Abstract
Microarrays and RNA-seq are at the forefront of high throughput transcriptome analyses. Since these methodologies are based on different principles there are concerns about the concordance of data between the two techniques. The concordance of RNA-seq and microarrays for genome-wide analysis of differential gene expression has not been rigorously assessed in clinically derived ligament tissues. To demonstrate the concordance between RNA-seq and microarrays and to assess potential benefits of RNA-seq over microarrays, we assessed differences in transcript expression in anterior cruciate ligament (ACL) tissues based on time-from-injury. ACL remnants were collected from patients with an ACL tear at the time of ACL reconstruction. RNA prepared from torn ACL remnants was subjected to Agilent microarrays (N = 24) and RNA-seq (N = 8). The correlation of biological replicates in RNA-seq and microarrays data was similar (0.98 vs. 0.97), demonstrating that each platform has high internal reproducibility. Correlations between the RNA-seq data and the individual microarrays were low, but correlations between the RNA-seq values and the geometric mean of the microarrays values were moderate. The cross-platform concordance for differentially expressed transcripts or enriched pathways was linearly correlated (r=0.64). RNA-Seq was superior in detecting low abundance transcripts and differentiating biologically critical isoforms. Additional independent validation of transcript expression was undertaken using microfluidic PCR for selected genes. There was a higher correlation between the PCR data and the RNA-seq as well as the microarrays data. These findings demonstrate that RNA-seq has advantages over microarrays for transcriptome profiling of ligament tissues when available and affordable.
Keywords: anterior cruciate ligament, time-from-injury, isoforms, periostin, transcripts
Graphical abstract
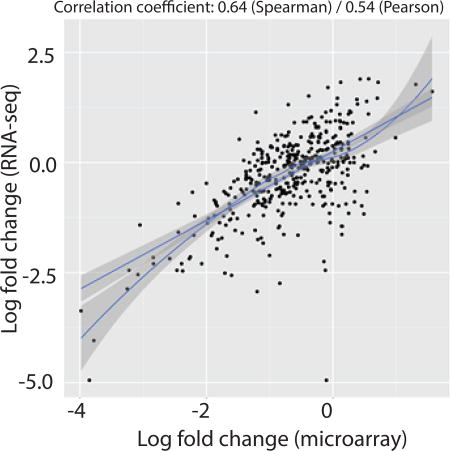
Transcriptome profiling of anterior cruciate ligament tears as a function of time-from-injury comparing microarrays to RNA-seq showed that the cross-platform concordance for differentially expressed transcripts or enriched pathways was linearly correlated (r=0.64). RNA-Seq was superior in detecting low abundance transcripts and differentiating biologically critical isoforms. Thus, RNA-seq is an extremely promising tool for the assessment of mRNA expression and identification of differentially expressed transcripts, comparable, and to some extent superior, to existing microarrays platforms in the analysis of ligamentous tissues.
INTRODUCTION
Microarrays and RNA-seq technologies have revolutionized gene discovery studies. The ability to simultaneously examine thousands of gene transcripts using a genome-wide transcriptome profiling approach puts these technologies in the forefront of high throughput screening. These technologies have a broad spectrum of applications including but not limited to identification of differentially expressed gene transcripts between healthy and diseased cells (and tissues), providing new insights into developmental processes, pharmacogenomics, and the examination of gene regulation1; 2. Because of their popularity in the scientific community, cost-effectiveness, and ease of analysis, microarrays remain the most extensively used approach in transcriptome profiling. Nevertheless, hybridization issues with degraded clinically derived RNA, low abundance transcripts, and the availability of probes for known genes on the chip are the drawbacks of microarrays technology3. Like microarrays, RNA-seq also has some potential pitfalls with the use of degraded RNA samples, however, unlike microarrays, there are several protocols available that can circumvent some, if not all, of the problems associated with RNA-seq such as to remove bases adapters and overrepresented or low-quality sequences4–6.
RNA-seq utilizes high throughput sequencing technology to directly sequence gene transcripts and is emerging as an alternative for whole-genome transcriptome profiling7. RNA-seq has considerable advantages for examining transcriptome profile structure such as the detection of novel transcripts and splice junctions, although it does pose novel algorithmic and logistical challenges for data analysis and storage8. It does not depend on genome annotation for prior probe selection and avoids the related biases introduced during hybridization of microarrays8. Despite the fact that many computational methods have been developed for alignment of reads, quantification of genes or gene transcripts and identification of differentially expressed genes, there is great variability in the maturity of the available computational tools8; 9.
Using the microarrays approach, we have recently identified a number of gene transcripts that showed repressed expression with time-from-injury in human anterior cruciate ligament (ACL) tears using the 24-sample microarrays data set10. We observed that the largest differences in expression of gene transcripts exist between acute (<3 month from injury) and chronic (>12 months from injury) groups with little differences between acute and intermediate (3–12 months from injury) and chronic and intermediate groups. The differentially expressed gene transcripts were enriched for numerous biological processes that were consistent with the initial repair activity in the injured ligament to repair. To the best of our knowledge, no study has compared RNA-seq and microarrays transcriptome profiles in any area of musculoskeletal research. Studies performed on other tissues have focused on the concordance between RNA-seq and microarrays8; 11; 12. Our study focused on both the consistencies, as well as differences, between these technologies and further investigated the reasons for any observed discrepancies. The purpose of the present study was to evaluate the relative concordance between the two assays and to determine the benefits of RNA-seq over the microarrays for evaluating injured ligament tissues.
METHODS
Study design
Informed written consent was obtained from study patients approved by the Institutional Review Board of Washington University. Patients with clinically diagnosed ACL tear were recruited (Table 1). Patients of any age, body mass index, and sex were included and patients with other concomitant intra-articular or associated extra-articular injuries and those undergoing revision ACL reconstruction were excluded. Previously we reported gene expression differences as a function of time-from-injury using RNA from 24 ACL tissues via microarrays. Here, we selected a subset of RNA samples (N = 8) from the 24-sample cohort and performed Illumina RNA-seq. As we already know that most important differences in gene expression exist between acute and chronic ACL tears10, we randomly selected samples from acute (N=5) and chronic (N=3) groups only for RNA-seq analysis. We compared the 8-sample RNA-seq data with the microarrays data using 24-sample cohort (14 acute, 4 chronic, while keeping the 6 intermediate samples in the model) as well as with microarrays data from the same 8 samples on which RNA-seq was performed.
Table 1.
Characteristics of study patients according to type of analysis
| Assay | Category | Acute | Intermediate | Chronic |
|---|---|---|---|---|
| Microarray | n | 14 | 6 | 4 |
| Average TFI (months) | 1.53 | 3.92 | 40.53 | |
| Mean age (Range) in years | 31.6 (13–63) | 37.8 (14.49) | 45.0 (20–63) | |
| Mean BMI | 26.3 | 24.6 | 28.2 | |
| Sex | 7 female, 7 male | 2 female, 4 male | 2 female, 2 male | |
| RNA-seq | n | 5 | - | 3 |
| Average TFI (Range) months | 1.41 | - | 50 | |
| Mean age (Range) years | 31.6 (16–50) | - | 45.7 (20–63) | |
| Mean BMI | 25.5 | - | 29.9 | |
| Sex | 2 female, 3 male | - | 1 female, 2 male |
TFI = time-from-injury; BMI = body mass index
Tissue collection, processing and RNA isolation
Fragments of torn ACL were collected at the time of ACL reconstruction surgery10. Tissues immersed in TRIzol reagent (Invitrogen) were homogenized using Polytron System (Kinematica AG). RNA was prepared using TRIzol-chloroform method followed by purification using Minispin columns (Qiagen)13. Quality and quantity of RNA samples was ascertained with the use of Agilent 2100 Bioanalyzer (Agilent Technologies). Mean RNA integrity number (RIN) was 4.7 (range 3.0 to 6.9). RNA was aliquoted after preparation and RIN did not change with time.
Microarrays hybridization
A total of 30-ng of RNA was amplified by WTA2 kit (Sigma-Aldrich) and 2.5-µg of complementary DNA was labeled with Kreatech ULS labeling kit (Kreatech Diagnostics). Labeled samples were purified with QIAquick polymerase-chain-reaction (PCR) purification columns (Qiagen) and quantitated on a Nanodrop spectrophotometer (NanoDrop Technologies). The labeled DNA was hybridized using Agilent Human 8×60K microarrays chips (Agilent Technologies) at 65°C for 20h followed by routine washing. The microarrays were scanned on an Agilent SureScan scanner to detect Cy5 fluorescence. Gridding and analysis of images were performed using Agilent Feature Extraction software v10.7.3.1.
Microarrays data analysis
Data were imported into the R/Bioconductor package Limma. The raw probe fluorescence signals were background subtracted, expressed in log2 format for normal distribution, and then quantile normalized to adjust gene expression signals for differences in hybridization efficiency. Genes with Limma quantile normalized expression levels <10% over the 95th percentile of negative background control probes in more than half the samples were pre-filtered to increase the signal to noise ratio among lower expressing genes and to reduce the number of genes tested and corrected for by the Benjamini-Hochberg method of adjusting p-values i.e. false-discovery rate (FDR). The Limma probe-level data were then averaged by probe identification and all probes were annotated with the R/Bioconductor package biomaRt14 to Ensembl Release 76 (GRch38.76). The quality and performance of the samples were then assessed with a spearman correlation matrix (Supplementary Fig. 1) of all probe-averaged quantile normalized logged intensities that passed the pre-filter as well as multidimensional scaling plots (Supplementary Fig. 2A) to assess the variance between and within conditions. Significance testing was performed with Limma’s generalized-linear model using moderated t-statistics and robustly trended estimates of residual standard errors to control for outlier sample variances due to varying degrees of hybridization and sample performance with factors controlling for other confounders (age, sex, body-mass-index, and variation in RNA integrity number). The residual standard errors were then plotted to confirm the trended fit conformed to the observed mean-variance relationship (Supplementary Fig. 2B). To create a comprehensive list of differentially expressed genes, we applied a ≥1.5 fold-change as the minimum threshold with secondary levels of FDR adjusted p-values ≤0.05 based on previous experience15–17.
RNA-seq library preparation
RNA quality was assessed prior to RNA-seq analysis with an Agilent Bioanalyzer. Library preparation was performed with 10-ng of total RNA. All samples were DNase-I treated to remove residual DNA and ds-cDNA was prepared using the SeqPlex RNA kit (Sigma-Aldrich) per manufacturer’s protocol. The cDNA was then blunt ended, an A base added to the 3’ ends, and then Illumina sequencing adapters were ligated to the ends. Ligated fragments were then amplified for 12 cycles using primers incorporating unique index tags. Fragments from all samples were then pooled and sequenced on an Illumina HiSeq-2500 across two lanes of sequencing in two different flow cells using single reads extending 50 bases, targeting 30M read per sample.
RNA-seq data analysis
RNA-seq reads were aligned to Homo sapiens Ensembl GRCh38.76 with STAR v2.0.4b18. Approximately, 28–43 million reads were obtained per sample. Low quality reads (<10%) were eliminated, resulting in 24–41 million mapped reads. In total, 11–25 million uniquely mapped read pairs per sample were obtained and aligned to the human reference genome as shown in (Supplementary Fig. 3). Gene counts were derived from the number of uniquely aligned unambiguous read by Subread:featureCount version 1.4.519. Transcript counts were produced by Sailfish version 0.6.320. Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction, known junction saturation, and read distribution over known gene models with RSeQC version 2.321. In order to determine that the total number of aligned reads properly represented all known exon-exon junctions, junction saturation curves (Supplementary Fig. 4A) and 3’/5’ end bias plots (Supplementary Fig. 4B) were created with RSeQC to verify that those reads aligned uniformly across known transcripts.
All gene-level and transcript counts were then imported into the R/Bioconductor package EdgeR22 and TMM (trimmed mean of M-values) normalized to adjust for differences in library size. Genes or transcripts not expressed in any sample were excluded from further analysis. Performance of the samples were assessed with a spearman correlation matrix (Supplementary Fig. 5A) and multidimensional scaling plots (Supplementary Fig. 5B) of the first 2 eigenvectors to confirm that time-from-injury was the largest source of variation and that within-group variance was less than those between groups. A plot of the tagwise and fitted trended dispersions estimates generated by EdgeR showed that the mean-variance relationship among biological replicates met the assumptions of the negative binomial model (Supplementary Fig. 5C). Generalized linear models with robust dispersion estimates were created to test for gene/transcript level differential expression between acute and chronic time-from-injury with additional blocking factors controlling for differences due to age, gender, body-mass-index, and sample quality based on variation in RNA integrity values. The fit of the trended and tagwise dispersion estimates were then plotted to confirm proper fit of the observed mean to variance relationship where the tagwise dispersions are equivalent to the biological coefficients of variation of each gene (Supplementary Fig. 5C). Differentially expressed genes and transcripts were then filtered for FDR adjusted p-values ≤0.05. The EdgeR analysis revealed that 49% of all differentially expressed genes with unadjusted p-value ≤0.01 could be derived from the time-from-injury component of the fitted additive generalized linear model alone for RNA-seq (Supplementary Fig. 6A) and 52% for microarrays (Supplementary Fig. 6B).
Gene isoforms
We selected POSTN as an example for isoform analysis for a the following specific reasons: (i) we have previously reported that its expression was highly down-regulated in chronic tears compared to acute tears10, which is consistent with RNA-seq data from the current study, (ii) this gene plays an important role in tissue repair and degeneration23–26, and (iii) there are six known transcript variants for this gene27. The process of accurately quantifying and validating isoforms (splice variants) of known genes is still an ongoing endeavor, but recent tools utilizing advanced expectation-maximization algorithms such as Sailfish, Salmon, RSEM, and Kallisto has made that process more efficient and accurate versus older methods such as Bayesian methods such as Cufflinks28. The examination of gene alternative splice isoforms has always been difficult at best or impossible with microarrays technology. Affymetrix now offers several new array types that attempt to query the expression of isoforms at the exon level, but de-convoluting the expression of genes with many isoforms has proven difficult due to the many to one relationship of the expressed exons to known or novel isoforms. Because RNA-seq reads can span known exons in the form of reads aligned across exon-exon or exon-retained intron junctions, it is now possible to de-convolute isoform expression patterns using advanced statistical modeling on the placement and numbers of unique and unambiguously aligned reads within exons with the number of reads aligned across known exon-exon or exon-retained intron junctions that define known expressed isoforms. Here, we used Sailfish20 to estimate the expression of isoforms using an expectation-maximization model on k-mers matched to a k-mer index derived from FASTA file comprised of all known isoforms of all known genes found in the Ensembl release 76 reference transcriptome.
Gene ontology
To highlight the biological interpretation of the large set of transcripts, grouping of genes/transcripts based on functional similarity was achieved using GeneGo MetaCore tools as described previously15. The altered biological processes (gene ontology distribution) were ranked based upon enrichment score and p-values. Gene ontology was performed for the differentially expressed genes between acute and chronic ACL remnants from 8-sample microarrays and RNA-seq analyses. In addition, we conducted gene ontology and network analysis on genes common to microarrays and RNA-seq to see if these genes are interlinked or they function in relation to each other using GeneGo MetaCore Direct Interaction path.
Microfluidic quantitative PCR
The expression of 12 transcripts differentially expressed by microarrays and RNA-seq was validated via microfluidic-based PCR 96.96 Dynamic Arrays (Fluidigm Corp.) as described previously17. Analysis of data was performed using 2−ΔCt method with peptidylprolyl isomerase A (PPIA, cyclophilin A) as the housekeeping gene. PPIA was selected as housekeeping gene based on our previous work as it showed relatively stable expression across all the samples10. We applied non-parametric two-tailed Mann-Whitney test (GraphPad) to observe the differences in transcript expression between any of the two time-from-injury categories. Data are presented as mean ± standard error of the mean.
Data deposition
The raw data is available on National Center of Biotechnology Information Gene Expression Ontology website (http:www.ncbi.nlm.nih.gov/projects/geo) with accession numbers GSE61385 (microarray) and GSE65469 (RNA-seq).
RESULTS
Gene transcripts differentially expressed by time-from-injury
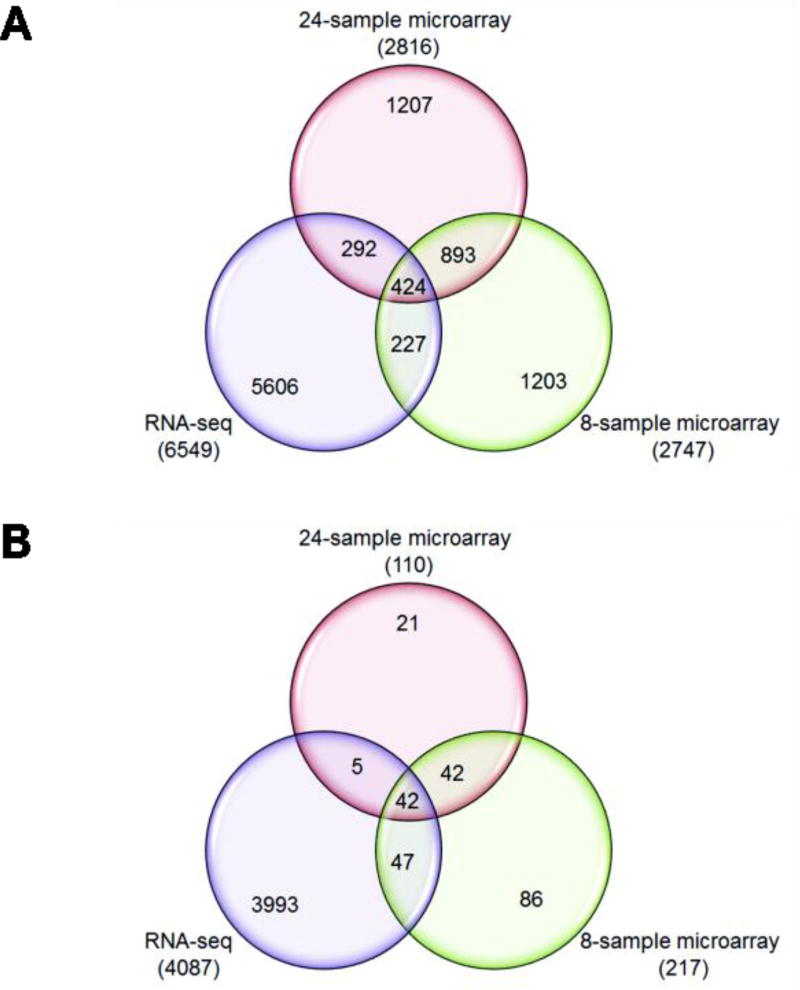
We detected 2816 gene transcripts significantly differentially expressed (at any fold-change) between acute and chronic tears in 24-sample microarrays (Supplementary Table 1), 2447 in 8-sample microarrays (Supplementary Table 2) and 6549 in RNA-seq samples (Supplementary Table 3). The number of genes differentially expressed between acute and chronic ACL tissues for each cohort is shown in Fig. 1A for any fold-change and in Fig. 1B at ≥1.5 log2 fold-change and P≤0.05. We noted that 424 gene transcripts were common to all three analyses at any fold-change (Supplementary Table 4) and 42 gene transcripts were common to all three analyses at ≥1.5 log2 fold-change (Table 2). Using a stringent criterion of FDR of <0.05, no significant gene transcripts were found for 24-sample microarray, only 61 in 8-sample microarrays (Supplementary Table 1) and 2112 in RNA-seq (Supplementary Table 2).
Figure 1.
Venn diagrams representing the number of differentially expressed gene transcripts for each comparison as well as their overlaps between the three comparisons are shown for any fold-change (A) or at a fold-change set at log2 fold of ≥ 1.5 (B at a P ≤ 0.05). The numbers of differentially expressed gene transcripts shown in parenthesis for each comparison and the numbers shown in overlapping areas represent the number of gene transcripts common to any two or three comparisons.
Table 2.
Gene transcripts common to microarray and RNA-seq analyses
| Gene symbol | Gene name | 24-sample microarray | 8-sample microarray | RNA-seq | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2 FC | P value | FDR | log2 FC | P value | FDR | log2 FC | P value | FDR | ||
| MMP9 | matrix metallopeptidase 9 | −1.87 | 0.037 | 0.324 | −1.83 | 0.004 | 0.155 | −6.48 | 0.000 | 0.000 |
| COL1A1 | collagen, type I, alpha 1 | −3.43 | 0.000 | 0.225 | −3.78 | 0.001 | 0.100 | −4.02 | 0.000 | 0.001 |
| HIST1H3G | histone cluster 1, H3g | −2.35 | 0.001 | 0.247 | −2.86 | 0.000 | 0.031 | −3.72 | 0.000 | 0.000 |
| CLEC11A | C-type lectin domain family 11, member A | −1.98 | 0.001 | 0.247 | −2.18 | 0.001 | 0.099 | −3.68 | 0.000 | 0.000 |
| LOXL2 | lysyl oxidase-like 2 | −1.98 | 0.001 | 0.247 | −2.95 | 0.000 | 0.031 | −3.67 | 0.000 | 0.000 |
| HIST1H2AI | histone cluster 1, H2ai | −2.37 | 0.001 | 0.247 | −2.80 | 0.000 | 0.054 | −3.60 | 0.000 | 0.000 |
| ARL4C | ADP-ribosylation factor-like 4C | −2.27 | 0.000 | 0.247 | −2.98 | 0.000 | 0.031 | −3.60 | 0.000 | 0.000 |
| HIST1H3F | histone cluster 1, H3f | −1.64 | 0.002 | 0.247 | −1.77 | 0.000 | 0.054 | −3.49 | 0.000 | 0.000 |
| ZNF469 | zinc finger protein 469 | −1.97 | 0.001 | 0.247 | −2.37 | 0.000 | 0.031 | −3.49 | 0.000 | 0.000 |
| HMOX1 | heme oxygenase 1 | −2.57 | 0.000 | 0.247 | −2.88 | 0.002 | 0.111 | −3.45 | 0.000 | 0.000 |
| HIST1H3B | histone cluster 1, H3b | −1.84 | 0.002 | 0.247 | −2.43 | 0.000 | 0.065 | −3.39 | 0.000 | 0.000 |
| POSTN | periostin, osteoblast specific factor | −3.99 | 0.000 | 0.225 | −3.98 | 0.000 | 0.026 | −3.22 | 0.000 | 0.000 |
| TGFBI | transforming growth factor, beta-induced | −2.44 | 0.000 | 0.247 | −3.25 | 0.000 | 0.060 | −3.14 | 0.000 | 0.000 |
| NDUFA4L2 | NDH dehydrogenase (ubiquinone) 1 alpha sub-complex, 4-like 2 | −1.67 | 0.006 | 0.267 | −2.02 | 0.000 | 0.036 | −3.03 | 0.000 | 0.000 |
| HIST1H4F | histone cluster 1, H4f | −1.68 | 0.004 | 0.257 | −1.75 | 0.006 | 0.179 | −2.77 | 0.000 | 0.000 |
| COL3A1 | collagen, type III, alpha 1 | −2.62 | 0.000 | 0.227 | −2.38 | 0.002 | 0.113 | −2.67 | 0.000 | 0.000 |
| VEGFA | vascular endothelial growth factor A | −1.54 | 0.025 | 0.306 | −2.46 | 0.000 | 0.031 | −2.66 | 0.000 | 0.000 |
| SERPINE1 | serpin peptidase inhibitor, clade E | −1.60 | 0.007 | 0.272 | −2.51 | 0.000 | 0.031 | −2.63 | 0.000 | 0.001 |
| COL1A2 | collagen, type I, alpha 2 | −2.96 | 0.000 | 0.227 | −3.08 | 0.000 | 0.077 | −2.57 | 0.000 | 0.006 |
| COL5A2 | collagen, type V, alpha 2 | −1.94 | 0.004 | 0.257 | −2.84 | 0.001 | 0.099 | −2.43 | 0.000 | 0.001 |
| MARCKSL1 | MARCKS-like 1 | −1.64 | 0.001 | 0.247 | −1.81 | 0.000 | 0.065 | −2.38 | 0.000 | 0.000 |
| COL6A3 | collagen, type VI, alpha 3 | −2.36 | 0.000 | 0.247 | −2.84 | 0.000 | 0.036 | −2.36 | 0.000 | 0.002 |
| FSCN1 | fascin actin-bundling protein 1 | −2.29 | 0.000 | 0.225 | −3.03 | 0.000 | 0.016 | −2.34 | 0.000 | 0.000 |
| MAFB | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B | −1.67 | 0.000 | 0.247 | −2.19 | 0.000 | 0.066 | −2.30 | 0.000 | 0.000 |
| PXDN | peroxidasin | −1.64 | 0.002 | 0.247 | −2.43 | 0.000 | 0.041 | −2.23 | 0.000 | 0.000 |
| COL12A1 | collagen, type XII, alpha 1 | −1.68 | 0.005 | 0.265 | −2.23 | 0.000 | 0.031 | −2.20 | 0.000 | 0.000 |
| COL6A1 | collagen, type VI, alpha 1 | −1.81 | 0.001 | 0.247 | −1.84 | 0.005 | 0.169 | −2.18 | 0.000 | 0.000 |
| HIST1H3D | histone cluster 1, H3d | −1.85 | 0.002 | 0.247 | −2.19 | 0.000 | 0.052 | −2.17 | 0.000 | 0.000 |
| SERPINH1 | serpin peptidase inhibitor, clade H, member 1 | −2.00 | 0.000 | 0.247 | −2.00 | 0.006 | 0.184 | −2.17 | 0.000 | 0.000 |
| HIST1H3H | histone cluster 1, H3h | −1.83 | 0.002 | 0.247 | −2.33 | 0.000 | 0.036 | −2.14 | 0.000 | 0.000 |
| FTLP3 | ferritin, light polypeptide 3 | −2.39 | 0.000 | 0.247 | −2.18 | 0.000 | 0.065 | −2.11 | 0.043 | 0.324 |
| AEBP1 | AE binding protein 1 | −2.38 | 0.000 | 0.227 | −2.25 | 0.002 | 0.113 | −2.11 | 0.000 | 0.001 |
| HIST1H2AE | histone cluster 1, H2ae | −1.54 | 0.002 | 0.247 | −1.71 | 0.001 | 0.098 | −2.03 | 0.000 | 0.000 |
| CEMIP | cell migration inducing protein, hyaluronan binding | −2.24 | 0.001 | 0.247 | −3.51 | 0.000 | 0.023 | −2.00 | 0.000 | 0.000 |
| CAMK2N1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 | −1.59 | 0.004 | 0.257 | −2.01 | 0.000 | 0.036 | −1.96 | 0.000 | 0.000 |
| PLAU | plasminogen activator, urokinase | −1.76 | 0.024 | 0.292 | −2.28 | 0.000 | 0.050 | −1.94 | 0.000 | 0.001 |
| EME1 | essential meiotic structure-specific endonuclease 1 | −1.56 | 0.002 | 0.247 | −2.22 | 0.000 | 0.031 | −1.77 | 0.004 | 0.080 |
| SLC16A3 | solute carrier family 16, member 3 | −2.06 | 0.001 | 0.247 | −2.57 | 0.000 | 0.050 | −1.68 | 0.000 | 0.014 |
| LGALS1 | lectin, galactoside-binding, soluble, 1 | −1.91 | 0.000 | 0.247 | −1.99 | 0.006 | 0.186 | −1.58 | 0.000 | 0.001 |
| EMILIN1 | elastin microfibril inter-facer 1 | −1.96 | 0.002 | 0.247 | −2.24 | 0.000 | 0.039 | −1.55 | 0.000 | 0.001 |
| G0S2 | G0/G1 switch 2 | 2.32 | 0.001 | 0.247 | 2.24 | 0.001 | 0.087 | 3.80 | 0.000 | 0.000 |
| PLIN4 | perilipin 4 | 2.21 | 0.000 | 0.225 | 2.76 | 0.000 | 0.050 | 6.48 | 0.000 | 0.000 |
FC = fold change; FDR = false discovery rate; −ve values indicate down-regulation compared to acute tears
Concordance between microarrays and RNA-seq
We investigated the concordance between 8-sample microarrays and RNA-seq based on the expression profile of gene transcripts differentially expressed between acute and chronic tears. We observed that RNA-seq data had a tighter distribution of fold-changes around zero and a characteristic fantail indicative of lower signal to noise ratios at the lowest levels of detectable gene expression (Supplementary Fig. 7A). Despite the higher ratio of attributable gene expression to the time-from-injury condition, the Limma microarrays data demonstrated lower signal to noise throughout the spectrum of gene expression as can be seen by the more diffuse cloud of signals in the MA plot (Supplementary Fig. 7B). A violin/box plot of the distribution of the observed log2 fold-changes across the two Lima microarrays analyses and single EdgeR RNA-seq dataset show that RNA-seq dataset has a much larger tighter and more elongated range of observed log2 fold-changes that indicates the RNA-seq data also exhibits signs of less log2 fold-change compression (Supplementary Fig. 7C). Further examination of the rank ordered observed p-values across all three datasets illustrate that the RNA-seq data not only exhibits less signs of log fold-change compression, but higher degrees of statistical sensitivity as well (Supplementary Fig. 7D).
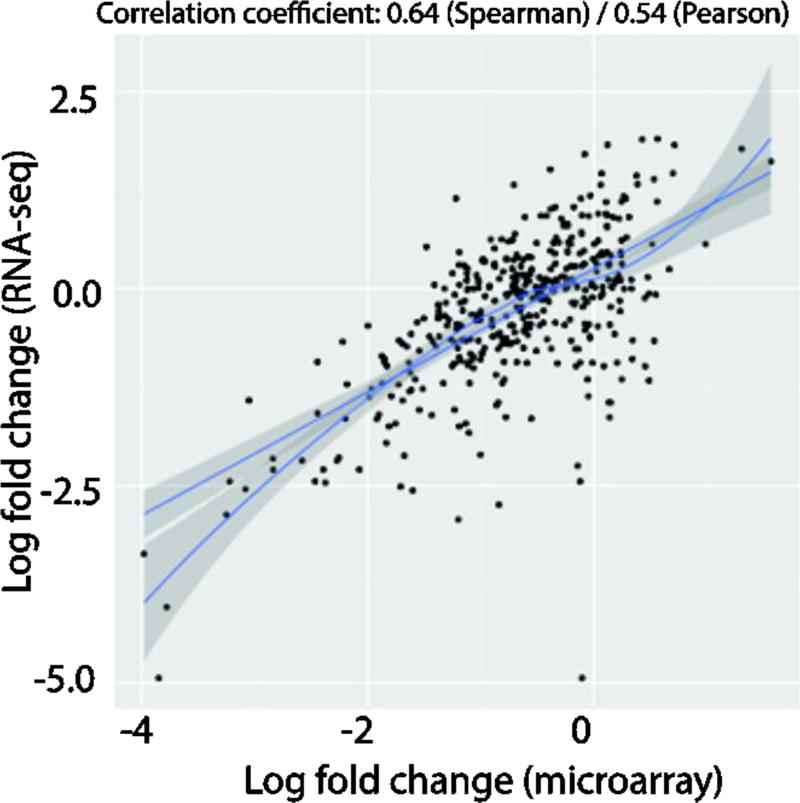
The correlation between log2 fold-changes between microarrays and RNA-seq datasets were computed by plotting an XY plot after filtering the gene-lists of both datasets for just those that were expressed greater than 8 log counts-per-million across all samples in the RNA-seq dataset. The results were then plotted with the RNA-seq data on the y-axis and the microarrays data on the x-axis and a least-sum-squares linear regression model and LOESS model was then fitted to the data. The measured Spearman and Pearson correlation coefficients across these high expressing genes were respectively 0.64 and 0.54, indicating that the cross-platform concordance was relatively moderate across the two similar RNA-seq and microarrays datasets, particularly when focused on highly expressed genes (Fig. 2). The correlation coefficients and XY plots become incomprehensible when low expressing genes are included (data not shown).
Figure 2.
XY plot of log2 fold-changes between N = 8 microarrays and RNA-seq datasets. The gene-lists of both datasets were filtered for just those that were expressed greater than 8 log counts-per-million across all samples in the RNA-seq dataset. The results were then plotted with the RNA-seq data on the y-axis and the microarrays data on the x-axis and a linear and LOESS model was then fitted to the data. Spearman and Pearson correlation coefficients were then measured, respectively.
An added advantage of RNA-seq is the ability to query the distribution of aligned reads across gene bodies. In this case, we have found that many of the differences between the two platforms can be attributed to the less than ideal RNA quality that is typical for operating room derived specimens, especially those coming from damaged tissues. RSeQC end bias analysis clearly shows that the reads sequenced and successfully aligned across all known genes by STAR have clear signs of RNA degradation indicated by drops in coverage across the 5 prime end and middle of a hypothetical normalized 100 base pair gene body (Supplementary Fig. 4B). This signature of degradation limits the depth of sequencing across the 5 prime end of known genes, but does not render them undetectable as long as there is some part of the degraded transcript that can be sequenced, but hybridization of these RNA fragments to probes targeting these regions in the reference genome are very negatively impacted due to a loss of hybridization efficiency. The high correlations between the two platforms for high expressing genes suggests that although the integrity of the samples were compromised, reasonable comparisons of relative expression can still be gained when limited to high expressing genes that may have less degradation or more tolerant of degradation due to sheer numbers of transcripts. Consequently, in future studies to identify biomarkers transferrable between two gene-expression measurement platforms, an emphasis should be placed on the above-median expressed genes when sample RNA integrity is less than optimal.
Gene isoforms
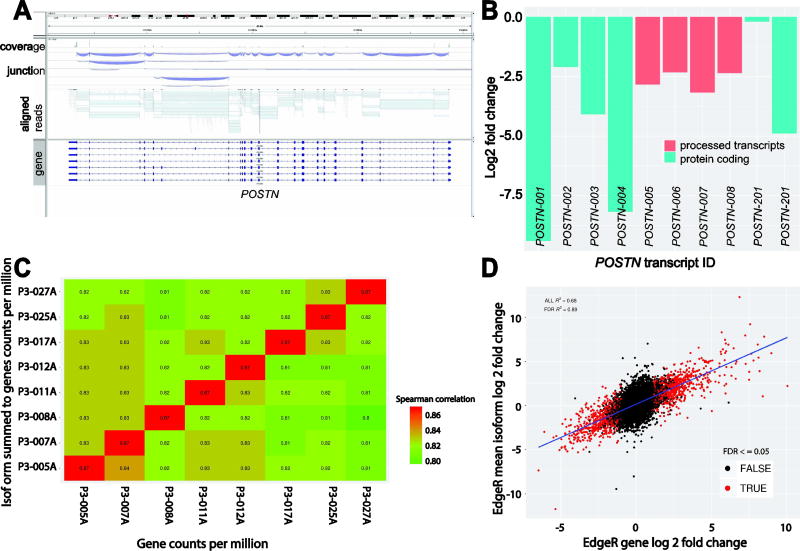
The complexity inherent in quantifying isoforms is readily apparent in the visualization of the aligned reads across POSTN with sashimi plots created by the Integrated Genomic Viewer (IGV). The plot clearly shows the convolution of reads aligned across exon-exon junctions of a single gene with multiple known isoforms (10 known isoforms; 6 of which are protein-coding and 4 are processed transcripts) and necessitates the use of advanced statistical modeling for the estimation of isoform level expression (Fig. 3A). Here we used Sailfish to quantify the expression of known Ensembl GRCh38 isoforms and measure their differential expression using the same statistical model and methods outlined for our gene level analysis with EdgeR. A closer examination of the EdgeR analysis of the Sailfish estimated counts per isoform revealed that POSTN-001 (−9.40 log2 fold-change) and POSTN-004 (−8.17 log2 fold-change) comprised the most differentially regulated isoforms driving the repression of the POSTN gene in chronic tears. A closer examination of the mean log2 fold-change observed by EdgeR across all the annotated isoforms of POSTN yield a value of −3.95 which closely follows the gene-level observed fold-change from EdgeR of −3.22 (Fig. 3B). The difference most likely comes from the uncertainty in accurately estimating the isoform expression of a complex transcript with any given statistical model rather than the absolute integer count value of reads aligned across all exons of a gene as was done with the gene-level analysis.
Figure 3.
(A) Display of aligned reads across POSTN with sashimi plots created by the Integrated Genomic Viewer (IGV) demonstrating the convolution of reads aligned across exon-exon junctions that make the use of statistical modeling necessary for the estimation of isoform level expression. (B) Barplot of the estimated log2 fold-changes observed by EdgeR on the estimated counts of reads per known isoform of POSTN. (C) The summed estimated counts for isoforms were compared to the counts-per-million of the genes used in our prior analyses in the form of a spearman correlation matrix. This matrix supports our prior expectation that the summation of expressed isoforms to the level of their parent genes in the data follows a positive trend with high correlation (87%). (D) The relative accuracy of isoform expression was further interrogated by averaging the observed isoform-level EdgeR log2 fold-changes for all isoforms of a given gene to their gene-level and then measuring the correlation of these changes to those of the same genes from a nearly identical EdgeR analysis of the genes previously described. An XY plot shows that the observed mean isoform and gene changes follow a linear trend with a Pearson correlation across all genes of 68% and 89% for all genes previously identified as having a Benjamini-Hochberg FDR adjusted P values ≤0.05. This supports the assumption that the Sailfish estimated counts for isoforms are reasonably accurate and differential expression analysis of these counts is statistically sound, especially when focused on isoforms of genes previously identified as statistically significant.
In order to pseudo-validate our isoform findings, we followed the logic that genes are the summation of their isoforms and that the observed gene-level log2 fold-change is approximately the mean of a given gene’s isoforms and used a method similar to the Bioconductor package tximport to sum the Sailfish estimated counts to the gene-level as illustrated by Soneson et. al.29 The summed estimated counts for isoforms were then expressed as counts-per-million (CPM) in order to adjust for differences in library size across samples and then compared to the counts-per-million of the genes used in our prior analyses in the form of a spearman correlation matrix (Fig. 3C). The spearman correlation matrix supports our prior expectation that the summation of expressed isoforms to the level of their parent genes in the data follows a positive trend with high correlation (87%). The relative accuracy of isoform expression was further bioinformatically interrogated by averaging the observed isoform-level EdgeR log2 fold-changes for all isoforms of a given gene to their gene-level and then measuring the correlation of these changes to those of the same genes from a nearly identical EdgeR analysis of the genes previously described. An XY plot shows that the observed mean isoform and gene changes follow a linear trend with a Pearson correlation across all genes of 68% and 89% for all genes previously identified as having a Benjamini-Hochberg FDR adjusted p-values less than or equal to 0.05 (Fig. 3D). This supports the assumption that the Sailfish estimated counts for isoforms are reasonably accurate and differential expression analysis of these counts is statistically sound, especially when focused on isoforms of genes previously identified as statistically significant. Where larger deviations between gene and isoform differential expression are observed, we have found that they are primarily limited to low expressing genes or genes with many isoforms where only one or two isoforms are driving the expression, such as is the case with POSTN. This may explain the differences we have observed in observed gene-level expression between the microarrays and RNA-seq datasets where the probes of the microarrays appear to favorably hybridize to the higher expressing isoforms of POSTN.
Gene transcripts common to microarrays and RNA-seq and their biological significance
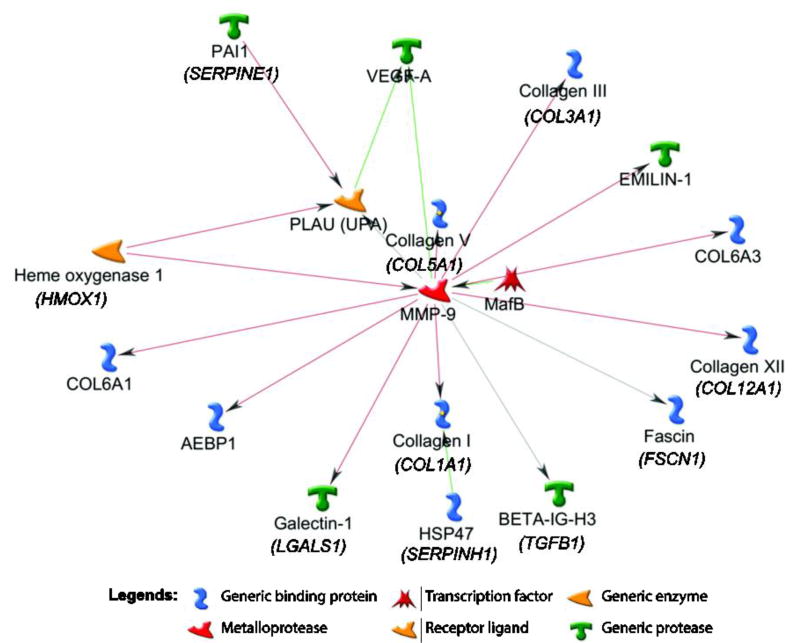
Further interrogation of the differentially expressed transcript in microarrays and the RNA-seq cohorts showed that 424 gene transcripts were found to recur across the datasets and majority of these were down-regulated in chronic tears. At a fold-change cutoff of ≥1.5 log2 fold 42 genes were common to the three platforms. Only two gene transcripts (G0S2 and PLIN4) were up-regulated while other 40 gene transcripts were down-regulated in chronic versus acute tears and were common across three analyses (Table 2, Fig. 4). Examples include COL1A1, COL1A2, COL3A1, COL5A2, COL6A1, COL6A3, COL12A1, TGFBI, POSTN, MMP9, VEGFA, PLUA, FSCN1, MAFB, HMOX1, AEBP1, and EMILIN1. Gene ontology of these 40 transcripts was represented by several important biological processes relevant to ACL healing (Table 3) such as extracellular matrix organization, blood vessel development, multicellular organismal metabolic process, regulation of cell-substrate adhesion, collagen metabolic process, cell motility, wound healing, blood vessel morphogenesis, cellular component organization or biogenesis, epithelial to mesenchymal transition, and response to hypoxia. Furthermore, network analysis showed that out of 40 transcripts down-regulated and common to microarrays and RNA-seq analyses, 18 were interlinked in a module clearly indicating that they correlate to a function, signifying their role in the same biological process (Fig. 5). Using lists of all gene transcripts differentially expressed between acute and chronic remnants identified by 8-sample microarrays and RNA-seq, we noted that only a small number of biological processes were common between the two platforms: only 7/50 processes were common between the two platforms for the gene transcripts elevated in chronic remnants (Supplementary Table 5) compared to 19/50 for the gene transcripts repressed in chronic remnants (Supplementary Table 6).
Figure 4.
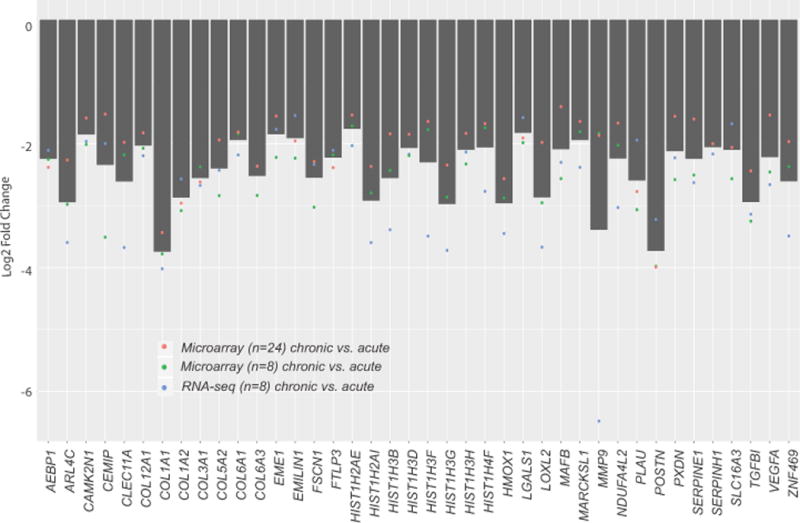
Barplot representing the mean log2 fold-change of 40 recurring genes across the N = 24 microarrays dataset, N = 8 microarrays dataset, and the N = 8 RNA-seq dataset. Individual points indicate the mean log2 fold-change for each dataset.
Table 3.
Biological processes repressed in chronic group compared to acute group*
| Biological process | P value | FDR | Gene |
|---|---|---|---|
| Extracellular matrix organization | 1.305E-12 | 1.140E-09 | LOXL2, PAI1, Collagen V, PLOD2, COL16A1, OSF-2, COL27A1, Collagen XII, EMILIN-1, PXDN, Lysyl oxidase, COL5A1, COL6A3 |
| Blood vessel development | 1.052E-08 | 4.476E-06 | FAP48, LOXL2, VEGF-A, PAI1, Collagen V, PLAU (UPA), Notch, SRPUL, NOTCH1 precursor, Lysyl oxidase, COL5A1 |
| Multicellular organismal metabolic process | 6.479E-08 | 1.225E-05 | CEL, Collagen V, COL16A1, Collagen XII, COL5A1, COL6A3 |
| Regulation of cell-substrate adhesion | 8.132E-07 | 7.689E-05 | VEGF-A, PAI1, PLAU (UPA), Notch, EMILIN-1, NOTCH1 precursor |
| Collagen metabolic process | 9.639E-07 | 8.634E-05 | Collagen V, COL16A1, Collagen XII, COL5A1, COL6A3 |
| Cell motility | 1.508E-06 | 1.128E-04 | Carbohydrate sulfotransferases, LOXL2, VEGF-A, Fascin, Collagen V, PLAU (UPA), Notch, Seprase, SRPUL, NOTCH1 precursor, COL5A1 |
| Wound healing | 2.381E-06 | 1.559E-04 | VEGF-A, PAI1, Collagen V, PLAU (UPA), Notch, Histone H3.1, NOTCH1 precursor, Lysyl oxidase, COL5A1, HIST1H3D |
| Blood vessel morphogenesis | 4.722E-06 | 2.232E-04 | FAP48, LOXL2, VEGF-A, PAI1, PLAU (UPA), Notch, SRPUL, NOTCH1 precursor |
| Cellular component organization or biogenesis | 5.737E-06 | 2.537E-04 | SC65, Carbohydrate sulfotransferases, LOXL2, VEGF-A, PAI1, Stathmin, Fascin, Collagen V, PLOD2, COL16A1, OSF-2, LOXL3, COL27A1, Collagen XII, Notch, UBE2C, Histone H3.1, EMILIN-1, PXDN, NOTCH1 precursor, Lysyl oxidase, COL5A1, COL6A3, HIST1H3D |
| Epithelial to mesenchymal transition | 6.726E-06 | 2.862E-04 | LOXL2, LOXL3, Notch, NOTCH1 precursor |
| Response to hypoxia | 1.108E-05 | 3.849E-04 | LOXL2, VEGF-A, PAI1, PLOD2, PLAU (UPA), Notch, NOTCH1 precursor |
Common across microarray (n=24 and n=8) and RNA-seq (n=8 analysis);
FDR = false discovery rate
Figure 5.
The molecular networks using 40 genes down-regulated in chronic remnants and common across microarrays and RNA-seq analyses were generated by GeneGo MetaCore are shown. Pathway analysis showed that these genes represent extracellular matrix organization and demonstrate significant interactions with each other pathways. Green arrows indicate activation, red arrows indicate inhibition, and gray arrows indicate unspecified interaction.
Validation of microarrays and RNA-Seq data
With the use of Fluidigm PCR, we validated expression of 14 gene transcripts based on biological interest or magnitude of their expression between acute and chronic ACL tears for 7 down-regulated in acute (Fig. 6A–G) and 7 up-regulated in acute (Fig. 6H–N). Expression pattern of all transcripts was highly concordant with microarrays and RNA-seq data. Therefore, it can be concluded that PCR data for the 14 gene tested showed 100% concordance (in expression pattern) with RNA-seq and microarrays data.
Figure 6.
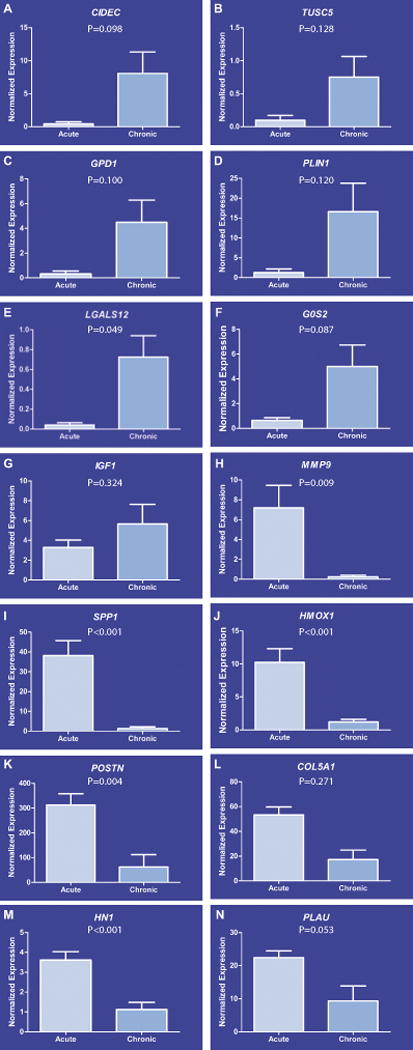
Validation of transcripts by microfluidic quantitative PCR. We validated the expression of 14 gene transcripts via microfluidic-based quantitative PCR based either on their biological significance or magnitude of their expression between acute and chronic groups. The expression pattern of 7 down-regulated in acute group (A–G) and 7 up-regulated in acute group (H–N) was highly concordant with microarrays and RNA-seq data sets.
DISCUSSION
This study comparing microarrays and RNA-seq to evaluate gene expression in ACL tears confirms that several transcripts representing important, distinct biological processes vary with time-from-injury. Overall, microarrays and RNA-seq show a significant difference between acute and chronic tears.
A number of studies have compared RNA-Seq and hybridization-based arrays in other tissues and have reported that RNA-seq has a broader dynamic range and detected low abundance transcripts and biologically critical isoforms, which were not possible with microarrays8; 30–34. Several investigators have proposed empirical protocols and statistical frameworks for the analysis of gene expression using RNA-seq, with a majority agreeing that RNA-seq data is negative-binomially distributed in nature and that count-based methods of differential expression analysis (EdgeR, DESeq2, or optionally Limma Voom) of the same gene across two or more conditions containing an appropriate number of biological replicates is the ideal method that yields the fewest false positives, highest true positives, and proper modeling of all known sources of biological or technical variation32; 35–38. The choice of tools or methods is largely determined by familiarity with the tools themselves, the ability to optimize the tools for their experimental conditions, and ease of use. Our decision to use STAR, subread:FeatureCounts, Sailfish, and EdgeR are largely guided by these same principles and analysis with similar tools such as Salmon with tximport for isoform and gene expression measurements, and DESeq2 or Limma Voom for differential expression analysis would yield very similar findings (data not shown, but available on request). Furthermore, numerous other studies have reviewed in detail the challenges and benefits associated with its technology and application7; 39–42 and have advocated the use of RNA-seq in transcriptome profiling of RNA samples.
Advantages of RNA-seq over microarrays include a higher number of differentially expressed gene transcripts than microarrays as it is based on sequencing rather than hybridization, which is independent of both probe availability and expression intensity7; 43 but is dependent on sequencing depth. The precision of expression measurements, especially for transcripts present in low abundance, is limited by the background levels of hybridization as well as hybridization properties44. Thus, although comparing hybridization results across arrays can detect gene expression differences among samples45, hybridization outcomes from a single sample may not provide a true assessment of the relative expression of different transcripts. Furthermore, arrays are generally limited to probing transcripts with relevant probes on the array and therefore accurate measurements of expression levels and the reliable detection of genes with low abundance are challenging to accomplish. This could be due to a number of reasons such as sub-optimal choice of probes, imperfect probe design, and incorrect probe annotations. However, a well-designed data analysis methodologies can resolve some, if not all, of these issues46.
While the focus of this study was to assess how RNA-seq might be used to characterize gene expression differences between samples, sequence data may help answer other questions that are difficult to address using arrays. In particular, array technologies can measure expression only of genes that have corresponding probes on the array, and, in most cases, probes are designed only to cover a very small portion of a gene. Consequently, it is not possible to detect novel transcribed regions or the presence of alternative splice forms of a gene. Both of these problems can potentially be overcome using the RNA-sequencing data. Since microarrays only yield gene level differences without any knowledge of the transcript variants (isoforms), RNA-seq could identify specific isoforms that each gene has with P values and fold-change, thereby providing new information on which specific isoforms are actually responsible for the gene expression differences. For instance, we knew that POSTN gene is significantly suppressed in chronic tears, but did not know which particular isoforms of POSTN drove the expression. RNA-seq informed us that isoforms 1, 3, 4 and 202 have −9.40, −4.08, −8.17 and −4.88-fold expression and other isoforms such as 201 remained unchanged or showed subtle differences in expression between acute and chronic tears (−0.19 fold). Estimating isoform-specific gene expression (especially where one isoform has greater influence on phenotype than others) may be important for deeper understanding of complex biological processes and for disease susceptibility genes mapping. Nevertheless, identification of splice variants using RNA-seq methodology assumes that an adequate number of reads span exon–exon junctions43. This may not hold true when a sample is sequenced in only a single lane, and additional data are perhaps needed to circumvent this issue. Furthermore, sample quality and proper library preparation are as important as sequencing depth when it comes to sequencing bias.
It is also worth mentioning that gene transcripts differentially expressed between acute and chronic remnants using the 24-sample cohort did not pass the most stringent criterion of 5% FDR and thus we had to limit the data to less stringent criterion of 5% P value. While reporting only P value-based significance level is not ideal, it is not entirely incorrect. In contrast, the 8-sample cohort yielded some (61 gene transcripts) and RNA-seq yielded numerous (2112 gene transcripts) that passed the 5% P values as well as 5% FDR thresholds. This observation represents a clear advantage of RNA-seq over the microarrays and it gives more reliable gene list, although additional validation by PCR, in situ hybridization or protein expression by immunostaining is still necessary.
This study compared the RNA-seq and microarrays using human ACL tear tissues measuring gene expression as a function of injury chronicity. We clearly observed several advantages associated with the use of RNA-seq over the microarrays in conjunction with current study. RNA-seq gave almost 40 times more gene transcripts than the microarrays for the same set of samples indicating that the former is more comprehensive. RNA-seq is more robust in identifying gene transcripts than microarrays, which are limited by the number of hybridized probes, background noise, and incorrect annotations. In addition, RNA-seq provided information about transcript variants and isoforms. RNA-seq is more sensitive than microarrays, as it can detect gene transcripts with very low expression, and is more accurate than microarrays across the spectrum of the expression.
We made two additional interesting observations when we compared microarrays with RNA-seq. First, RNA-seq data had a tighter distribution of fold-changes around zero and a characteristic fantail indicative of lower signal to noise ratios at the lowest levels of detectable gene expression. Second, RNA-seq data exhibited less signs of log fold-change compression and but higher degrees of statistical sensitivity. This supports the observed higher sensitivity and dynamic range of RNA-seq over conventional microarrays gene expression approaches as evidenced by the large differences in significant gene expression between the two platforms allowing for increasing levels of statistical stringency. The differences between the two microarrays datasets are a result of the exclusion of the intermediate condition and the absence of related factors for age, sex, body-mass-index, and RNA integrity number. When coefficients of a statistical model are changed, so do the resulting proportion of variances to the remaining factors thereby inducing differences of statistical results even in the case where many of the samples within the statistical models are identical. Had these ACL tissues come from genetically identical mice, it most likely would not have been necessary to include additional factors in the statistical modeling and the differences between acute and chronic across the two microarrays analyses would be negligible.
Our data showed relatively moderate cross-platform concordance across the two similar RNA-seq and microarrays datasets. This suggests that the above-median expressed genes may have a good transferability between the two platforms and that the differences in the observed log2 fold-changes and statistical significance may be a result of the limitations of hybridization based technologies and degraded tissue specimens as evidenced by nearly no correlation across low to medium expressors and only moderate correlation across high expressing genes.
We identified a discrete set of biological processes based on differentially expressed gene transcripts by 8-sample microarrays and RNA-seq. Although there was a larger overlap in the biological processes from gene transcripts repressed in chronic remnants between the two platforms, there is no reason to give preference to the biological processes from one platform over the other. However, here discuss some the biological processes that were enriched for gene transcripts common to all platforms.
A number of important gene transcripts were common across all analysis, with and most (40 out of 42) were repressed in chronic tears than acute tears, which is consistent with our prior study10. Similarly, the biological processes represented by these repressed genes demonstrated that pathways associated with tissue repair and matrix synthesis were inhibited in chronic tears, again as previously reported10. Finally, the gene transcripts repressed in chronic tears were not independent but rather work in concert as evidenced by the biological processes and network analysis.
This study has some limitations. While our analysis covered mRNA expression, we did not examine other species of RNA (e.g. micro-RNAs and long non-coding RNAs). Our analysis of isoform detection was limited primarily to the POSTN gene. In addition, the data on transcript isoforms is largely computational and were not validated by PCR. Further studies will test the expression of specific isoforms by PCR and western blot in cells and tissues. In this study, we did not use single-end 150 bp or paired-end sequencing to evaluate the differences in sequencing types. However, available literature suggests that there is a saturation point where increased depth of sequencing does not increase the knowledge gained from gene or isoform level differential expression analysis. Junction saturation plots of all known exon-exon junctions are very good at showing at what point a particular set of samples would benefit from further sequencing. The characteristic signature in this case would be a plateau of saturation. In the case of paired-end sequencing, the extra information to be gained over single-end sequencing would largely benefit the estimation of isoform level expression since longer split reads would span more exon-exon junctions that could be used to determine what isoforms of a given gene are expressed over other isoforms9; 47. If one of the primary goals of a study is to evaluate splice variants, chimeric fusion genes, or expressed single nucleotide polymorphisms (allele specific expression), then both higher depths of sequencing and paired-end sequencing would be the best choice. None of these three factors were primary goals in this study and were therefore not evaluated.
In conclusion, RNA-seq appears to be an extremely promising tool for the assessment of mRNA expression as well as identification of differentially expressed gene transcripts, comparable, and to some extent superior, to existing microarrays platforms in the analysis of ligamentous tissues. As the costs of sequencing are falling rapidly, RNA-seq is becoming widely embraced in the research community for accurate gene profiling. Future research into musculoskeletal biology should strongly consider the use of RNA-seq where appropriate and feasible.
Supplementary Material
Supplementary Figure 1. Spearman correlation matrix of 24 microarrays samples. Raw log intensities were background subtracted, pre-filtered to exclude low expressers, and quantile normalized. Correlations seem to indicate moderate degrees of variance with a few sample outliers.
Supplementary Figure 2. (A) A 2-dimensional plot of the first two dimensions representing the variation from one sample to another showed a moderate ability to distinguish the three categories of time-from-injury indicating moderate to low signal to noise ratios between samples of each condition. (B) SA plot of fit of residual standard errors versus average expression showed a robustly trended fit of residual standard errors shows that Limma accounts for large mean-variance deviations that might otherwise obscure or confound results.
Supplementary Figure 3. RNA-seq alignment and count summary showed total sequenced reads, total number of aligned, reads total number of uniquely aligned reads, total number of uniquely aligned unambiguous reads to genes as determined by Subread:featureCounts, the total number of estimated reads aligned to transcripts as estimated by Sailfish, the fraction of uniquely aligned unambiguous reads to ribosomal and mitochondrial ribosomal genes, and a histogram of gene and transcript counts at 5, 50, and 100 counts-per-million mapped reads. Quality assurance shows that all samples pass minimum thresholds of 20 million reads per sample and total percent of aligned reads greater than 85 percent.
Supplementary Figure 4. (A) Junction saturation plot of known exon-exon junctions for 8 RNA-seq samples derived by RSeQC. All known exon-exon junctions annotated by GRCh38.76 were queried for the number of spanning reads at progressive levels of random sampling from the STAR aligned bam files. As sampling approaching 100%, ideal samples will plateau indicating sufficient sequencing depth to interrogate all known transcripts. (B) Coverage plot of all known genes normalized to 100 bp gene models derived by RSeQC. All genes annotated by GRCh38.76 were normalized to 100 base-pair theoretical models and queried for the depth and position of aligned reads within the normalized models. The plots illustrate the nature of the SeqPlex library kit’s ability to amplify and prepare libraries from even low mass samples to produce RNA-seq data that has uneven but uniform coverage of genes across samples of varying conditions.
Supplementary Figure 5. (A) Spearman correlation matrix of the numbers of uniquely aligned reads to genes normalized to log2 counts-per-million (log2 CPM). Counts of uniquely aligned reads to GRCh38.76 annotated genes are enumerated by Subread:featureCounts and normalized to log2 CPM by EdgeR. The non-parametric spearman correlation of the normalized counts shows high correlation within time-from-injury conditions with moderate drop in correlation between conditions. (B) Multidimensional scaling plot of 8 RNA-seq samples. Distinct clusters with one outlier indicated the variation between groups was greater than those between biological replicates. (C) Tagwise and robustly trended dispersion plot of the empirically observed dispersions from the Poisson model created by EdgeR is also shown. Plot of biological coefficient of variation versus relative abundance in log2CPM. Variance decreases as abundance increases as would be expected under the negative binomial (over-dispersed Poisson) model.
Supplementary Figure 6. (A) Pie graph of the percentage of significant genes for known conditions and confounding factors for (A) RNA-seq and (B) microarrays (N = 8) data. Each coefficient of the fitted EdgeR generalized linear model yielded significant genes with P < 0.05. The percentage of genes respective to each condition and categorical factor was attributed as a known source of differential expression.
Supplementary Figure 7. Plots of log2 fold-change versus average expression values where genes that were found to be significant were highlighted in red. Significance stringency cutoff was platform and analysis specific. (A) RNA-seq dataset had an FDR adjusted p-value stringency cutoff of less than 0.01 and (B) The microarrays dataset has a cutoff of an FDR adjusted p-value less than 0.05. (C) A violin/box plot of the distribution of the observed log2 fold-changes across the two Lima microarrays analyses and single EdgeR RNA-seq dataset show that RNA-seq dataset has a much tighter and elongated range of observed log2 fold-changes implying that the RNA-seq data also exhibit signs of less log2 fold-change compression. (D) Additional examination of the rank ordered observed p-values across all three datasets illustrates that the RNA-seq data not only exhibit less signs of log2 fold-change compression, but also higher degrees of statistical sensitivity.
Acknowledgments
We thank Washington University Genome Technology Access Center for help with transcriptome and quantitative PCR assays.
Funding
These studies were supported by these grants from the National Institutes of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health (NIH): R01AR063757 (PI: L. J. Sandell), P30-AR057235 (Musculoskeletal Research Center, PI: L. J. Sandell), 1K99AR064837 (PI: M. F. Rai). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the NIAMS.
Footnotes
Competing interests
No competing interests exist for the current study.
Financial conflict of interest
L. J. Sandell owns stock or stock options in ISTO Technologies and receives royalties from Merck/Millipore for a type IIA collagen N-propeptide enzyme-linked immunosorbent assay. L. J. Sandell is Editor in Chief of Journal of Orthopaedic Research. R. H. Brophy, E. D. Tycksen, and M. F. Rai have nothing to disclose.
Author contributions
Study design: M. F. Rai, R. H. Brophy, and L. J. Sandell
Study conduct: M. F. Rai, E. D. Tycksen and R. H. Brophy
Data analysis: M. F. Rai, and E. D. Tycksen
Manuscript writing: M. F. Rai, E. D. Tycksen, L. J. Sandell and R. H. Brophy
References
- 1.Kerr G, Ruskin HJ, Crane M, et al. Techniques for clustering gene expression data. Computers in biology and medicine. 2008;38:283–293. doi: 10.1016/j.compbiomed.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Passador-Gurgel G, Hsieh WP, Hunt P, et al. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet. 2007;39:264–268. doi: 10.1038/ng1944. [DOI] [PubMed] [Google Scholar]
- 3.Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003;22:6497–6507. doi: 10.1038/sj.onc.1206865. [DOI] [PubMed] [Google Scholar]
- 4.Sigurgeirsson B, Emanuelsson O, Lundeberg J. Sequencing degraded RNA addressed by 3' tag counting. PLoS One. 2014;9:e91851. doi: 10.1371/journal.pone.0091851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallego Romero I, Pai AA, Tung J, et al. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 2014;12:42. doi: 10.1186/1741-7007-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen EA, Souaiaia T, Herstein JS, et al. Effect of RNA integrity on uniquely mapped reads in RNA-Seq. BMC Res Notes. 2014;7:753. doi: 10.1186/1756-0500-7-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S, Fung-Leung WP, Bittner A, et al. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber M, Grabherr MG, Guttman M, et al. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 10.Brophy RH, Tycksen ED, Sandell LJ, et al. Changes in Transcriptome-Wide Gene Expression of Anterior Cruciate Ligament Tears Based on Time From Injury. Am J Sports Med. 2016;44:2064–2075. doi: 10.1177/0363546516643810. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Gong B, Bushel PR, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32:926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trost B, Moir CA, Gillespie ZE, et al. Concordance between RNA-sequencing data and DNA microarray data in transcriptome analysis of proliferative and quiescent fibroblasts. R Soc Open Sci. 2015;2:150402. doi: 10.1098/rsos.150402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rai MF, Sandell LJ, Cheverud JM, et al. Relationship of age and body mass index to the expression of obesity and osteoarthritis-related genes in human meniscus. Int J Obes (Lond) 2013;37:1238–1246. doi: 10.1038/ijo.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durinck S, Moreau Y, Kasprzyk A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 15.Rai MF, Patra D, Sandell LJ, et al. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis Rheum. 2013;65:2090–2101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai MF, Patra D, Sandell LJ, et al. Relationship of gene expression in the injured human meniscus to body mass index: a biologic connection between obesity and osteoarthritis. Arthritis Rheumatol. 2014;66:2152–2164. doi: 10.1002/art.38643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai MF, Sandell LJ, Zhang B, et al. RNA Microarray Analysis of Macroscopically Normal Articular Cartilage from Knees Undergoing Partial Medial Meniscectomy: Potential Prediction of the Risk for Developing Osteoarthritis. PLoS One. 2016;11:e0155373. doi: 10.1371/journal.pone.0155373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 20.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32:462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014;42:e91. doi: 10.1093/nar/gku310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway SJ, Izuhara K, Kudo Y, et al. The role of periostin in tissue remodeling across health and disease. Cell Mol Life Sci. 2014;71:1279–1288. doi: 10.1007/s00018-013-1494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attur M, Yang Q, Shimada K, et al. Elevated expression of periostin in human osteoarthritic cartilage and its potential role in matrix degradation via matrix metalloproteinase-13. FASEB J. 2015;29:4107–4121. doi: 10.1096/fj.15-272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chijimatsu R, Kunugiza Y, Taniyama Y, et al. Expression and pathological effects of periostin in human osteoarthritis cartilage. BMC Musculoskelet Disord. 2015;16:215. doi: 10.1186/s12891-015-0682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeser RF, Olex AL, McNulty MA, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012;64:705–717. doi: 10.1002/art.33388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morra L, Moch H. Periostin expression and epithelial-mesenchymal transition in cancer: a review and an update. Virchows Arch. 2011;459:465–475. doi: 10.1007/s00428-011-1151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanitz A, Gypas F, Gruber AJ, et al. Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 2015;16:150. doi: 10.1186/s13059-015-0702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X, Fu N, Guo S, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottomly D, Walter NA, Hunter JE, et al. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One. 2011;6:e17820. doi: 10.1371/journal.pone.0017820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Ferguson J, Ng SM, et al. Effector CD4+ T cell expression signatures and immune-mediated disease associated genes. PLoS One. 2012;7:e38510. doi: 10.1371/journal.pone.0038510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirbu A, Kerr G, Crane M, et al. RNA-Seq vs dual- and single-channel microarray data: sensitivity analysis for differential expression and clustering. PLoS One. 2012;7:e50986. doi: 10.1371/journal.pone.0050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome biology. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law CW, Chen Y, Shi W, et al. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium SM-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloom JS, Khan Z, Kruglyak L, et al. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burbidge HM, Goulden BE, Jones BR. An experimental evaluation of castellated laryngofissure and bilateral arytenoid lateralisation for the relief of laryngeal paralysis in dogs. Australian veterinary journal. 1991;68:268–272. doi: 10.1111/j.1751-0813.1991.tb03239.x. [DOI] [PubMed] [Google Scholar]
- 41.Bradford JR, Hey Y, Yates T, et al. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010;11:282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC biology. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 44.Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 45.Allison DB, Cui X, Page GP, et al. Microarray data analysis: from disarray to consolidation and consensus. Nature reviews Genetics. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 46.Draghici S, Khatri P, Eklund AC, et al. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katz Y, Wang ET, Airoldi EM, et al. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Spearman correlation matrix of 24 microarrays samples. Raw log intensities were background subtracted, pre-filtered to exclude low expressers, and quantile normalized. Correlations seem to indicate moderate degrees of variance with a few sample outliers.
Supplementary Figure 2. (A) A 2-dimensional plot of the first two dimensions representing the variation from one sample to another showed a moderate ability to distinguish the three categories of time-from-injury indicating moderate to low signal to noise ratios between samples of each condition. (B) SA plot of fit of residual standard errors versus average expression showed a robustly trended fit of residual standard errors shows that Limma accounts for large mean-variance deviations that might otherwise obscure or confound results.
Supplementary Figure 3. RNA-seq alignment and count summary showed total sequenced reads, total number of aligned, reads total number of uniquely aligned reads, total number of uniquely aligned unambiguous reads to genes as determined by Subread:featureCounts, the total number of estimated reads aligned to transcripts as estimated by Sailfish, the fraction of uniquely aligned unambiguous reads to ribosomal and mitochondrial ribosomal genes, and a histogram of gene and transcript counts at 5, 50, and 100 counts-per-million mapped reads. Quality assurance shows that all samples pass minimum thresholds of 20 million reads per sample and total percent of aligned reads greater than 85 percent.
Supplementary Figure 4. (A) Junction saturation plot of known exon-exon junctions for 8 RNA-seq samples derived by RSeQC. All known exon-exon junctions annotated by GRCh38.76 were queried for the number of spanning reads at progressive levels of random sampling from the STAR aligned bam files. As sampling approaching 100%, ideal samples will plateau indicating sufficient sequencing depth to interrogate all known transcripts. (B) Coverage plot of all known genes normalized to 100 bp gene models derived by RSeQC. All genes annotated by GRCh38.76 were normalized to 100 base-pair theoretical models and queried for the depth and position of aligned reads within the normalized models. The plots illustrate the nature of the SeqPlex library kit’s ability to amplify and prepare libraries from even low mass samples to produce RNA-seq data that has uneven but uniform coverage of genes across samples of varying conditions.
Supplementary Figure 5. (A) Spearman correlation matrix of the numbers of uniquely aligned reads to genes normalized to log2 counts-per-million (log2 CPM). Counts of uniquely aligned reads to GRCh38.76 annotated genes are enumerated by Subread:featureCounts and normalized to log2 CPM by EdgeR. The non-parametric spearman correlation of the normalized counts shows high correlation within time-from-injury conditions with moderate drop in correlation between conditions. (B) Multidimensional scaling plot of 8 RNA-seq samples. Distinct clusters with one outlier indicated the variation between groups was greater than those between biological replicates. (C) Tagwise and robustly trended dispersion plot of the empirically observed dispersions from the Poisson model created by EdgeR is also shown. Plot of biological coefficient of variation versus relative abundance in log2CPM. Variance decreases as abundance increases as would be expected under the negative binomial (over-dispersed Poisson) model.
Supplementary Figure 6. (A) Pie graph of the percentage of significant genes for known conditions and confounding factors for (A) RNA-seq and (B) microarrays (N = 8) data. Each coefficient of the fitted EdgeR generalized linear model yielded significant genes with P < 0.05. The percentage of genes respective to each condition and categorical factor was attributed as a known source of differential expression.
Supplementary Figure 7. Plots of log2 fold-change versus average expression values where genes that were found to be significant were highlighted in red. Significance stringency cutoff was platform and analysis specific. (A) RNA-seq dataset had an FDR adjusted p-value stringency cutoff of less than 0.01 and (B) The microarrays dataset has a cutoff of an FDR adjusted p-value less than 0.05. (C) A violin/box plot of the distribution of the observed log2 fold-changes across the two Lima microarrays analyses and single EdgeR RNA-seq dataset show that RNA-seq dataset has a much tighter and elongated range of observed log2 fold-changes implying that the RNA-seq data also exhibit signs of less log2 fold-change compression. (D) Additional examination of the rank ordered observed p-values across all three datasets illustrates that the RNA-seq data not only exhibit less signs of log2 fold-change compression, but also higher degrees of statistical sensitivity.