Abstract
Aspergillus fumigatus is the most prevalent filamentous fungal pathogen of humans, causing either severe allergic bronchopulmonary aspergillosis or often fatal invasive pulmonary aspergillosis (IPA) in individuals with hyper- or hypo-immune deficiencies, respectively. Disease is primarily initiated upon the inhalation of the ubiquitous airborne conidia—the initial inoculum produced by A. fumigatus—which are complete developmental units with an ability to exploit diverse environments, ranging from agricultural composts to animal lungs. Upon infection, conidia initially rely on their own metabolic processes for survival in the host’s lungs, a nutritionally limiting environment. One such nutritional limitation is the availability of aromatic amino acids (AAAs) as animals lack the enzymes to synthesize tryptophan (Trp) and phenylalanine and only produce tyrosine from dietary phenylalanine. However, A. fumigatus produces all three AAAs through the shikimate–chorismate pathway, where they play a critical role in fungal growth and development and in yielding many downstream metabolites. The downstream metabolites of Trp in A. fumigatus include the immunomodulatory kynurenine derived from indoleamine 2,3-dioxygenase (IDO) and toxins such as fumiquinazolines, gliotoxin, and fumitremorgins. Host IDO activity and/or host/microbe-derived kynurenines are increasingly correlated with many Aspergillus diseases including IPA and infections of chronic granulomatous disease patients. In this review, we will describe the potential metabolic cross talk between the host and the pathogen, specifically focusing on Trp metabolism, the implications for therapeutics, and the recent studies on the coevolution of host and microbe IDO activation in regulating inflammation, while controlling infection.
Keywords: Aspergillus fumigatus, tryptophan metabolism, IDO, kynurenines, toxins, non-ribosomal peptides, peripheral tolerance, Th17 cells
Introduction
Aspergillus fumigatus is a saprophytic fungus that has a worldwide distribution. The asexual spores (called conidia) are ubiquitous and individuals inhale hundreds of spores daily. While most inhaled conidia are cleared by individuals with a healthy immune system, A. fumigatus can act as an opportunistic human pathogen in individuals with altered immune functions. Disease presentation can vary on the status of the host’s immune system; A. fumigatus can cause allergic bronchopulmonary aspergillosis, a severe allergenic response, in the hyper-immune, or the fatal invasive growth invasive pulmonary aspergillosis (IPA) in the hypo-immune, or in individuals with other susceptibilities such as patients unable to mount the necessary oxidative defenses such as in individuals with chronic granulomatous disease (CGD) (1).
The manifestation of disease is dependent not only on the host’s immune status but also fungal factors including strain heterogeneity (2). A. fumigatus growth in the mammalian lung, following survival of resident pulmonary defenses, requires the fungus to adapt to a hypoxic and nutritionally scarce environment. Aspergillus mutants unable to synthesize primary metabolites necessary for growth are generally impaired in virulence. For example, deletion of cpcA, a transcription factor that globally modulates amino acid biosynthesis in the fungus led to a less virulent phenotype in a murine model of IPA (3). Additional studies have shown that mutants in sulfur utilization (4), uracil/uridine synthesis (5), zinc uptake, iron acquisition, and many more (6, 7) are also decreased in virulence.
To complicate disease progression further, there is an alarming rise in antifungal resistance strains of A. fumigatus (8, 9). Therefore, an understanding of A. fumigatus and host metabolic pathways is important in identifying nutrient limitations. One critical metabolic pathway is the biosynthesis of aromatic amino acids [AAAs, tryptophan (Trp), phenylalanine, and tyrosine], which are required not only for growth of A. fumigatus but are also precursors for several toxins (Table 1). The host relies on dietary sources for all AAAs while A. fumigatus synthesizes all three. However, the host and A. fumigatus both possess AAA catabolic enzymes. In particular, one key enzyme important in immune homeostasis is indoleamine 2,3-dioxygenase (IDO), which converts Trp to kynurenine and related metabolites in both organisms. Historically, host IDOs activity has been described as an effective antimicrobial control for pathogens that are natural Trp auxotrophs such as Staphylococcus aureus, Chlamydia spp., and Toxoplasma gondii, presumably by Trp starvation (10). However, A. fumigatus can synthesize its own Trp and thus the Trp starvation may not be an effective pathogen control for those microbes able to synthesize their own Trp pools. Although, IDOs also play additional roles in host defenses through modifying kynurenine levels and subsequent cytokine responses as described below. In this review, we will summarize the recent studies describing the anabolic and the catabolic pathways of Trp metabolism, the implications for therapeutics, and the host–pathogen interaction.
Table 1.
Aspergillus fumigatus non-ribosomal peptides containing aromatic amino acids (AAAs) in their peptide structure.
| Toxin | AAAa | Interaction with hostb-i |
|---|---|---|
| Fumigaclavine Cb | Tryptophan (Trp) | Downregulation of Th1 cytokines including TNF-α, IL-1β, and IL-17A. Induction of host cell apoptosis Decrease activation of caspase-1 |
| Fumiquinazoline Cc | Anthranilate Trp |
Cytotoxic |
| Fumisoquind | Tyrosine | Nothing reported |
| Fumitremorgine Tryprostatin Af Verruculogeng |
Trp | Neurotoxic and produces tremors in mice Tryprostatin causes the inhibition of microtubule assembly Verruculogen causes modification of the electrophysiological properties of HNEC |
| Gliotoxinh | Phenylalanine | Virulent in a steroid murine model of IPA Induction of host cell apoptosis and causes epithelial cell damage Inhibition of phagocytosis and oxidative bursts |
| Hexadehydroastechromei | Trp | Overexpression resulted in a significantly higher virulence in a neutropenic murine model of IPAb |
IPA, invasive pulmonary aspergillosis; HNEC, human nasal epithelial cells.
aOnly the AAA is designated. Other amino acids are also in the structure of these metabolites.
Trp Synthesis and Potential Therapeutic Targeting
Chorismic acid derived from the shikimic acid pathway is a key intermediate in producing Trp, phenylalanine (Phe), and tyrosine (Tyr) in microorganisms including A. fumigatus (Figure 1). Trp and Phe are classified as essential amino acids, whereas mammals acquire them from diet, whereas Tyr is synthesized via the hydroxylation of Phe (23, 24). The absence of the AAA biosynthetic enzymes and the low bioavailability of Trp in humans makes the Trp biosynthetic enzymes attractive targets for antifungals (25).
Figure 1.
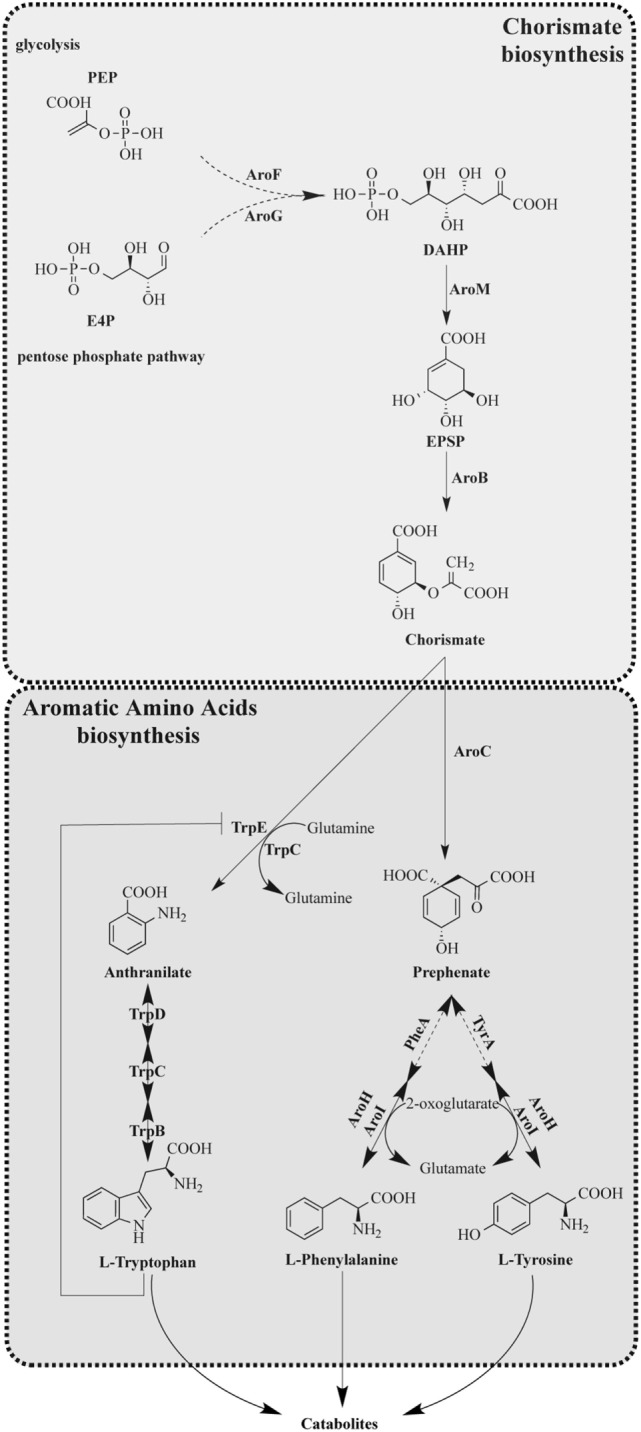
Tryptophan anabolism of Aspergillus fumigatus. [Modified from that of Wang et al. (28)]. Solid arrows indicate characterized reaction as being present in A. fumigatus with product detected. Dashed arrows indicate uncharacterized reactions; however, putative orthologs are present in A. fumigatus.
Fungal Trp Anabolic Pathway
Aromatic amino acid synthesis has been extensively studied in Saccharomyces cerevisiae and provides the basis for the functional characterization of orthologous enzymes in filamentous fungi (23, 24, 26–28). The shikimic acid pathway is a 7-enzymatic step reaction that initiates with two substrates, phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P), which are intermediates of glycolysis and pentose phosphate pathways, respectively (29) (Figure 1). The first step of the shikimic acid pathway is catalyzed by 3-deoxy-d-arabinoheptulosonate 7-phosphate (DAHP) synthase to convert PEP and E4P to DAHP. In S. cerevisiae and A. nidulans, there are two DAHP synthases, Aro3 and Aro4, which are allosterically inhibited by phenylalanine and tyrosine, respectively (24). Steps 2–6 in filamentous fungi such as A. nidulans and A. fumigatus are completed by the pentafunctional enzyme AroM, or Aro1 in the model organism S. cerevisiae (30). The shikimate pathway culminates in the production of chorismic acid synthesized by the enzyme chorismate synthase (AroB) from 5-enolpyruvylshikimate-3-phosphate (EPSP) (31).
The synthesis of Trp from chorismate is initiated by an anthranilate synthase (AS), which converts chorismate to anthranilate, followed by three enzymatic steps as presented in Figure 1 with the respective functions outlined in Table 2. AS(s) in S. cerevisiae have been characterized, and it consists of two subunits: anthranilate synthase subunit I (AAS-I), which binds chorismate and is subject to feedback inhibition by Trp and anthranilate synthase subunit II (AAS-II) which is a glutamine amidotransferase (32). The A. nidulans trpC, an AAS-II encoding gene, was characterized in 1977 (33) and found exchangeable with an A. fumigatus trpC in 1994 (34). Wang et al. (28) recently characterized trpE, the AAS-I encoding gene in A. fumigatus. Wang et al. (28) explored the functions of two putative AAS-Is termed trpE and icsA by creating null mutants. The deletion of trpE led to a Trp auxotrophic strain, whereas the deletion of icsA did not. To ensure that icsA did not serve a redundant role for Trp synthesis, the group overexpressed icsA in a trpE deletion and showed that the overexpression of icsA does not reverse the Trp auxotrophy concluding that TrpE is the only AS in A. fumigatus. Interestingly, the group showed that IcsA is an active enzyme in A. fumigatus as the precursor-chorismate pool is altered in the absence or overproduction of IcsA; however, the product is not known (28). Sasse et al. also confirmed that deletion of trpE (they termed trpA) results in an A. fumigatus Trp auxotrophy (31).
Table 2.
Aspergillus fumigatus tryptophan (Trp) metabolism genes and putative protein function.
| Protein | Ortholog in Saccharomyces cerevisiae | Gene name in A. fumigatus | Protein functiona | Ortholog in mammals |
|---|---|---|---|---|
| Chorismate biosynthesis | ||||
| AroF | Aro3 | Afu1g02110 | DAHP synthase | – |
| AroG | Aro4 | Afu7g04070 | ||
| AroM | Aro1 | Afu1g13740 | EPSP synthase | – |
| AroB | Aro2 | Afu1g06940 | Chorismate synthase | – |
| Aromatic amino acid (AAA) biosynthesis | ||||
| TrpE | Trp2 | Afu6g12580 | Anthranilate synthase | – |
| TrpC | Trp3 | Afu1g13090 | ||
| TrpD | Trp4 | Afu4g11980 | Anthranilate phosphoribosyltransferase | – |
| TrpC | Trp1 | Afu1g13090 | Phosphoribosylanthranilate isomerase | – |
| TrpB | Trp5 | Afu2g13250 | Trp synthase | – |
| IcsA | – | Afu6g12110 | Isochorismate synthase | – |
| AroC | Aro7 | Afu5g13130 | Chorismate mutase | – |
| PheA | Pha2 | Afu5g05690 | Prephenate dehydratase | – |
| TyrA | Tyr1 | Afu2g10450 | Prephenate dehydrogenase | – |
| AroH | Aro8 | Afu2g13630 | AAA transaminase | – |
| AroI | Aro9 | Afu5g02290 | ||
| Trp degradation | ||||
| IdoA | Bna2 | Afu3g14250 | Indoleamine 2,3-dioxygenases | IDO1 |
| IdoB | Bna2 | Afu4g09830 | IDO2 | |
| IdoC | Bna2 | Afu7g02010 | TDO | |
| FmdS | Bna7 | Afu1g09960 | Kynurenine formamidase | AFMID |
| Bna4 | Bna4 | Afu6g07340 | Kynureninase | |
| Bna5 | Bna5 | Afu4g09840 | KYNU | |
| AroH | Aro8 | Afu2g13630 | AAA transaminase | LAAO |
| AroI | Aro9 | Afu5g02290 | ||
| AadA | – | Afu3g02240 | Trp carboxylase | AADC |
| MaoN | – | Afu3g00100 | Monoamine oxidase | MAOA |
| AldA | Ald4 | Afu2g00720 | Aldehyde dehydrogenase | ALDH |
| Ald5 | Afu7g01000 | |||
| Afu6g11430 | ||||
aPrediction of protein function based on AspGD (http://www.aspgd.org/) and KEGG (http://www.genome.jp/kegg/kegg2.html).
DAHP synthase, 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase; EPSP synthase, enolpyruvylshikimate-3-phosphate synthase; AFMID, arylformamidase; KYNU, kynureninase; LAAO, l-amino-acid oxidase; AADC, aromatic-l-amino-acid decarboxylase, MAOA, monoamine oxidase; ALDH2, aldehyde dehydrogenase family.
The Shikimate Pathway As Potential Antifungal Targets
Currently, there are four major classes of antifungals: azoles and amphotericin B targeting ergosterol, 5-fluorocytosine targeting DNA synthesis, and echinocandins targeting cell wall synthesis. These antifungals either exhibit high toxicity to the mammalian cell (particularly amphotericin B and 5-fluorocytosine) or lose efficacy due to the emergence of drug resistant strains (azoles and echinocandins) (9). With A. fumigatus being a eukaryotic pathogen and sharing many proteins with mammalian hosts, there are limitations to developing effective and safe antifungals and therefore a great need for treatments that are fungal specific. Since Trp is a human-essential amino acid and the enzymes in the biosynthesis are fungal specific, several studies have suggested utilizing and finding drugs to target the enzymes of this pathway (35–38).
Targeting essential amino acid pathways have already shown potential for new classes of antifungals. Several groups have explored inhibitors of genes or enzymes involved in methionine biosynthesis. Azoxybacillin, a compound isolated from B. cereus targets methionine biosynthesis by interfering with expression of homoserine transacetylase and sulfite reductase encoding genes (39–41). Whereas azoxybacillin displayed a broad spectrum antifungal activity in vitro, in vivo activity was low possibly due to bioavailability in the host (41). R1-331, a natural product from Streptomyces akiyoshiensis, is an effective inhibitor of homoserine dehydrogenase involved in both methionine and threonine biosynthesis (42, 43). Yamaguchi et al. show that R1-331 was active against medically important fungi such as Candida albicans and Cryptoccocus neoformans and proved to be effective in the treatment of systemic murine candidiasis (42, 44).
Compounds targeting AAA pathways are limited with the most famous being the herbicide Roundup, where the active ingredient glyphosate inhibits EPSP synthase, one of the first enzymes initiating the shikimate pathway (45) (Figure 1). Glyphosate has shown to inhibit growth of several fungi including Candida maltose (46), Pneumocystis (47), and Cryptococcus neoformans where glyphosate delayed fungal melanization in vitro and in vivo and prolonged mice survival during infection (48). Another inhibitor of AAA pathway is a fluorinated anthranilate moiety, 6-FABA, which targets the TrpE enzyme and showed bactericidal activity when used on Mycobacterium tuberculosis (49). The studies of these inhibitors suggest that the Trp biosynthetic pathway could be fruitful in future antifungal drug design.
The value of AAA pathways as drug targets is supported by the findings that AAA auxotrophic mutants are less virulent in animal infection models. Sasse et al. explored the possibility of these pathways as potential drug target by testing the virulence of several AAA auxotrophic mutants in a murine IPA model (31). This study demonstrated that AroM (Figure 1) was required for A. fumigatus viability. The group also constructed a conditional AroB repression strain that was attenuated virulence. Both a Trp auxotroph (TrpE mutant) and Tyr/Phe auxotroph (AroC mutant) were severely attenuated in virulence for pulmonary infection. Interestingly, the group also unveiled a putative difference in AAA distribution within the host by conducting a systemic infection showing that in a bloodstream infection the TrpE and the AroC mutants although less virulent, can establish some infection (31). Taken together, these results suggest that inhibitors of AAA biosynthetic pathways can potentially be used against A. fumigatus as a standalone treatment in a localized pulmonary infection or as an additive treatment in a systemic infection. The result of the bloodstream infection observed by Sasse et al. also suggests that there are mechanisms for the fungus to sense Trp in its environment and utilize it. In S. cerevisiae, the Trp specific permease, Tat2, is required for Trp uptake in yeast (50) and its closest homolog in A. fumigatus (Afu7g04290) is upregulated during fungal encounters with neutrophils (51) and dendritic cells (52). In A. nidulans, the G-protein coupled receptor (Gpr) H may be responsible for sensing Trp and glucose and GprH is conserved in A. fumigatus (53, 54). Perhaps, for a systemic infection, inhibition of specific permeases or development of a GprH antagonist would be useful in reducing infections by Aspergillus.
Catabolic Trp Metabolites in Fungi and Host
Although the anabolic Trp pathway is absent in mammals, the common catabolic pathways exist in the mammalian host with possession of the same enzymes as A. fumigatus (Figure 2). Through the expression of Trp degradation enzymes, immune cells are both controlling inflammation and combating microbial infection. In addition to Trp degradation pathways conserved with animals, A. fumigatus can also direct Trp pools to secondary metabolites that may impact host health and response (Table 1).
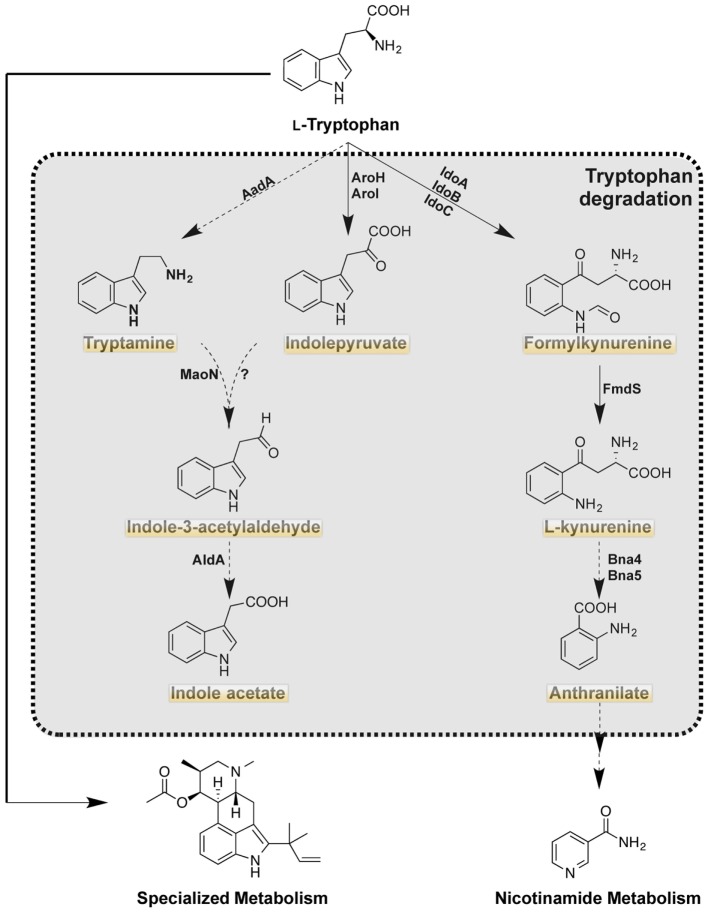
Figure 2.
Tryptophan catabolism of Aspergillus fumigatus. Highlighted products are putatively produced in the mammalian host as the orthologous enzymes are present in the host (http://www.genome.jp/kegg). Solid arrows indicate characterized reaction as being present in A. fumigatus with product detected. Dashed arrows indicate uncharacterized reactions, however putative orthologs are present in A. fumigatus.
Fungal Trp Catabolism
There are three putative pathways (Figure 2; Table 2) for the degradation of Trp in A. fumigatus. The kynurenine branch is catalyzed by IDOs that convert Trp into formylkynurenine. In A. fumigatus, there are 3 putative ido genes: idoA, idoB, and idoC, the orthologs of A. oryzae idoα, idoβ, and idoγ, respectively (55). Enzymatic studies of Aspergillus oryzae IDO enzymes suggest two of the three enzymes, IDOα, and IDOβ, may participate in Trp degradation as they have a higher affinity of its substrate. However the recent study by Wang et al., suggested IDOb might be the more dominant enzyme than IDOa as determined by gene expression of A. fumigatus grown on Trp amended media (28). Additionally, idoC gene expression was slightly induced by the addition of Trp, but the authors note that IDOc had a closer relationship to bacterial IDOs than to fungal IDOs (28, 55). In S. cerevisiae, formylkynurenine is further oxidized to the immunomodulatory product kynurenine by a kynurenine formamidase denoted as Bna7, which has been described in A. nidulans and is predicted to be involved in NAD (+), biosynthesis (56). The kynurenine branch in fungi is involved in the de novo biosynthesis of NAD (+), a coenzyme that is required for oxidation-reduction reactions (57).
Tryptophan can also be metabolized via the indole pyruvate pathway, initiated through the transamination of Trp by aromatic aminotransferases (termed Aro8 and Aro9 in S. cerevisiae). These aromatic aminotransferases are also involved in the synthesis of Phe and Tyr in S. cerevisiae, and their orthologs are present in A. fumigatus (28). In S. cerevisiae, the deletion of both Aro8 and Aro9 results in Phe and Tyr auxotrophies (24, 58, 59). In Candida spp., where filamentation and pigment production play a role in virulence, the products of these enzymes have been described to influence both phenotypes. The deletion of aro8 in C. glabrata results in a reduced pigment production and leads to an increased sensitivity to hydrogen peroxide (27). The aro8 and aro9 mutants of C. albicans results in a decreased conversion of Trp to indole acetaldehyde, which is formed via decarboxylation of indole pyruvate. Filamentation of C. albicans increased with the exposure to indole acetaldehyde (60).
Indole acetaldehyde can also be produced via the third putative product of Trp degradation: tryptamine. Tryptamine is most famously known as the active compound in psilocybin and for its similarity to serotonins (61). Although the production of tryptamine has yet to be described in A. fumigatus, the downstream product of the tryptamine and indole pyruvate pathway, indole acetic acid has been described in several Aspergillus spp. including A. fumigatus (62, 63). Downstream metabolism of kynurenine, indole pyruvate, and tryptamine has not been explored further, but A. fumigatus does possess putative enzymes for the re-synthesis of anthranilate, the precursor to Trp (as denoted in Figures 1 and 2).
AAA Incorporation into Aspergillus Toxins
Many filamentous fungi, including A. fumigatus, produce bioactive small molecules that can have detrimental impacts on human health. Subsets of these toxins are small peptides synthesized by non-ribosomal peptide synthetases (NRPS). Several pathogenic Aspergillus species synthesize AAA derived peptides including gliotoxin (Phe and serine) (64), fumiquinazoline (Trp, Anthranilate, and Alanine) (13), fumigaclavine (Trp) (65), fumitremorgin (Trp and Proline) (66), hexadehydroastechrome (Trp and Alanine) (21), fumisoquin (Tyr, Serine, and Methionine) (14), DPP-IV inhibitor WYK-1 (Trp, Tyr, and Leucine) (67), cyclopiazonic acid (Trp) (68) and benzomalvin (Phe and Anthranilate) (69). Table 1 summarizes the known AAA derived secondary metabolites of A. fumigatus and their effect on the host.
Although, fumiquinazolines have yet to be assessed for virulence in an animal model, they are known to have cytotoxic properties (13). Fumigaclavines have been described to have immunosuppressive properties in several studies including the suppression of antifungal cytokines such as TNFα, IL-17, and IFN-γ (12). Fumitremorgin, verruculogen, and tryprostatin—all related products of the fumitremorgin pathway—induce tremorgenic activity in mice and act on the central nervous system (15, 66, 70, 71). Mutants in the hexadehydroastechrome pathway (21) and gliotoxin (20, 21) pathways have altered virulence in murine IPA models. Decreased virulence of the gliP mutant (GliP is the NRPS required for gliotoxin synthesis) is dependent on host immune status [reviewed in Ref. (7)]. Overexpression of hasA encoding the hexadehydroastechrome transcription factor and thus leading to increased hexadehydroastechrome production was more virulent than wild type A. fumigatus in a neutropenic model of IPA (21). Although the exact mechanism underlying the increased virulence of the OE::hasA strain is unknown, iron homeostasis and cross talk between metabolic pathways may contribute to the increased virulence of OE::hasA (22). These studies highlight the potential contribution of AAA derived toxins in virulence of A. fumigatus.
Host Trp Catabolism via IDO
The function of host IDO during mammalian infection was originally thought to center on the anti-proliferative effects of pathogenic microorganism via deprivation of Trp exerted by the host. IDO is up-regulated by interferon gamma (IFNγ) and depletes Trp (the least abundant essential amino acid) to inhibit pathogen expansion (72, 73), as demonstrated in the constraint of chlamydial growth (74). Numerous studies have since implicated IDO activity as important in fungal infections and have reported the relative outcomes of IDO expression on disease progression (Table 3). Accumulating data continues to support that IDO participates in the host–pathogen interaction in human epithelial cells; therefore, the co-evolution of host and microbe Trp metabolism has been investigated (75). The current consensus is that IDO activation is pivotal in regulating inflammatory processes directly via Trp depletion and indirectly via the IDO-mediated release of Trp catabolic secondary metabolites (namely, kynurenines).
Table 3.
Summary of studies where IDO enzyme activity was found to be implicated in fungal infections.
| Fungus | Mouse model | IDO | Outcome | Reference |
|---|---|---|---|---|
| Candida albicans | Gastrointestinal inf. | Upregulation | Protection of the host against fungus | (76) |
| C. albicans | Gastrointestinal inf. | Upregulation | Protection of the host against fungus | (77) |
| C. albicans | Gastrointestinal inf. | Upregulation | Protection of the host against fungus | (78) |
| C. albicans | In vitro | Downregulation | Not done | (79) |
| Aspergillus fumigatus | Keratitis | Upregulation | Protection of the host against fungus | (80) |
| A. fumigatus | Allergy | Overexpression | Protection of the host against fungus | (81) |
| A. fumigatus | IPA | Upregulation | Protection of the host against fungus | (82) |
| A. fumigatus | IPA in chronic granulomatous disease mice | Upregulation | Protection of the host against fungus | (83) |
| A. fumigatus | IPA in CF mice | Upregulation | Protection of the host against fungus | (84) |
| Paracoccidioides brasiliensis | Pulmonary infection | Upregulation | Protection of the host against fungus | (85) |
| P. brasiliensis | Pulmonary infection | Upregulation | Protection of the host against fungus | (86) |
| P. brasiliensis | Pulmonary infection | Upregulation | Protection of the host against fungus | (87) |
| Histoplasma capsulatum | Pulmonary infection | Downregulation | Protection of the host against fungus | (88) |
| H. capsulatum | Pulmonary infection | Upregulation | Protection of the host against fungus | (89) |
Dietary Trp is catabolized by two different IDO protein isoforms, IDO1 and IDO2 that are expressed by immune cells, and TDO (Trp 2,3-dioxygenase) that is mainly expressed in the liver. Cells involved in the innate processes of the anti-microbial defense, such as dendritic cells (DCs), neutrophils, and macrophages express IDO1 upon microbial encounter mainly via toll-like receptor stimulation. How fungi specifically induce IDO expression is not known; however, induction by other pathogens is associated with pathogen associated molecular patterns, including lipopolysaccharides and CpG oligodeoxynucleotides (78, 90–92), underlining a role for kynurenine metabolism in microbial-induced inflammatory processes.
IDO-Mediated Tolerance: Impacts on Antimicrobial Responses
Evolutionary studies have shown that the host immune defense against microbes is characterized by three different mechanisms: avoidance, resistance, and tolerance (93). Modules of immunity provide resistance to limit pathogen burden and tolerance and host damage caused by the immune reaction per se. However, the inflammatory reaction, although largely considered beneficial for its antimicrobial functions, may also contribute to pathogenicity. Thus, rescue from infection pathology may not only depend on microbial colonization (and inactivation of resistance mechanisms) but also on the resolution of tissue inflammatory pathology through tolerogenic responses to pathogens (94).
Studies using a mouse model of mucosal or invasive C. albicans infection found that systemic inhibition of IDO in vivo reduced gastrointestinal inflammation and unexpectedly, elevated the levels of fungal colonization compared to control mice (77). Notably, tolerogenic responses toward C. albicans were abrogated when IDO was antagonized in vivo, as shown in various models of inflammatory disorders (95, 96). As with C. albicans, IDO and kynurenine production during A. fumigatus infection contributes to fungal pathogen eradication and the regulation of an unacceptable level of tissue damage (97). Indeed, IDO can increase kynurenine host levels to induce adaptive Treg expansion while limiting Th17 polarization (83, 96). In this context, the Th17 pathway, which downregulates Trp catabolism, may instead favor pathology and better explain the paradoxical correlation between fungal infection and chronic inflammation (98).
Another example of this paradox was demonstrated in the context of CGD, in which an NADPH oxidase defect results in reduced host production of antimicrobial ROS and extreme susceptibility to Aspergillus infections (1, 83). Although human studies have excluded a role for IDO in CGD (99), further investigations into the IDO pathway are warranted as such studies have failed to demonstrate functional IDO activity at sites of chronic inflammation. Measures of IDO functional activity during IPA have, however, been made in mouse models, and implicate defective IDO activity as a key mediator of chronic inflammation in CGD (83). An exaggerated Th17 pulmonary response was associated with reduced fungal clearance in mouse models of CGD that develop IPA. Here, reduced IDO function was directly related to NADPH/ROS deficiency, as ROS is essential for IDO catalytic activity in mammals (100). ROS deficiency as a result of reduced NADPH function, significantly enhanced IL-17 inflammation and fungal germination in the lung, thus further reducing neutrophil-mediated antimicrobial activities (83).
Since regulation of homeostasis and peripheral tolerance are extremely important in prevention of invasive Aspergillosis or allergy to Aspergillus antigens (97, 101), the role of IDO has been extensively studied in this model of fungal infection (81, 82, 102). These studies highlight the induction of the IDO metabolic pathway at different site of fungal colonization as keratinocytes or lung as well as the important anti-inflammatory activity of IDO in the tissue microenvironment (80–82, 84).
Aryl Hydrocarbon Receptor (AhR) Activation by IDO Metabolites in Mammals: Biological Consequences
The AhR is a ligand-activated transcription factor first identified for its role during embryonic development and induction of xenobiotic metabolizing enzymes as a response to environmental toxins, such as dioxin (103). More recently, AhR has been shown to play a critical role in immunity by acting as an immune modulator during fungal infection (85). The connection between the AhR and the immune response lies in part in the endogenous Ahr ligands, which comprise many Trp metabolites, including kynurenine (104). Microbial Trp-derived metabolites can activate the AhR, leading to adjustments in the immune response that may hinder disease development (105). The AhR–IDO axis has been recently demonstrated in fungal infection, highlighting a role for IDO-derived metabolites to trigger AhR target genes (85, 106). For example, one AhR target gene, Il22, has been widely studied in the context of fungal/microbial infections (105, 107–109). AhR activation by IDO metabolites can also mediate the expansion of peripheral Treg with anti-inflammatory properties. Using IDO-deficient mice, increased pulmonary disease caused by Paracoccidioides brasiliensis was associated with decreased Treg expansion and reduced AhR protein expression (85). In murine models of IPA, distinct Treg populations capable of mediating anti-inflammatory effects expand following exposure to Aspergillus conidia (97). Late in infection, tolerogenic adaptive Treg (with shared phenotypic identity with the Treg controlling autoimmune diseases or diabetes) produce IL-10 and TGFβ, inhibit Th2 cells, and prevent an allergic reaction to Aspergillus (97).
Conclusion
The interplay of Trp metabolic pathways and fungal/host interactions is intriguing with many unanswered questions of the exact nature of crosstalk of shared metabolites and consequences of activation of Trp degradative pathways. In Aspergillus infections in particular, not only does the pathogen synthesize and degrade Trp but it also can utilize this amino acid (and its precursor anthranilate or the other two AAAs Tyr and Phe) to yield several potentially damaging toxins (Table 1). Also, as both host and Aspergillus share catabolic IDO pathways, it is unclear which organism may generate immunomodulatory Trp degradation products and if they respond to each other’s products (e.g., kynurenine). Development of A. fumigatus IDO mutants for investigation of disease development could yield valuable information on this front. The research on the expression host IDOs exhibit the importance of an extremely coordinated immune response to mount the right inflammatory response for clearance of spores. However, while an increased IDO expression in the host can control inflammation, the suppression of the IDO-regulated antifungal Th17 responses can favor fungal growth. In this context, it will be critical to explore the entire IDO-mediated innate response, including the specific T cell regulatory subsets affected by IDO activity.
Although the Trp catabolic pathway is shared between host and pathogen, the anabolic pathway is unique to A. fumigatus. The antifungals currently used in treatment are becoming increasingly ineffective with emerging drug resistant strains; therefore, drugs targeting essential fungal specific pathways are needed. A proposal for fungal treatment has been highlighted through the studies of essential amino acids. As AAA mutants are auxotrophs and decreased in virulence (28, 31), investigations of drugs targeting these pathway enzymes could lead to novel antifungal compounds. Indeed, a few compounds have exhibited some efficacy in targeting Trp metabolic pathways in M. tuberculosis and several fungi and efforts to identify additional inhibitors are warranted.
Author Contributions
TC, TZ, and NK have made a substantial, direct, and intellectual contribution to the work and LR provided intellectual insights to the revision. All authors have approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank support from Dalai Lama Trust MSN178745 to TC, The Italian Grant “Programma per Giovani Ricercatori - Rita Levi Montalcini 2013” to TZ, and NIH 5R01AI065728-10 to NK.
References
- 1.Segal BH, Romani LR. Invasive aspergillosis in chronic granulomatous disease. Med Mycol (2009) 47:S282–90. 10.1080/13693780902736620 [DOI] [PubMed] [Google Scholar]
- 2.Keller NP. Heterogeneity confounds establishment of “a” model microbial strain. MBio (2017) 8:e00135–17. 10.1128/mBio.00135-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krappmann S, Bignell EM, Reichard U, Rogers T, Haynes K, Braus GH. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol Microbiol (2004) 52:785–99. 10.1111/j.1365-2958.2004.04015.x [DOI] [PubMed] [Google Scholar]
- 4.Amich J, Dumig M, O’keeffe G, Binder J, Doyle S, Beilhack A, et al. Exploration of sulfur assimilation of Aspergillus fumigatus reveals biosynthesis of sulfur-containing amino acids as a virulence determinant. Infect Immun (2016) 84:917–29. 10.1128/IAI.01124-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Enfert C, Diaquin M, Delit A, Wuscher N, Debeaupuis JP, Huerre M, et al. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect Immun (1996) 64:4401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes JC. Aspergillus fumigatus: growth and virulence. Med Mycol (2006) 44(Suppl 1):S77–81. 10.1080/13693780600779419 [DOI] [PubMed] [Google Scholar]
- 7.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev (2009) 22:447–65. 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanglard D. Emerging threats in antifungal-resistant fungal pathogens. Front Med (2016) 3:11. 10.3389/fmed.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis (2017) 17(12):e383–92. 10.1016/S1473-3099(17)30316-X [DOI] [PubMed] [Google Scholar]
- 10.Mackenzie CR, Heseler K, Muller A, Daubener W. Role of indoleamine 2,3-dioxygenase in antimicrobial defence and immuno-regulation: tryptophan depletion versus production of toxic kynurenines. Curr Drug Metab (2007) 8:237–44. 10.2174/138920007780362518 [DOI] [PubMed] [Google Scholar]
- 11.Zhu YX, Yao LY, Jiao RH, Lu YH, Tan RX. Enhanced production of fumigaclavine C in liquid culture of Aspergillus fumigatus under a two-stage process. Bioresour Technol (2014) 152:162–8. 10.1016/j.biortech.2013.10.089 [DOI] [PubMed] [Google Scholar]
- 12.Guo W, Hu S, Elgehama A, Shao F, Ren R, Liu W, et al. Fumigaclavine C ameliorates dextran sulfate sodium-induced murine experimental colitis via NLRP3 inflammasome inhibition. J Pharmacol Sci (2015) 129:101–6. 10.1016/j.jphs.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Lim FY, Ames B, Walsh CT, Keller NP. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell Microbiol (2014) 16:1267–83. 10.1111/cmi.12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baccile JA, Spraker JE, Le HH, Brandenburger E, Gomez C, Bok JW, et al. Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat Chem Biol (2016) 12:419–24. 10.1038/nchembio.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki S, Kikkawa K, Yamazaki M. Abnormal behavioral effects elicited by a neurotropic mycotoxin, fumitremorgin A in mice. J Pharmacobiodyn (1984) 7:935–42. 10.1248/bpb1978.7.935 [DOI] [PubMed] [Google Scholar]
- 16.Frisvad JC, Larsen TO. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section fumigati. Front Microbiol (2015) 6:1485. 10.3389/fmicb.2015.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui T, Kondoh M, Cui CB, Mayumi T, Osada H. Tryprostatin A, a specific and novel inhibitor of microtubule assembly. Biochem J (1998) 333(Pt 3):543–8. 10.1042/bj3330543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khoufache K, Puel O, Loiseau N, Delaforge M, Rivollet D, Coste A, et al. Verruculogen associated with Aspergillus fumigatus hyphae and conidia modifies the electrophysiological properties of human nasal epithelial cells. BMC Microbiol (2007) 7:5. 10.1186/1471-2180-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Holte D, Zoller J, Umemiya S, Simke LR, Baran PS. Total synthesis of verruculogen and fumitremorgin A enabled by ligand-controlled C-H borylation. J Am Chem Soc (2015) 137:10160–3. 10.1021/jacs.5b07154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, Galvez EM, et al. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot Cell (2007) 6:1562–9. 10.1128/EC.00141-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J Am Chem Soc (2013) 135:2064–7. 10.1021/ja311145n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiemann P, Lechner BE, Baccile JA, Velk TA, Yin WB, Bok JW, et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front Microbiol (2014) 5:530. 10.3389/fmicb.2014.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingens F. Regulation of aromatic amino acid biosynthesis in microorganisms. Acta Microbiol Acad Sci Hung (1976) 23:161–6. [PubMed] [Google Scholar]
- 24.Braus GH. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev (1991) 55:349–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagliamonte A, Gessa R, Biggio G, Vargiu L, Gessa GL. Daily changes of free serum tryptophan in humans. Life Sci (1974) 14:349–54. 10.1016/0024-3205(74)90065-4 [DOI] [PubMed] [Google Scholar]
- 26.Pereira SA, Livi GP. Aromatic amino-acid biosynthesis in Candida albicans: identification of the ARO4 gene encoding a second DAHP synthase. Curr Genet (1996) 29:441–5. 10.1007/BF02221512 [DOI] [PubMed] [Google Scholar]
- 27.Brunke S, Seider K, Almeida RS, Heyken A, Fleck CB, Brock M, et al. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol Microbiol (2010) 76:25–47. 10.1111/j.1365-2958.2010.07052.x [DOI] [PubMed] [Google Scholar]
- 28.Wang PM, Choera T, Wiemann P, Pisithkul T, Amador-Noguez D, Keller NP. TrpE feedback mutants reveal roadblocks and conduits toward increasing secondary metabolism in Aspergillus fumigatus. Fungal Genet Biol (2016) 89:102–13. 10.1016/j.fgb.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins AR, Lamb HK, Moore JD, Charles IG, Roberts CF. The pre-chorismate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects. J Gen Microbiol (1993) 139:2891–9. 10.1099/00221287-139-12-2891 [DOI] [PubMed] [Google Scholar]
- 30.Duncan K, Edwards RM, Coggins JR. The pentafunctional AroM enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J (1987) 246:375–86. 10.1042/bj2460375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasse A, Hamer SN, Amich J, Binder J, Krappmann S. Mutant characterization and in vivo conditional repression identify aromatic amino acid biosynthesis to be essential for Aspergillus fumigatus virulence. Virulence (2016) 7:56–62. 10.1080/21505594.2015.1109766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graf R, Mehmann B, Braus GH. Analysis of feedback-resistant anthranilate synthases from Saccharomyces cerevisiae. J Bacteriol (1993) 175:1061–8. 10.1128/jb.175.4.1061-1068.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kafer E. The anthranilate synthetase enzyme complex and the trifunctional trpC gene of Aspergillus. Can J Genet Cytol (1977) 19:723–38. 10.1139/g77-079 [DOI] [PubMed] [Google Scholar]
- 34.Borgia PT, Dodge CL, Eagleton LE, Adams TH. Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans. FEMS Microbiol Lett (1994) 122:227–31. 10.1111/j.1574-6968.1994.tb07172.x [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, et al. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog (2007) 3:e24. 10.1371/journal.ppat.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco B, Prado V, Lence E, Otero JM, Garcia-Doval C, Van Raaij MJ, et al. Mycobacterium tuberculosis shikimate kinase inhibitors: design and simulation studies of the catalytic turnover. J Am Chem Soc (2013) 135:12366–76. 10.1021/ja405853p [DOI] [PubMed] [Google Scholar]
- 37.Ianiri G, Idnurm A. Essential gene discovery in the basidiomycete Cryptococcus neoformans for antifungal drug target prioritization. MBio (2015) 6:e02334–14. 10.1128/mBio.02334-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaltdorf M, Srivastava M, Gupta SK, Liang C, Binder J, Dietl AM, et al. Systematic identification of anti-fungal drug targets by a metabolic network approach. Front Mol Biosci (2016) 3:22. 10.3389/fmolb.2016.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aoki Y, Kondoh M, Nakamura M, Fujii T, Yamazaki T, Shimada H, et al. A new methionine antagonist that has antifungal activity: mode of action. J Antibiot (Tokyo) (1994) 47:909–16. 10.7164/antibiotics.47.909 [DOI] [PubMed] [Google Scholar]
- 40.Aoki Y, Kamiyama T, Fujii T, Yamamoto M, Ohwada J, Arisawa M. Design of an antifungal methionine inhibitor not antagonized by methionine. Biol Pharm Bull (1995) 18:1267–71. 10.1248/bpb.18.1267 [DOI] [PubMed] [Google Scholar]
- 41.Aoki Y, Yamamoto M, Hosseini-Mazinani SM, Koshikawa N, Sugimoto K, Arisawa M. Antifungal azoxybacilin exhibits activity by inhibiting gene expression of sulfite reductase. Antimicrob Agents Chemother (1996) 40:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi M, Yamaki H, Shinoda T, Tago Y, Suzuki H, Nishimura T, et al. The mode of antifungal action of (S)2-amino-4-oxo-5-hydroxypentanoic acid, RI-331. J Antibiot (Tokyo) (1990) 43:411–6. 10.7164/antibiotics.43.411 [DOI] [PubMed] [Google Scholar]
- 43.Jacques SL, Ejim LJ, Wright GD. Homoserine dehydrogenase from Saccharomyces cerevisiae: kinetic mechanism and stereochemistry of hydride transfer. Biochim Biophys Acta (2001) 1544:42–54. 10.1016/S0167-4838(00)00202-8 [DOI] [PubMed] [Google Scholar]
- 44.Yamaki H, Yamaguchi M, Nishimura T, Shinoda T, Yamaguchi H. Unique mechanism of action of an antifungal antibiotic RI-331. Drugs Exp Clin Res (1988) 14:467–72. [PubMed] [Google Scholar]
- 45.Tan S, Evans R, Singh B. Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids (2006) 30:195–204. 10.1007/s00726-005-0254-1 [DOI] [PubMed] [Google Scholar]
- 46.Bode R, Melo C, Birnbaum D. Mode of action of glyphosate in Candida maltosa. Arch Microbiol (1984) 140:83–5. 10.1007/BF00409776 [DOI] [PubMed] [Google Scholar]
- 47.Chin K, Wyder MA, Kaneshiro ES. Glyphosate reduces organism viability and inhibits growth in vitro of Pneumocystis. J Eukaryot Microbiol (1999) 46:139S–41S. [PubMed] [Google Scholar]
- 48.Fernandes JD, Martho K, Tofik V, Vallim MA, Pascon RC. The role of amino acid permease and tryptophan biosynthesis in Cryptococcus neoformans survival. PLoS One (2015) 10:e0132369. 10.1371/journal.pone.0132369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdel-Rahman HM, El-Koussi NA, Hassan HY. Fluorinated 1,2,4-triazolo[1,5-a]pyrimidine-6-carboxylic acid derivatives as antimycobacterial agents. Arch Pharm (Weinheim) (2009) 342:94–9. 10.1002/ardp.200800113 [DOI] [PubMed] [Google Scholar]
- 50.Forsberg H, Gilstring CF, Zargari A, Martinez P, Ljungdahl PO. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol Microbiol (2001) 42:215–28. 10.1046/j.1365-2958.2001.02627.x [DOI] [PubMed] [Google Scholar]
- 51.Sugui JA, Kim HS, Zarember KA, Chang YC, Gallin JI, Nierman WC, et al. Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS One (2008) 3:e2655. 10.1371/journal.pone.0002655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morton CO, Varga JJ, Hornbach A, Mezger M, Sennefelder H, Kneitz S, et al. The temporal dynamics of differential gene expression in Aspergillus fumigatus interacting with human immature dendritic cells in vitro. PLoS One (2011) 6:e16016. 10.1371/journal.pone.0016016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grice CM, Bertuzzi M, Bignell EM. Receptor-mediated signaling in Aspergillus fumigatus. Front Microbiol (2013) 4:26. 10.3389/fmicb.2013.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown NA, Dos Reis TF, Ries LN, Caldana C, Mah JH, Yu JH, et al. G-protein coupled receptor-mediated nutrient sensing and developmental control in Aspergillus nidulans. Mol Microbiol (2015) 98:420–39. 10.1111/mmi.13135 [DOI] [PubMed] [Google Scholar]
- 55.Yuasa HJ, Ball HJ. The evolution of three types of indoleamine 2,3 dioxygenases in fungi with distinct molecular and biochemical characteristics. Gene (2012) 504:64–74. 10.1016/j.gene.2012.04.082 [DOI] [PubMed] [Google Scholar]
- 56.Fraser JA, Davis MA, Hynes MJ. The formamidase gene of Aspergillus nidulans: regulation by nitrogen metabolite repression and transcriptional interference by an overlapping upstream gene. Genetics (2001) 157:119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohashi K, Kawai S, Murata K. Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD+ biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot Cell (2013) 12:648–53. 10.1128/EC.00339-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urrestarazu A, Vissers S, Iraqui I, Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet (1998) 257:230–7. 10.1007/s004380050643 [DOI] [PubMed] [Google Scholar]
- 59.Bulfer SL, Brunzelle JS, Trievel RC. Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase. Protein Sci (2013) 22:1417–24. 10.1002/pro.2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao RP, Hunter A, Kashpur O, Normanly J. Aberrant synthesis of indole-3-acetic acid in Saccharomyces cerevisiae triggers morphogenic transition, a virulence trait of pathogenic fungi. Genetics (2010) 185:211–20. 10.1534/genetics.109.112854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tylš F, Páleníček T, Horáček J. Psilocybin – summary of knowledge and new perspectives. Eur Neuropsychopharmacol (2014) 24:342–56. 10.1016/j.euroneuro.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 62.Bilkay IS, Karakoç S, Aksoz N. Indole-3-acetic acid and gibberellic acid production in Aspergillus niger. Turk J Biol (2010) 34:313–8. 10.3906/biy-0812-15 [DOI] [Google Scholar]
- 63.Pattaeva M, Bakhtiyor R. Growth and phytohormones production by thermophilic Aspergillus fumigatus 2 and thermotolerant Aspergillus terreus 8 strains in salt stress. Br J Appl Sci Technol (2015) 8:305–12. 10.9734/BJAST/2015/12292 [DOI] [Google Scholar]
- 64.Macdonald JC, Slater GP. Biosynthesis of gliotoxin and mycelianamide. Can J Biochem (1975) 53:475–8. 10.1139/o75-066 [DOI] [PubMed] [Google Scholar]
- 65.Mulinti P, Allen NA, Coyle CM, Gravelat FN, Sheppard DC, Panaccione DG. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr Microbiol (2014) 68:1–5. 10.1007/s00284-013-0434-2 [DOI] [PubMed] [Google Scholar]
- 66.Yamazaki M, Suzuki S. Toxicology of tremorgenic mycotoxins, fumitremorgin A and B. Dev Toxicol Environ Sci (1986) 12:273–82. [PubMed] [Google Scholar]
- 67.Imamura K, Tsuyama Y, Hirata T, Shiraishi S, Sakamoto K, Yamada O, et al. Identification of a gene involved in the synthesis of a dipeptidyl peptidase IV inhibitor in Aspergillus oryzae. Appl Environ Microbiol (2012) 78:6996–7002. 10.1128/AEM.01770-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry (2009) 48:8746–57. 10.1021/bi901123r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol (2017) 13:895–901. 10.1038/nchembio.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamazaki M, Fujimoto H, Kawasaki T. Chemistry of tremorogenic metabolites. Fumitremorgin A from Aspergillus fumigatus. Chem Pharm Bull (Tokyo) (1980) 28:245–54. 10.1248/cpb.28.245 [DOI] [PubMed] [Google Scholar]
- 71.Cole RJ. Fungal tremorogens. Prikl Biokhim Mikrobiol (1993) 29:44–50. [PubMed] [Google Scholar]
- 72.Colangelo EJ. Cervicocranium and the aviator’s protective helmet. Aviat Space Environ Med (1975) 46:1263–4. [PubMed] [Google Scholar]
- 73.Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L. Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes Infect (2009) 11:133–41. 10.1016/j.micinf.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 74.Thomas SM, Garrity LF, Brandt CR, Schobert CS, Feng GS, Taylor MW, et al. IFN-gamma-mediated antimicrobial response. Indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. J Immunol (1993) 150:5529–34. [PubMed] [Google Scholar]
- 75.Aldajani WA, Salazar F, Sewell HF, Knox A, Ghaemmaghami AM. Expression and regulation of immune-modulatory enzyme indoleamine 2,3-dioxygenase (IDO) by human airway epithelial cells and its effect on T cell activation. Oncotarget (2016) 7:57606–17. 10.18632/oncotarget.11586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Luca A, Montagnoli C, Zelante T, Bonifazi P, Bozza S, Moretti S, et al. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol (2007) 179:5999–6008. 10.4049/jimmunol.179.9.5999 [DOI] [PubMed] [Google Scholar]
- 77.Bozza S, Fallarino F, Pitzurra L, Zelante T, Montagnoli C, Bellocchio S, et al. A crucial role for tryptophan catabolism at the host/Candida albicans interface. J Immunol (2005) 174:2910–8. 10.4049/jimmunol.174.5.2910 [DOI] [PubMed] [Google Scholar]
- 78.Bonifazi P, Zelante T, D’angelo C, De Luca A, Moretti S, Bozza S, et al. Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol (2009) 2:362–74. 10.1038/mi.2009.17 [DOI] [PubMed] [Google Scholar]
- 79.Cheng SC, Van De Veerdonk F, Smeekens S, Joosten LA, Van Der Meer JW, Kullberg BJ, et al. Candida albicans dampens host defense by downregulating IL-17 production. J Immunol (2010) 185:2450–7. 10.4049/jimmunol.1000756 [DOI] [PubMed] [Google Scholar]
- 80.Jiang N, Zhao G, Lin J, Hu L, Che C, Li C, et al. Indoleamine 2,3-dioxygenase is involved in the inflammation response of corneal epithelial cells to Aspergillus fumigatus infections. PLoS One (2015) 10:e0137423. 10.1371/journal.pone.0137423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paveglio SA, Allard J, Foster Hodgkins SR, Ather JL, Bevelander M, Campbell JM, et al. Airway epithelial indoleamine 2,3-dioxygenase inhibits CD4+ T cells during Aspergillus fumigatus antigen exposure. Am J Respir Cell Mol Biol (2011) 44:11–23. 10.1165/rcmb.2009-0167OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Luca A, Bozza S, Zelante T, Zagarella S, D’angelo C, Perruccio K, et al. Non-hematopoietic cells contribute to protective tolerance to Aspergillus fumigatus via a TRIF pathway converging on IDO. Cell Mol Immunol (2010) 7:459–70. 10.1038/cmi.2010.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Romani L, Fallarino F, De Luca A, Montagnoli C, D’angelo C, Zelante T, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature (2008) 451:211–5. 10.1038/nature06471 [DOI] [PubMed] [Google Scholar]
- 84.Iannitti RG, Carvalho A, Cunha C, De Luca A, Giovannini G, Casagrande A, et al. Th17/Treg imbalance in murine cystic fibrosis is linked to indoleamine 2,3-dioxygenase deficiency but corrected by kynurenines. Am J Respir Crit Care Med (2013) 187:609–20. 10.1164/rccm.201207-1346OC [DOI] [PubMed] [Google Scholar]
- 85.Araujo EF, Feriotti C, Galdino NAL, Preite NW, Calich VLG, Loures FV. The IDO-AhR axis controls Th17/Treg immunity in a pulmonary model of fungal infection. Front Immunol (2017) 8:880. 10.3389/fimmu.2017.00880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Araujo EF, Medeiros DH, Galdino NA, Condino-Neto A, Calich VL, Loures FV. Tolerogenic plasmacytoid dendritic cells control Paracoccidioides brasiliensis infection by inducting regulatory T cells in an IDO-dependent manner. PLoS Pathog (2016) 12:e1006115. 10.1371/journal.ppat.1006115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Araujo EF, Loures FV, Bazan SB, Feriotti C, Pina A, Schanoski AS, et al. Indoleamine 2,3-dioxygenase controls fungal loads and immunity in paracoccidioidomicosis but is more important to susceptible than resistant hosts. PLoS Negl Trop Dis (2014) 8:e3330. 10.1371/journal.pntd.0003330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hage CA, Horan DJ, Durkin M, Connolly P, Desta Z, Skaar TC, et al. Histoplasma capsulatum preferentially induces IDO in the lung. Med Mycol (2013) 51:270–9. 10.3109/13693786.2012.710857 [DOI] [PubMed] [Google Scholar]
- 89.George MM, Subramanian Vignesh K, Landero Figueroa JA, Caruso JA, Deepe GS, Jr. Zinc induces dendritic cell tolerogenic phenotype and skews regulatory T cell-Th17 balance. J Immunol (2016) 197:1864–76. 10.4049/jimmunol.1600410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorski JP, Howard JB. Effect of methylamine on the structure and function of the fourth component of human complement, C4. J Biol Chem (1980) 255:10025–8. [PubMed] [Google Scholar]
- 91.Jung ID, Lee C-M, Jeong Y-I, Lee JS, Park WS, Han J, et al. Differential regulation of indoleamine 2,3-dioxygenase by lipopolysaccharide and interferon gamma in murine bone marrow derived dendritic cells. FEBS Lett (2007) 581:1449–56. 10.1016/j.febslet.2007.02.073 [DOI] [PubMed] [Google Scholar]
- 92.Fallarino F, Volpi C, Zelante T, Vacca C, Calvitti M, Fioretti MC, et al. IDO mediates TLR9-driven protection from experimental autoimmune diabetes. J Immunol (2009) 183:6303–12. 10.4049/jimmunol.0901577 [DOI] [PubMed] [Google Scholar]
- 93.Rivas FV, Chervonsky AV, Medzhitov R. ART and immunology. Trends Immunol (2014) 35:451. 10.1016/j.it.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 94.Soares MP, Gozzelino R, Weis S. Tissue damage control in disease tolerance. Trends Immunol (2014) 35:483–94. 10.1016/j.it.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 95.Romani L. Cell mediated immunity to fungi: a reassessment. Med Mycol (2008) 46:515–29. 10.1080/13693780801971450 [DOI] [PubMed] [Google Scholar]
- 96.Zelante T, Bozza S, De Luca A, D’angelo C, Bonifazi P, Moretti S, et al. Th17 cells in the setting of Aspergillus infection and pathology. Med Mycol (2009) 47(Suppl 1):S162–9. 10.1080/13693780802140766 [DOI] [PubMed] [Google Scholar]
- 97.Montagnoli C, Fallarino F, Gaziano R, Bozza S, Bellocchio S, Zelante T, et al. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J Immunol (2006) 176:1712–23. 10.4049/jimmunol.176.3.1712 [DOI] [PubMed] [Google Scholar]
- 98.De Luca A, Pariano M, Cellini B, Costantini C, Villella VR, Jose SS, et al. The IL-17F/IL-17RC axis promotes respiratory allergy in the proximal airways. Cell Rep (2017) 20:1667–80. 10.1016/j.celrep.2017.07.063 [DOI] [PubMed] [Google Scholar]
- 99.De Ravin SS, Zarember KA, Long-Priel D, Chan KC, Fox SD, Gallin JI, et al. Tryptophan/kynurenine metabolism in human leukocytes is independent of superoxide and is fully maintained in chronic granulomatous disease. Blood (2010) 116:1755–60. 10.1182/blood-2009-07-233734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Macchiarulo A, Camaioni E, Nuti R, Pellicciari R. Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease. Amino Acids (2009) 37:219–29. 10.1007/s00726-008-0137-3 [DOI] [PubMed] [Google Scholar]
- 101.Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol (2007) 37:2695–706. 10.1002/eji.200737409 [DOI] [PubMed] [Google Scholar]
- 102.Jiang N, Zhao GQ, Lin J, Hu LT, Che CY, Li C, et al. Expression of indoleamine 2,3-dioxygenase in a murine model of Aspergillus fumigatus keratitis. Int J Ophthalmol (2016) 9:491–6. 10.18240/ijo.2016.04.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mackowiak B, Wang H. Mechanisms of xenobiotic receptor activation: direct vs. indirect. Biochim Biophys Acta (2016) 1859:1130–40. 10.1016/j.bbagrm.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos (2015) 43:1522–35. 10.1124/dmd.115.064246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity (2013) 39:372–85. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 106.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature (2014) 511:184–90. 10.1038/nature13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goupil M, Cousineau-Cote V, Aumont F, Senechal S, Gaboury L, Hanna Z, et al. Defective IL-17- and IL-22-dependent mucosal host response to Candida albicans determines susceptibility to oral candidiasis in mice expressing the HIV-1 transgene. BMC Immunol (2014) 15:49. 10.1186/s12865-014-0049-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity (2014) 40:25–39. 10.1016/j.immuni.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu X, Weiss ID, Zhang HH, Singh SP, Wynn TA, Wilson MS, et al. Conventional NK cells can produce IL-22 and promote host defense in Klebsiella pneumoniae pneumonia. J Immunol (2014) 192:1778–86. 10.4049/jimmunol.1300039 [DOI] [PMC free article] [PubMed] [Google Scholar]