) cultured to mid-logarithmic phase was stained with FM 4-64FX (catalog no. F34653; Invitrogen) at 5 μg/ml. Bacteria were centrifuged at 4°C, resuspended in 100 μl of 10 mM sodium phosphate (pH 7.4), and coincubated with N-terminally fluorescein isothiocyanate (FITC)-conjugated peptides at 100 μg/ml at 37°C for 20 min. Approximately 10 μl of the mixture was used to coat a slide and dried at room temperature. Samples were fixed with 4% paraformaldehyde and observed using a Zeiss LSM 780 NLO confocal microscope.
SOD activity analysis.
The experiment is based on the observation that SOD promotes the dismutation of superoxide anions, which interact with WST-8 and generate the formazan dye. MDRAb cultured to the stationary phase was centrifuged and resuspended in 200 μl of 10 mM sodium phosphate (pH 7.4). The cytoplasmic content was collected using ultrasonic disruption steps and diluted to 1,500 μg/ml. A Beyotime SOD assay kit (catalog no. S0101; Shanghai, People's Republic of China) was used according to the manufacturer's instructions. Briefly, the cytoplasmic content was coincubated with 100-, 300-, and 500-μg/ml peptide solutions at 37°C for 10 min. Then, 100 μl of the WST-8/enzyme working solution and 20 μl of the reaction starting solution were added. The mixture was incubated at 37°C for 30 min, and the absorbance was determined at 450 nm. The inhibition ratio (IR) of SOD was determined as (Acon − Apep)/(Acon − Ablank) × 100%. The SOD activity unit was calculated as IR/(1 − IR). This assay was conducted in duplicate and repeated three times.
Catalase activity analysis.
This experiment is based on the observation that catalase eliminates hydrogen peroxide, which interacts with peroxidase and generates N-(4-antipyryl)-3-chloro-5-sulfonate-p-benzoquinonemonoimine. A Beyotime catalase assay kit (catalog no. S0051) was used. A 10-μl aliquot of the cytoplasmic content (1,500 μg/ml) was coincubated with 100- and 400-μg/ml peptide solutions at 37°C for 10 min. The mixture was diluted 1:20 and coincubated with 250 mM hydrogen peroxide at 25°C for 5 min. Then, 450 μl of the stop solution were added, which was then diluted 1:5 and incubated with 200 μl of the color development solution. The absorbance was determined at 520 nm using a microplate reader. This assay was conducted in duplicate and repeated three times.
Bacterial ROS detection.
The ROS content in MDRAb exposed to peptides was determined using a Beyotime ROS assay kit (catalog no. S0033). MDRAb (2 × 108 CFU/ml) cells cultured to mid-logarithmic phase were stained with 2′,7′-dichloro-dihydro-fluorescein diacetate (DCFH-DA) and coincubated with peptides at 6.25 and 12.5 μg/ml at 37°C for 30 min. The fluorescence intensity was measured at 525 nm upon excitation at 488 nm using a Tecan Infinite M1000 Pro microplate reader (Männedorf). This experiment was conducted in duplicate and repeated three times.
AbOmpA preparation.
AbOmpA was prepared by Detai Biologics (Nanjing, People's Republic of China), as previously described (53). Briefly, the gene encoding AbOmpA (1,109 bp) was optimized from 92 to 1,054 bp (see Fig. S9A in the supplemental material). The sequence was inserted into a pET30a vector at the NdeI and HindIII sites (see Fig. S9B in the supplemental material), which was then transformed into E. coli BL21(DE3) cells. Cultures from single colonies were grown at 37°C overnight in the presence of 50 μg/ml kanamycin (K103026; Aladdin, Shanghai, People's Republic of China). The pET-30a-OmpA expression plasmid was extracted from bacterial cultures and verified by gene sequencing. Seed cultures were further diluted 1:100 with Luria-Bertani broth, followed by incubation at 37°C with a shaking speed of 200 rpm. When the optical density at 600 nm reached 0.5, 500 μM isopropyl-β-d-thiogalactoside (IPTG; catalog no. I5502; Sigma) was added, and the bacteria were cultured at 15°C for 16 h. Cells were collected and lysed by sonication. The bacterial contents were further dissolved in an 8 M urea solution. AbOmpA was purified with Ni-IDA resin (C600029; Sangon Biotech, Shanghai, People's Republic of China), eluted with gradient concentrations of imidazole (I5513; Sigma), and analyzed using SDS-PAGE and Coomassie blue staining (see Fig. S9C in the supplemental material). The eluent was dialyzed with an 8,000 Da tube (C006622; Sangon Biotech), purified with a polymixin B Sepharose column (A610319, Sangon Biotech), and finally sterilized with a 0.22-μm-pore size filter.
AbOmpA inhibition assay.
Human laryngeal epithelial HEp-2 cells obtained from the cell bank of Chinese Academy of Sciences (Shanghai, People's Republic of China) were cultured in Dulbecco modified Eagle medium (SH30022.01B; HyClone) supplemented with 10% fetal bovine serum (16000-044; Gibco) and seeded into 96-well plates at a density of 4,000 CFU/well. After adherence, the cells were washed with phosphate-buffered saline and coincubated with AbOmpA at 5 μg/ml in the absence or presence of peptides at 37°C for 9 h. Peptides were prepared in sterile water at concentrations of 10, 30, and 50 μg/ml. A Cell Counting Kit-8 (CCK-8, catalog no. CK04; Dojindo) was used to detect cell survival. The absorbance was determined at 450 nm using a microplate reader. This experiment was conducted in duplicate and repeated three times.
Statistical analysis.
Statistically significant differences (P) between groups were calculated using one-way analysis of variance and the Student-Newman-Keuls multiple-comparison test with SPSS 16.0 software. A P value of <0.05 was defined as statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate L. Wang and Q. Wei (Biomedical Analysis Center, TMMU) for assisting with the LSCM and SEM, respectively.
This study was supported by grants from the National Natural Science Fund of China (81703395, 81502960, 81725019, and 81371688), the Funds from PLA (AWS16J014 and AWS13J002), and the Program for Scientific and Technological Innovation Leader of Chongqing (CSTCKJCXLJRC06).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01504-17.
REFERENCES
- 1.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zilberberg MD, Kollef MH, Shorr AF. 2016. Secular trends in Acinetobacter baumannii resistance in respiratory and blood stream specimens in the United States, 2003 to 2012: a survey study. J Hosp Med 11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 3.Kuo LC, Lai CC, Liao CH, Hsu CK, Chang YL, Chang CY, Hsueh PR. 2007. Multidrug-resistant Acinetobacter baumannii bacteraemia: clinical features, antimicrobial therapy, and outcome. Clin Microbiol Infect 13:196–198. doi: 10.1111/j.1469-0691.2006.01601.x. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Shen M, Zhang N, Wang S, Xu Y, Chen S, Chen F, Yang K, He T, Wang A, Su Y, Cheng T, Zhao J, Wang J. 2016. Reduction impairs the antibacterial activity but benefits the LPS neutralization ability of human enteric defensin 5. Sci Rep 6:22875. doi: 10.1038/srep22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 8.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 9.Wang A, Wang S, Shen M, Chen F, Zou Z, Ran X, Cheng T, Su Y, Wang J. 2009. High level expression and purification of bioactive human alpha-defensin 5 mature peptide in Pichia pastoris. Appl Microbiol Biotechnol 84:877–884. doi: 10.1007/s00253-009-2020-x. [DOI] [PubMed] [Google Scholar]
- 10.Wang A, Chen F, Wang Y, Shen M, Xu Y, Hu J, Wang S, Geng F, Wang C, Ran X, Su Y, Cheng T, Wang J. 2013. Enhancement of antiviral activity of human alpha-defensin 5 against herpes simplex virus 2 by arginine mutagenesis at adaptive evolution sites. J Virol 87:2835–2845. doi: 10.1128/JVI.02209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Shen M, Gohain N, Tolbert WD, Chen F, Zhang N, Yang K, Wang A, Su Y, Cheng T, Zhao J, Pazgier M, Wang J. 2015. Design of a potent antibiotic peptide based on the active region of human defensin 5. J Med Chem 58:3083–3093. doi: 10.1021/jm501824a. [DOI] [PubMed] [Google Scholar]
- 12.Wanniarachchi YA, Kaczmarek P, Wan A, Nolan EM. 2011. Human defensin 5 disulfide array mutants: disulfide bond deletion attenuates antibacterial activity against Staphylococcus aureus. Biochemistry 50:8005–8017. doi: 10.1021/bi201043j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SA, Liu J, Xiang Y, Wang YJ, Lee WH, Zhang Y. 2014. Therapeutic potential of the antimicrobial peptide OH-CATH30 for antibiotic-resistant Pseudomonas aeruginosa keratitis. Antimicrob Agents Chemother 58:3144–3150. doi: 10.1128/AAC.00095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansour SC, de la Fuente-Nunez C, Hancock RE. 2015. Peptide IDR-1018: modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J Pept Sci 21:323–329. doi: 10.1002/psc.2708. [DOI] [PubMed] [Google Scholar]
- 15.Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. 2011. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature 469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 16.Lin MF, Tsai PW, Chen JY, Lin YY, Lan CY. 2015. OmpA binding mediates the effect of antimicrobial peptide LL-37 on Acinetobacter baumannii. PLoS One 10:e0141107. doi: 10.1371/journal.pone.0141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol 8:216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, Choi CH, Kim JW, Lee JC. 2010. Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J Microbiol 48:387–392. doi: 10.1007/s12275-010-0155-1. [DOI] [PubMed] [Google Scholar]
- 19.Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, Hyun SH, Kim SA, Lee SK, Lee JC. 2005. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol 7:1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim C, Slavinskaya Z, Merrill AR, Kaufmann SH. 2006. Human alpha-defensins neutralize toxins of the mono-ADP-ribosyltransferase family. Biochem J 399:225–229. doi: 10.1042/BJ20060425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim C, Gajendran N, Mittrucker HW, Weiwad M, Song YH, Hurwitz R, Wilmanns M, Fischer G, Kaufmann SH. 2005. Human alpha-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc Natl Acad Sci U S A 102:4830–4835. doi: 10.1073/pnas.0500508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehrer RI, Jung G, Ruchala P, Andre S, Gabius HJ, Lu W. 2009. Multivalent binding of carbohydrates by the human alpha-defensin, HD5. J Immunol 183:480–490. doi: 10.4049/jimmunol.0900244. [DOI] [PubMed] [Google Scholar]
- 23.Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, Lu W. 2012. Functional determinants of human enteric alpha-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem 287:21615–21627. doi: 10.1074/jbc.M112.367995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Hancock RE. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J Biol Chem 274:29–35. doi: 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- 25.Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. 2009. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect Immun 77:5035–5043. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng SY, You RI, Lai MJ, Lin NT, Chen LK, Chang KC. 2017. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci Rep 7:11477. doi: 10.1038/s41598-017-11832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maisetta G, Di Luca M, Esin S, Florio W, Brancatisano FL, Bottai D, Campa M, Batoni G. 2008. Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 29:1–6. doi: 10.1016/j.peptides.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Mangoni ML, Papo N, Barra D, Simmaco M, Bozzi A, Di Giulio A, Rinaldi AC. 2004. Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochem J 380:859–865. doi: 10.1042/bj20031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilmes M, Cammue BP, Sahl HG, Thevissen K. 2011. Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat Prod Rep 28:1350–1358. doi: 10.1039/c1np00022e. [DOI] [PubMed] [Google Scholar]
- 30.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. 1989. Interaction of human defensins with Escherichia coli: mechanism of bactericidal activity. J Clin Invest 84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. 2009. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem 284:29180–29192. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. 2002. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 33.Flamm RK, Rhomberg PR, Sader HS. 2017. In vitro activity of the novel lactone ketolide nafithromycin (WCK 4873) when tested against contemporary clinical bacteria from a global surveillance program. Antimicrob Agents Chemother 61:e01230-. doi: 10.1128/AAC.01230-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaumburg F, Bletz S, Mellmann A, Becker K, Idelevich EA. 2017. Susceptibility of MDR Pseudomonas aeruginosa to ceftolozane/tazobactam and comparison of different susceptibility testing methods. J Antimicrob Chemother 72:3079–3084. doi: 10.1093/jac/dkx253. [DOI] [PubMed] [Google Scholar]
- 35.Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, Cheng FC, Loury DJ, Fiddes JC. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother 41:1738–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlusselhuber M, Jung S, Bruhn O, Goux D, Leippe M, Leclercq R, Laugier C, Grotzinger J, Cauchard J. 2012. In vitro potential of equine DEFA1 and eCATH1 as alternative antimicrobial drugs in rhodococcosis treatment. Antimicrob Agents Chemother 56:1749–1755. doi: 10.1128/AAC.05797-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieczorek M, Jenssen H, Kindrachuk J, Scott WR, Elliott M, Hilpert K, Cheng JT, Hancock RE, Straus SK. 2010. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem Biol 17:970–980. doi: 10.1016/j.chembiol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Ericksen B, Wu Z, Lu W, Lehrer RI. 2005. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother 49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas P. 2009. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J 276:6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x. [DOI] [PubMed] [Google Scholar]
- 40.Kluver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, Adermann K. 2005. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 44:9804–9816. doi: 10.1021/bi050272k. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder BO, Ehmann D, Precht JC, Castillo PA, Küchler R, Berger J, Schaller M, Stange EF, Wehkamp J. 2015. Paneth cell α-defensin 6 (HD-6) is an antimicrobial peptide. Mucosal Immunol 8:661. doi: 10.1038/mi.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehrer RI, Lu W. 2012. α-Defensins in human innate immunity. Immunol Rev 245:84. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 43.Asthana N, Yadav SP, Ghosh JK. 2004. Dissection of antibacterial and toxic activity of melittin: a leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J Biol Chem 279:55042–55050. doi: 10.1074/jbc.M408881200. [DOI] [PubMed] [Google Scholar]
- 44.Dougherty DA. 1996. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 45.Zou G, de Leeuw E, Li C, Pazgier M, Li C, Zeng P, Lu WY, Lubkowski J, Lu W. 2007. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem 282:19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt NW, Tai KP, Kamdar K, Mishra A, Lai GH, Zhao K, Ouellette AJ, Wong GC. 2012. Arginine in α-defensins: differential effects on bactericidal activity correspond to geometry of membrane curvature generation and peptide-lipid phase behavior. J Biol Chem 287:21866–21872. doi: 10.1074/jbc.M112.358721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam KL, Wang H, Siaw TA, Chapman MR, Waring AJ, Kindt JT, Lee KY. 2012. Mechanism of structural transformations induced by antimicrobial peptides in lipid membranes. Biochim Biophys Acta 1818:194–204. doi: 10.1016/j.bbamem.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Yang ST, Shin SY, Hahm KS, Kim JI. 2006. Different modes in antibiotic action of tritrpticin analogs, cathelicidin-derived Trp-rich and Pro/Arg-rich peptides. Biochim Biophys Acta 1758:1580–1586. doi: 10.1016/j.bbamem.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Baez A, Shiloach J. 2014. Effect of elevated oxygen concentration on bacteria, yeasts, and cells propagated for production of biological compounds. Microb Cell Fact 13:181. doi: 10.1186/s12934-014-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siddiqui H, Tasneem S, Farooq S, Sami A, Atta Ur R, Choudhary MI. 2017. Harmaline and its derivatives against infectious multidrug-resistant Escherichia coli. Med Chem 13:465–476. doi: 10.2174/1573406413666170125113832. [DOI] [PubMed] [Google Scholar]
- 51.Bhargava N, Sharma P, Capalash N. 2014. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun 82:3417–3425. doi: 10.1128/IAI.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kudryashova E, Seveau S, Lu W, Kudryashov DS. 2015. Retrocyclins neutralize bacterial toxins by potentiating their unfolding. Biochem J 467:311–320. doi: 10.1042/BJ20150049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol 10:309–319. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.