ABSTRACT
The emergence of colistin-resistant Klebsiella pneumoniae (CoRKp) is a public health concern, since this antibiotic has become the last line of treatment for infections caused by multidrug-resistant (MDR) Gram negatives. In this study, we have investigated the molecular basis of colistin resistance in 13 MDR K. pneumoniae strains isolated from 12 patients in a teaching hospital in Sousse, Tunisia. Whole-genome sequencing (WGS) was used to decipher the molecular mechanism of colistin resistance and to identify the resistome of these CoRKp isolates. It revealed a genome of ca. 5.5 Mbp in size with a G+C content of 57%, corresponding to that commonly observed for K. pneumoniae. These isolates belonged to the 5 different sequence types (ST11, ST15, ST101, ST147, and ST392), and their resistome was composed of acquired β-lactamases, including extended-spectrum beta-lactamase and carbapenemase genes (blaCTX-M-15, blaOXA-204, blaOXA-48, and blaNDM-1 genes), aminoglycoside resistance genes [aac(6′)Ib-cr, aph(3″)-Ib, aph(6)-Id, and aac(3)-IIa], and fosfomycin (fosA), fluoroquinolone (qnr-like), chloramphenicol, trimethoprim, and tetracycline resistance genes. All of the isolates were identified as having a mutated mgrB gene. Mapping reads with reference sequences of the most common genes involved in colistin resistance revealed several modifications in mgrB, pmr, and pho operons (deletions, insertions, and substitutions) likely affecting the function of these proteins. It is worth noting that among the 12 patients, 10 were treated with colistin before the isolation of CoRKp. No plasmid encoding mcr-1 to mcr-5 genes was found in these isolates. This study corresponds to the first molecular characterization of a collection of CoRKp strains in Tunisia and highlights that the small-transmembrane protein MgrB is a main mechanism for colistin resistance in K. pneumoniae.
KEYWORDS: WGS, resistome, colistin resistance, BasR, BasS, MgrB, PhoP, PhoQ
INTRODUCTION
Over the past 2 decades, carbapenemase-producing Klebsiella pneumoniae (CPKp) isolates have become a major global health concern, because they may cause severe infections with high mortality rates (1). The capacity of K. pneumoniae to accumulate enzymatic and nonenzymatic resistance mechanisms against most clinically available antibiotics, including beta-lactams, carbapenems, fluoroquinolones, and aminoglycosides, is increasing worldwide, leading the medical community to face limiting therapeutic options to deal with infections caused by this microorganism (2). While the volume from the pharmaceutical pipeline is waning, with a relative paucity of new antibiotics active against such strains, we are assisting in a resurgence in the use of colistin, an old class of cyclic polypeptide antibiotics that was discovered in 1947 (3). The target of the antimicrobial action of colistin is the negatively charged lipopolysaccharide (LPS) of the external membrane, allowing penetration through the outer membrane by displacing Ca2+ and Mg2+. Products of pmr (polymyxin resistance) genes promote the covalent modification of lipopolysaccharide, reducing the attachment of colistin to the external surface of Gram-negative bacteria, and leads to bacterial cell death (4, 5). Nevertheless, reports of colistin-resistant K. pneumoniae (CoRKp) are increasing worldwide, likely as a result of selective pressure following exposure to colistin treatment (4, 6–8).
Colistin resistance mechanisms involve alteration of the lipid A biosynthetic pathway and modification of LPS surface (9, 10). These modifications reduce the electrostatic affinity between the cationic colistin and anionic LPS by modification with 4-amino-4-deoxy-l-arabinose (Ara4N) and phosphoethanolamine (PEtN) (10, 11). Modification of Ara4N is achieved by the pmrHFIJKLM operon and pmrC (10, 11). The most common molecular mechanisms sustaining this resistance showed that PmrAB, PmrD, PhoPQ, and the small transmembrane protein MgrB regulate expression of the pmrHFIJKLM operon (4, 7, 10–12). In addition, the crrB gene also has been described to be involved in colistin resistance in K. pneumoniae (9). Finally, the recent report of the plasmid-mediated colistin resistance mcr-1-like genes raised great medical concern worldwide (13). Since its initial description, the mcr-1 gene has been identified worldwide in Enterobacteriaceae (mostly Escherichia coli) and recovered from human and animal samples (14).
While resistance to colistin has been reported worldwide, there is still a lack in many studies, especially from Tunisia (15), of an explanation of the exact mechanisms responsible for this resistance. Whole-genome sequencing of 13 CoRKp isolates was applied to gain insights into the molecular mechanisms sustaining colistin resistance, their resistomes, and their genetic relatedness.
(This work was presented in part at the 36th Interdisciplinary Meeting on Anti-Infectious Chemotherapy [RICAI], Paris, France, 2016.)
RESULTS AND DISCUSSION
Demographic, clinical, and microbiological data.
During the 4-year study period, 2,826 K. pneumoniae isolates were recovered, of which 491 were carbapenem resistant (17.5%) and 13 were colistin resistant, as revealed by reduced inhibition zones around the colistin disks on routine antibiograms (0.4%). These 13 CoRKp isolates were isolated in urine samples of 12 patients admitted to different wards in a teaching hospital in Tunisia over a 4-year period (January 2012 to March 2016). These isolates exhibited a multidrug-resistant (MDR) profile (resistance to beta-lactams, including carbapenems for 7/13 isolates, and to aminoglycosides, to all of the tested fluoroquinolones, tetracycline, and trimethoprim-sulfamethoxazole) (Table 1).
TABLE 1.
Demographic, clinical, and microbiological features for colistin-resistant Klebsiella pneumoniae isolatesa
| Patient | Date of admission to hospitald (yr.mo) | Sample type | Ward | Treatment started prior to isolation of CoRKp | Isolate | Date of isolation (day.mo.yr) | MICb (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CSTc | ERT | DOR | MEM | IMP | TMO | TGC | |||||||
| 1 | C | Urine | Urology C | CST | C2 | 12.02.11 | 16 | 0.16 | 0.064 | 0.032 | 0.25 | 2.5 | 1 |
| 2 | 12.02 | Urine | Nephrology | LVX + CIP + CST | C3 | 12.03.27 | 8 | 1 | 0.25 | 0.50 | 0.75 | 512 | 3 |
| 3 | C | Urine | Urology C | CST | C4 | 12.03.22 | 32 | 0.032 | 0.032 | 0.023 | 0.094 | 3 | 1.5 |
| 4 | C | Urine | Nephrology-peritoneal dialysis | NA | C5 | 13.12.12 | 16 | 0.50 | 0.25 | 0.25 | 0.38 | 192 | 2 |
| 5 | 13.11 | Urine | Nephrology | IMP + FOF + OFX + CTX | C6 | 14.01.10 | 8 | >32 | 24 | >32 | >32 | >1024 | 1 |
| Urine | C7 | 8 | >32 | >32 | >32 | >32 | >1024 | 0.75 | |||||
| 6 | 15.02 | Superficial Pus | Medical ICU | IMP + TG + CST | C8 | 15.03.10 | >128 | 0.016 | 0.064 | 0.032 | 0.19 | 3 | 1 |
| 7 | 15.08 | Urine | Urology | CTX + OFX + CST | C9 | 15.08.31 | >128 | 6 | 3 | 4 | 1.5 | 128 | 3 |
| 8 | 15.09 | Catheter | Surgical ICU | IMP + CTX + OFX + GEN + MTR + CST | C10 | 15.10.05 | 8 | >32 | 6 | 12 | 4 | >1024 | 0.75 |
| 9 | 15.10 | Blood | Medical ICU | IMP + VNC + TEC + TZP + TIG + CST | C11 | 15.10.14 | 16 | >32 | 5 | 12 | 4 | >1024 | 1.5 |
| 10 | 15.11 | Urine | Medical ICU | IMP + VNC + RIF + CIP + CST | C12 | 15.11.25 | 16 | >32 | 6 | 8 | 4 | >1024 | 1 |
| 11 | 15.10 | Blood | Surgical ICU | IMP + VNC + RIF + AMC + AN + CST | C13 | 15.11.25 | 8 | 0.032 | 0.032 | 0.023 | 0.125 | 8 | 0.5 |
| 12 | NA | Urine | Nephrology C | SXT + TIG + CST | C14 | 16.03.15 | 16 | 0.016 | 0.032 | 0.023 | 0.9 | 8 | 0.5 |
NA, not available; CST, colistin; DOR, doripenem; ERT, ertapenem; IMP, imipenem; MEM, meropenem; TGC, tigecycline; TMO, temocillin.
MIC was determined using Etest (bioMérieux-France) and interpreted according to updated EUCAST breakpoint tables for interpretation of MICs and zone diameters, 2015, version 5.0 (www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf).
MICs for colistin were also determined by broth microdilution in cation-adjusted Mueller-Hinton broth (MHB-CA) according to CLSI guidelines.
C, consultation.
The patients were of different ages (between 16 and 79 years old), presented different comorbidities, and were from different wards. Ten out of the 12 patients were previously treated with colistin, as this molecule is used in our hospital as the last-line therapeutic option for severe Gram-negative infection (Table 1). The majority of patients in the present study were admitted to intensive care units (ICUs), known to have chronic kidney disease, or under peritoneal dialysis. These patients (10/12) received colistin regimens adjusted to their renal function. Whether previous colistin exposure contributed to the development of colistin resistance was not addressed in this report. However, resistance to colistin due to selective pressure following prolonged exposure to colistin, especially with subinhibitory concentrations, has been reported (7, 16–18). On the other hand, colistin resistance may appear without any prior colistin exposure simply as the result of spontaneous mutations of genomic DNA or through horizontal gene transfer (19).
Resistome and genetic relatedness of CoRKp isolates.
The draft genome of the 13 CoRKp isolates resulted in a mean of 3,671,577 reads, with an average length of 14,989 bp. The mean number of mapped contigs was 233, with an N50 of 150,349 bp. Genomic GC content showed little variation around an average value of 57%. The genomes ranged in size between 5,376,511 and 5,833,838 bp (with an average size of 5,591,208 bp), which is in accordance with the genome of K. pneumoniae (20).
Using Rast software, 5,285 coding sequences (CDSs) were identified in the genomes encoding several subsystems (591) involved in essential metabolism of the bacteria, including Gram-negative cell wall components (82 CDSs), RNA (61 CDSs), and DNA protein metabolism (256 CDSs and 142 CDSs, respectively). Accessory features were also present, such as those conferring resistance to antibiotics and toxic compounds, e.g., beta-lactams, fosfomycin, fluoroquinolones, arsenic, copper, mercury, etc.
In silico multilocus sequence typing (MLST) revealed 5 distinct sequence types (ST): ST01, ST147, ST11, ST15, and ST392 (Table 2).
TABLE 2.
Genomic features and epidemiological relatedness of the 13 Klebsiella pneumonia isolates reported in the present study
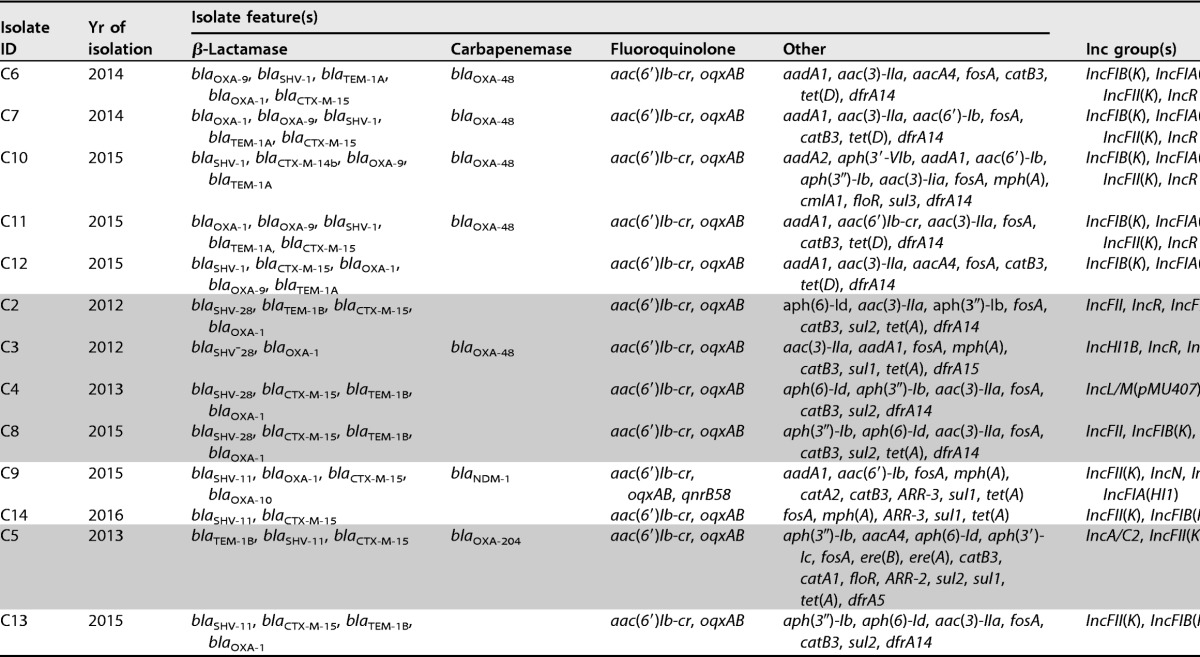
A wide variety of antibiotic resistance genes against aminoglycoside, β-lactam, fluoroquinolone, macrolide, chloramphenicol, sulfonamide, fosfomycin, phenicol, tetracycline, co-trimoxazole, and rifampin antibiotics were found in the CoRKp genomes (Table 2). Diverse drug resistance genes were found within the same ST type, highlighting the considerable genomic plasticity of clinical K. pneumoniae isolates. The resistome was in accordance with the antibiotic susceptibility testing results (Table 1). Indeed, all isolates harbored at least one beta-lactam resistance gene (blaCTX-M-15 or blaTEM-1B), and seven isolates carried carbapenemases (blaOXA-48 [n = 5], blaOXA-204 [n = 1], and blaNDM-1 [n = 1]). blaOXA-48 was identified in clones ST101 and ST15, blaOXA-204 in ST147, and blaNDM in ST11 (Table 2). The 13 isolates belonged to 5 STs. ST101, ST15, ST11, and ST147 have been described worldwide, as well as in Tunisia, and are associated with the carbapenemases OXA-48, KPC, OXA-204, and NDM and the extended-spectrum beta-lactamase (ESBL) CTX-M-15 (21–25). The single ST392 strain has never been described in Tunisia but was sporadically observed in three distinct countries related to KPC-2 in China (26), to NDM in Colombia (27), and to OXA-244 in Spain (28). In our study, ST-2 was associated with CTX-M-15 but not any carbapenemase.
All of the strains carried a fosfomycin resistance gene (fosA) and plasmid-mediated quinolone resistance (PMQR) genes: the bifunctional aminoglycoside resistance and fluoroquinolone-inactivating determinants aac(6′)lb-cr and qnrB58, present only in clone ST11 that harbored the blaNDM-1 gene.
Analysis of chromosomally encoded resistance genes revealed the presence of the quinolone efflux pumps oqxAB, the multidrug efflux pump, i.e., acrB and mexCD, and mutations in the quinolone resistance genes (gyrase and topoisomerases). Several mutations were found in the quinolone resistance-determining region (QRDR) of gyrA, leading to amino acid changes F83Y, A87N and F83I, and A87D in five isolates of ST101 and one of ST147 and of ST392, respectively. In addition, in all isolates, a single gyrB mutation leading to amino acid change V189A and several other amino acid changes were detected in parC and parE, likely being polymorphisms. Mutations in the QRDR, i.e., gyrA, gyrB, parC, and parE, are likely at the origin of the high level of fluoroquinolone resistance, but several PMQR genes [qnrB and aac(6′)lb-cr] and chromosomally located fluoroquinolone resistance genes have also been described (oqxAB, acrB, and mexCD).
Colistin resistance mechanisms.
Plasmid-mediated colistin resistance genes mcr-1 to mcr-5 were not identified in the isolates of the present study (14). In order to uncover the molecular mechanisms sustaining the colistin resistance, we have targeted the role of alterations of the MgrB protein as well as those of the two-component regulatory systems (TCRS) phoPQ and pmrAB, crrB, a gene recently described to be responsible for the colistin resistant phenotype in K. pneumoniae (9), and the operon arnBCADTEF.
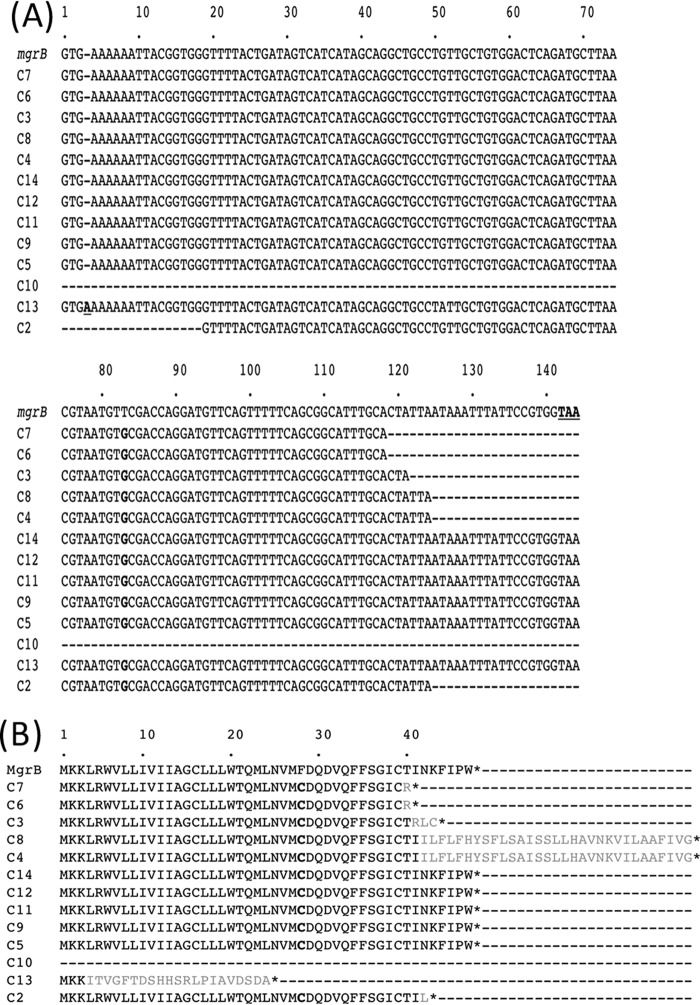
Sequence analysis revealed that all of the mgrB genes were mutated, resulting in amino acid sequence changes of the small regulatory transmembrane protein MgrB (Fig. 1A and B and Table 3): (i) missense and frameshift mutations resulting in amino acid changes or premature ending of the protein, respectively; (ii) complete deletions of the mgrB gene in isolate C10; (iii) and insertional inactivation caused by insertion sequences (IS) belonging to several IS families (IS1382-like, IS5-like, and IS1-like families). Due to these insertions, the length and the sequence of the resulting MgrB protein was either reduced (40, 42, and 43 amino acids [aa]) or increased (72 aa) compared to that of the wild-type (WT) MgrB protein (47 aa). For C13, and due to the frameshift, the resulting MgrB protein is only 24 aa long, with only 3 aa corresponding to the wild-type sequence. The T83G amino acid substitution was identified in all MgrB proteins derived from all five unrelated ST types.
FIG 1.
DNA and deduced protein sequence alignments of mgrB alleles. (A) DNA sequence alignments for wild-type mgrB gene from colistin-susceptible K. pneumoniae strain NH53 (accession number KF852760) (8) and mgrB genes from the present study. The insertion of an A in C13 is boldfaced and underlined. The substitutions (T83G) are boldfaced. The dashes indicate deletions of 20 to 26 amino acids or a complete deletion in isolate C10. (B) Alignment of wild-type MgrB from colistin-susceptible K. pneumoniae strain NH53 (accession number KF852760) (8) and MgrB from the isolates of the present study. Changes in amino acid sequences compared to the WT MgrB protein sequence are indicated in boldface, and additional amino acids compared to the sequence of WT MgrB protein are indicated in gray. MgrB protein of the C13 isolate corresponds to a short nonfunctional protein due to the frameshift mutation in its corresponding gene. MgrB proteins of C4, C8, C3, C7, C6, and C2 corresponded of nonfunctional proteins (truncated or longer) due to insertional inactivation by diverse IS within their corresponding mgrB gene. For isolate C10, the complete mgrB allele was deleted (indicated by dashes).
TABLE 3.
Mutations reported in the present study within genes involved in colistin resistance (mgrB, pmrAB, phoPQ, eptA)
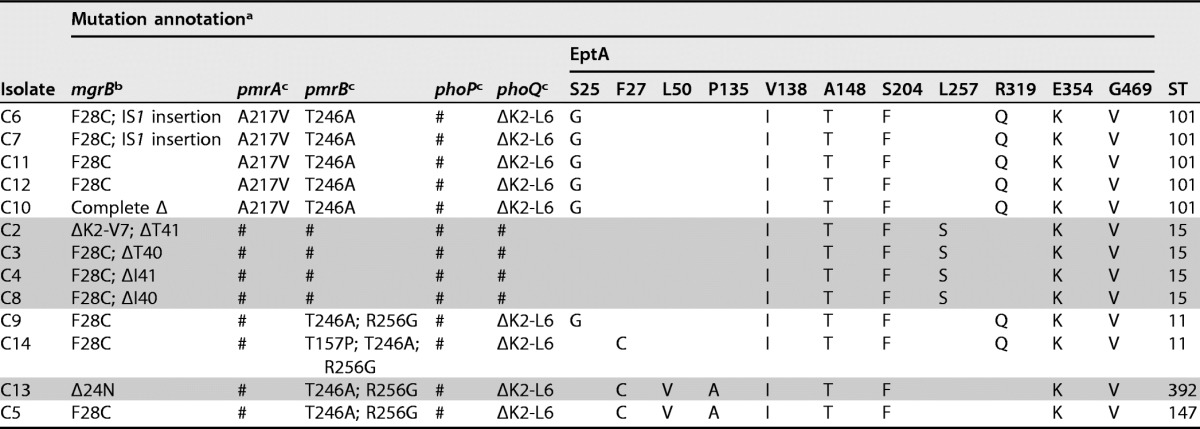
a #, identical to the wild-type strain; Δ, deletion.
b Aligned with Klebsiella pneumoniae strain NH53 MgrB (GenBank accession no. KF852760.1) (8).
Our results showed that colistin resistance in K. pneumoniae was directly linked to mutations identified within the mgrB gene, as previously suggested (29), and that insertional inactivation mediated by mobile DNA, even though described in the literature as the most prevalent mechanism involved in colistin resistance in K. pneumoniae (5, 8, 29–32), is not the only mechanism leading to MgrB inactivation. Indeed, complementation with a WT mgrB allele of T83G-MgrB strains (C6, C11, and C14) resulted in restoration of colistin susceptibility, suggesting that the observed mutations are at the origin of colistin resistance (Table 4).
TABLE 4.
Complementation assays
| Isolate | MgrB mutation | Colistin MICs (μg/ml) |
|---|---|---|
| C6 | F28C; IS1 insertion | 8 |
| C6 + pMgrB | 0.5 | |
| C11 | F28C | 16 |
| C11 + pMgrB | 2 | |
| C14 | F28C | 16 |
| C14 + pMgrB | 1 |
In order to see whether the inserted ISs were already present in the chromosome of the different K. pneumoniae isolates, blast searches were performed against the entire contigs. Several copies of IS5-like elements were present in all isolates, while IS1382-like and IS1-like were present only in the respective isolates where they have been found, suggesting an insertion from a plasmid.
Sequence analysis of pmrA, pmrB, phoP, phoQ, operon arnBCADTEF, and eptA (pmrC) genes known to be involved in LPS synthesis revealed several polymorphisms in nucleotide sequences compared to the wild-type sequence of Klebsiella pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession no. CP000647) (Table 3). These polymorphisms were linked to the different ST types. Within strains belonging to ST15, no variation compared to wild-type protein sequences was detected for pmrAB or phoPQ, but 6 amino acid changes were observed in all EptA proteins (V138I, A148T, S204F, L257S, E354K, and G469V). The crrB gene was identical to that of the WT sequence of K. pneumoniae (9).
PmrA variant A217V, PmrB variant T246A, and PhoPQ variant ΔK2-L6 were found in all isolates belonging to ST101 and did not correspond to any mutations previously identified as contributing to colistin resistance.
For strains of ST11, ST147, and ST392, PmrA and PhoP were identical to the wild-type proteins of K. pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession no. CP000647), while a novel PmrB variant (T246A, R256G) and PhoQ (ΔK2-L6) were detected. Finally, a PmrB variant, T157P, initially described by Jayol et al. (33) as contributing to colistin resistance, has been described in isolate C14. Whether these changes influence colistin resistance is not known, as some have never been described and likely correspond to polymorphisms. In order to study the real contribution of these novel variants in colistin resistance, further experiments are necessary, such as complementation assays with wild-type alleles and transcriptional studies.
Plasmids and conjugation.
All of the K. pneumoniae isolates possessed several plasmids varying in size, as revealed by Kieser plasmid analysis (34) (see Fig. S2 in the supplemental material). Most of the β-lactam resistance-carrying plasmids could be transferred by conjugation (data not shown) (35), but no colistin resistance determinant could be transferred. PCR experiments confirmed the presence of the blaCTX-M, blaOXA-48-like, and blaNDM genes in the transconjugants, but we were not able to reconstruct these plasmids using our Illumina-generated sequencing data. Nevertheless, using in silico analysis, the presence of several Inc groups, namely, FII, FII(K), FIB(K), FIA(HI1), HI1B, L/M, A/C2, N, and R types (36), were found in the K. pneumoniae isolates (Table 2). Some of these Inc groups have been clearly incriminated in horizontal gene transfer of plasmids encoding antibiotic resistance determinants, such as IncL/M and the blaOXA-48 gene, Inc FII and blaCTX-M-15, and IncR and blaNDM (37, 38).
Conclusions.
This work corresponds, to the best our knowledge, to the first genomic investigation of colistin resistance in K. pneumoniae from a clinical setting from Tunisia, and it showed emergence of colistin resistance as early as 2012, with a subsequent polyclonal dissemination. Here, we were able to determine the major role of MgrB in colistin resistance in K. pneumoniae. The genomic data of our study may serve for future comparative genomic and molecular epidemiological studies aiming at deciphering the genomic basis of the emergence of resistance mechanisms and the dynamics of the spread of MDR K. pneumoniae strains.
MATERIALS AND METHODS
Bacterial strains and clinical data.
The Sahloul university hospital is a 629-bed teaching hospital with specialty services, including an operating room and five intensive care units. CoRKp strains were from the Sahloul university teaching hospital (629 beds) and were identified through review of the clinical microbiology laboratory database from January 2012 to March 2016. The clinical data were retrieved from the medical records of each patient, and the data on colistin use during the hospitalization were obtained from pharmacy records.
Identification and antimicrobial susceptibility testing.
Thirteen CoRKp clinical isolates were retained from 12 patients and included in this study. Isolates were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) as previously described (39). Routine antibiograms were determined by disk diffusion on Mueller-Hinton agar plates at 37°C. The MICs of β-lactams, including ertapenem, imipenem, meropenem, and doripenem, in addition to tigecycline, were determined by Etest (bioMérieux-France).
For strains showing resistance to colistin (CR), MICs were determined by broth microdilution (BMD) in cation-adjusted Mueller-Hinton broth (MHB-CA) and interpreted according to updated CLSI guidelines (http://www.captodayonline.com/new-clsi-editions-m100-s24-em100/). Colistin sulfate (obtained from Sigma-Aldrich [St. Louis, MO]) was tested with Tween 80 (with a final concentration of 0.002%) over a range of dilutions from 0.125 to 128 μg/ml. All results were interpreted according to updated EUCAST breakpoint tables for interpretation of MICs and zone diameters of 2015, version 5.0 (www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf).
Plasmid identification and transformation.
Plasmid DNA of all carbapenem-resistant K. pneumoniae (CR-KP) isolates was extracted using the Kieser method as previously described (34). Plasmids of ca. 154, 66, 48, and 7 kb of Escherichia coli NCTC 50192 were used as plasmid size markers. Plasmid DNA was analyzed by agarose gel electrophoresis.
Detection of carbapenemase activity and β-lactamase genes.
The carbapenemase activity was searched for using the Carba NP test as previously described (30). Class A ESBLs and carbapenem-hydrolyzing resistance genes were sought by PCR as previously described (35).
Whole-genome sequencing.
The genomic DNA was extracted from overnight cultures in LB agar (Bio-Rad, Marnes-la-Coquette, France) using an UltraClean microbial DNA isolation kit (Mo Bio Laboratories). Genomic DNA quantification was performed using a Qubit fluorometer (Life Technologies, Carlsbad, CA) and adjusted to 0.2 ng/μl. Library preparation was performed using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) and Illumina MiSeq 2000 sequencer with v3 chemistry using 2- by 150-bp paired-end reads for sequencing. The 150-bp paired-end reads were de novo assembled using CLC Genomic Workbench with a minimum contig length of 200 bp.
Antibiotic resistance-related genes were predicted using the ResFinder database, version 2.1 (40), with the following parameters: “all databases” was used for antimicrobial configuration, type of reads was “assembled genomes/contigs,” and we used thresholds of 98% identity and 80% coverage between sequences. This data set of resistance genes was complemented with BLASTp searches against the ARDB (Antibiotic Resistance Genes Database), version 1.1 (41),the using “resistance gene complete” database with 40% identity and E value of 0.0001. Multilocus sequence typing (MLST) was performed in silico according to the K. pneumoniae MLST database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). For the detection and typing of the presence of known replicon types of plasmids in the isolates studied, Plasmid Finder and pMLST (http://cge.cbs.dtu.dk/services/PlasmidFinder/) were used (40).
MgrB complementation assay.
The wild-type mgrB allele was cloned from K. pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession no. CP000647) into cloning vector pCRscriptCm in the same orientation as the plasmid-borne promoter pLAC. The resulting plasmid was named pMgrB. Three representative isolates (C6, C11, and C14) were prepared to be electrocompetent, as previously described (35), and subsequently pMgrB was electroporated into them (35). Electroporants were selected on 100 μg/ml chloramphenicol containing trypticase soy agar plates.
Accession number(s).
The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession numbers MSYN00000000 to MSZF00000000.
ACKNOWLEDGMENTS
We thank the many dedicated staff members who helped with data collection and technical issues: the members of the French National Reference Center for Antibiotic Resistance, Carbapenemase-Producing Enterobacteriaceae, and the staff of the Laboratory of Clinical Microbiology, University Hospital of Sahloul, Sousse, Tunisia. We thank in particular Kalboussi Nesrine (Pharmacy Department of University Hospital of Sahloul, Sousse, Tunisia).
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (EA7361), Université Paris Sud. R.A.B. and T.N. are members of the Laboratory of Excellence LERMIT, supported by a grant from ANR (ANR-10-LABX-33). In addition, this study was partially supported by the University Hospital of Sahloul, Sousse, Tunisia.
We have no conflicts of interest to declare.
REFERENCES
- 1.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falagas ME, Michalopoulos A. 2006. Polymyxins: old antibiotics are back. Lancet 367:633–634. doi: 10.1016/S0140-6736(06)68241-X. [DOI] [PubMed] [Google Scholar]
- 4.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Rojas R, McConnell MJ, Jimenez-Mejias ME, Dominguez-Herrera J, Fernandez-Cuenca F, Pachon J. 2013. Colistin resistance in a clinical Acinetobacter baumannii strain appearing after colistin treatment: effect on virulence and bacterial fitness. Antimicrob Agents Chemother 57:4587–4589. doi: 10.1128/AAC.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother 58:4399–4403. doi: 10.1128/AAC.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain J-M. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H-Y, Chen Y-F, Peng H-L. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 14.Jeannot K, Bolard A, Plesiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49(5):526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Mezghani Maalej S, Rekik Meziou M, Mahjoubi F, Hammami A. 2012. Epidemiological study of Enterobacteriaceae resistance to colistin in Sfax (Tunisia). Médecine Mal Infect 42:256–263. [DOI] [PubMed] [Google Scholar]
- 16.Kempf I, Fleury MA, Drider D, Bruneau M, Sanders P, Chauvin C, Madec J-Y, Jouy E. 2013. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int J Antimicrob Agents 42:379–383. doi: 10.1016/j.ijantimicag.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Halaby T, al Naiemi N, Kluytmans J, van der Palen J, Vandenbroucke-Grauls CMJE. 2013. Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob Agents Chemother 57:3224–3229. doi: 10.1128/AAC.02634-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J-Y, Choi M-J, Choi HJ, Ko KS. 2015. Preservation of acquired colistin resistance in gram-negative bacteria. Antimicrob Agents Chemother 60:609–612. doi: 10.1128/AAC.01574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olaitan AO, Morand S, Rolain J-M. 2016. Emergence of colistin-resistant bacteria in humans without colistin usage: a new worry and cause for vigilance. Int J Antimicrob Agents 47:1–3. doi: 10.1016/j.ijantimicag.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou H-Y. 2012. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol 194:1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben Tanfous F, Alonso CA, Achour W, Ruiz-Ripa L, Torres C, Ben Hassen A. 2016. First description of KPC-2-producing Escherichia coli and ST15 OXA-48-positive Klebsiella pneumoniae in Tunisia. Microb Drug Resist 23:365–375. [DOI] [PubMed] [Google Scholar]
- 22.Grami R, Mansour W, Ben Haj Khalifa A, Dahmen S, Chatre P, Haenni M, Aouni M, Madec J-Y. 2016. Emergence of ST147 Klebsiella pneumoniae producing OXA-204 carbapenemase in a university hospital, Tunisia. Microb Drug Resist 22:137–140. doi: 10.1089/mdr.2014.0278. [DOI] [PubMed] [Google Scholar]
- 23.Charfi K, Mansour W, Ben Haj Khalifa A, Mastouri M, Aouni M, Mammeri H. 2015. Emergence of OXA-204 β-lactamase in Tunisia. Diagn Microbiol Infect Dis 82:314–317. doi: 10.1016/j.diagmicrobio.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Mansour W, Grami R, Ben Haj Khalifa A, Dahmen S, Châtre P, Haenni M, Aouni M, Madec J-Y. 2015. Dissemination of multidrug-resistant blaCTX-M-15/IncFIIk plasmids in Klebsiella pneumoniae isolates from hospital- and community-acquired human infections in Tunisia. Diagn Microbiol Infect Dis 83:298–304. doi: 10.1016/j.diagmicrobio.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Messaoudi A, Haenni M, Mansour W, Saras E, Bel Haj Khalifa A, Chaouch C, Naija W, Boujâafar N, Bouallègue O, Madec J-Y. 2017. ST147 NDM-1-producing Klebsiella pneumoniae spread in two Tunisian hospitals. J Antimicrob Chemother 72:315–316. doi: 10.1093/jac/dkw401. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D, Han L. 2013. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect 19:E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 27.Rojas LJ, Wright MS, De La Cadena E, Motoa G, Hujer KM, Villegas MV, Adams MD, Bonomo RA. 2016. Initial assessment of the molecular epidemiology of bla NDM-1 in Colombia. Antimicrob Agents Chemother 60:4346–4350. doi: 10.1128/AAC.03072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oteo J, Hernandez JM, Espasa M, Fleites A, Saez D, Bautista V, Perez-Vazquez M, Fernandez-Garcia MD, Delgado-Iribarren A, Sanchez-Romero I, Garcia-Picazo L, Miguel MD, Solis S, Aznar E, Trujillo G, Mediavilla C, Fontanals D, Rojo S, Vindel A, Campos J. 2013. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother 68:317–321. doi: 10.1093/jac/dks383. [DOI] [PubMed] [Google Scholar]
- 29.Poirel L, Jayol A, Bontron S, Villegas M-V, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 30.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Y-H, Lin T-L, Pan Y-J, Wang Y-P, Lin Y-T, Wang J-T. 2015. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother 59:2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Camacho E, Gomez-Gil R, Tobes R, Manrique M, Lorenzo M, Galvan B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoner FM, Alvarez-Tejado M, Garcillan-Barcia MP, De la Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 33.Jayol A, Poirel L, Brink A, Villegas M-V, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 35.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Fernandez A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier P-E, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis Off Publ Infect Dis Soc Am 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 40.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Pop M. 2009. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]