Abstract
Eosinophilic esophagitis (EoE) is an emerging disease that is distinguished from gastroesophageal reflux disease (GERD) by the expression of a unique esophageal transcriptome and the interplay of early life environmental factors with distinct genetic susceptibility elements at 5q22 (TSLP) and 2p23 (CAPN14). Rare genetic syndromes have uncovered the contribution of barrier disruption, mediated in part by defective desmosomes and dysregulated transforming factor beta (TGF-β) production and signaling, to EoE pathophysiology. Experimental modeling has defined a cooperative role of activated eosinophils, mast cells, and the cytokines IL-5 and IL-13, mediated by allergic sensitization to multiple foods. Understanding these processes is opening the way to better treatment based on disrupting allergic inflammatory and T helper type 2 cytokine–mediated responses including anti-cytokine therapeutics and dietary therapy.
Keywords: Allergy, Desmosome, Genetics, Inflammation
Eosinophilic esophagitis (EoE) is a chronic, T helper type 2 (Th2)–associated inflammatory disease characterized by predominant and marked eosinophilic inflammation of the esophagus (a peak count of ≥15 eosinophils per high-power field of esophageal biopsy tissue [eos/hpf]); the diagnosis has been traditionally limited to patients who have persistent esophageal eosinophilia after a documented proton-pump inhibitor (PPI) trial1 but it has recently been recommended that PPI responsiveness is not part of the diagnostic criteria but rather an appropriate and effective treatment for some patients (Molina-Infante J Gut 2016 and Lucendo United European Gastro Journal 2017). The disease is associated with upper gastrointestinal symptoms that vary with age and can include fibrostenotic complications. EoE is triggered by allergen exposure, typically food allergens, and is responsive to topical glucocorticoids and dietary elimination therapy (Figure 1). The pathogenesis of EoE is being extensively studied, and there have been recent advances concerning the genetic and environmental contributors, as well as the cellular and molecular etiology. This has led to numerous new therapies targeting these molecular pathways, which are currently being tested for disease treatment. Herein, we will focus on recent advances concerning the pathogenesis of EoE.
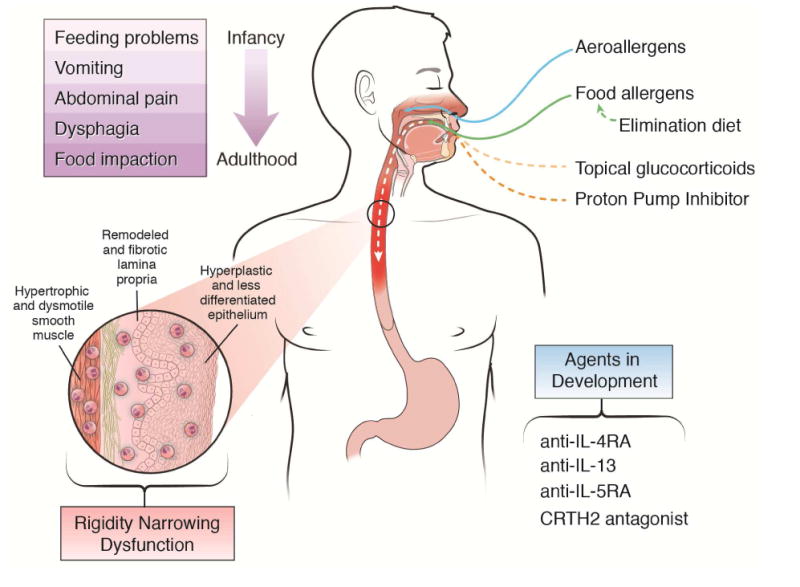
Figure 1. Clinical, pathologic, and therapeutics of EoE.
Allergens drive eosinophilic esophagitis (EoE); however, current (glucocorticoid and dietary therapy) and future interventions can treat the disease. The presenting symptoms are shown, leading to esophageal inflammation, remodeling, rigidity, and dysfunction.
Genetic etiology
The prevalence of EoE is approximately 1/2000 and has a known male predominance, with a male-to-female ratio approaching 3:1.2,3 EoE has a strong hereditability pattern, with familial associations having relative risk ratios as high as 64-fold amongst brothers.4 Proband concordinace in monozygotic twins is 58%, substantiating a genetic etiology.5 Several different studies, including candidate-gene identification and genome-wide association studies (GWAS) have identified multiple genes that are likely contributing to the development of EoE. These genes include thymic stromal lymphopoietin (TSLP), calpain 14 (CAPN14), EMSY, LRRC32, STAT6 and ANKRD27 (Table 1). However, it is important to note that dizygotic twins have a 36% concordance, whereas non-twin siblings have a 2.4% concordance; the stark difference demonstrates the substantial influence of a shared twin environment, likely via epigenetic mechanisms, at least partially.5 Consistent with this, the strongly associated EoE genes CCL26 (encoding eotaxin-3, a potent eosinophil chemoattractant and activating factor induced by IL-3) and CAPN14 (encoding CAPN14) are under epigenetic regulation.6,7
Table 1. EoE genetic risk loci (statistically significant and replicated).
| Genetic risk loci | Genes encoded | Odds ratio for most associated SNP at each locus | Genetic mechanism | Pathogenic mechanism |
|---|---|---|---|---|
| 2p23 | CAPN14 | 1.98 | Promoter variant leads to genotype-dependent expression of CAPN14, likely involving epigenetic mechanism | CAPN14 is a proteolytic enzyme specific to the esophagus that is induced by IL-13 and involved in epithelial homeostasis and repair |
| 5q22 | TSLP WDR36 | 0.74 | Multiple risk alleles associated with genotype-dependent expression of TSLP | TSLP induces Th2 cell development and activates eosinophils and basophils |
| 11q13 | LRRC32 EMSY | 2.49 | Not yet described | LRRC32 is a TGF-beta binding protein. EMSY is involved in transcriptional regulation. |
| 12q13 | STAT6 | 1.5 | STAT6 is the primary mediator of IL-4 and IL-13 signaling | STAT6 is a downstream signaling mediator of IL-4Rα and important for Th2 development |
| 19q13 | ANKRD27 PDCD5 RGS9BP | 1.6 | Not yet described | ANKRD27 inhibits the SNARE complex; PDCD5 is involved in apoptotic pathways. RGS9BP is not expressed in the esophagus or by immune cells. |
Abbreviations: EoE, eosinophilic esophagitis; SNP, single-nucleotide polymorphism; TSLP, thymic stromal lymphopoietin. Risk shown is positive and hence adjusted for being a common or rare allele.
A section of the human genome, known as the EoE transcriptome, has a conserved expression in the esophagus of patients with EoE; this region is not dysregulated in patients with gastroesophageal reflux disease (GERD).8 The most highly expressed gene, compared to controls, is the IL-13–induced gene CCL26.8,9 The EoE transcriptome is distributed throughout the genome, but the strongest “hot spot” for transcriptional changes occurs at 1q21, which encodes for the epidermal differentiation complex (EDC). This region contains genes that are involved in squamous epithelial cell differentiation, such as filaggrin; these genes are notably downregulated in EoE, consistent with a loss of epithelial cell differentiation and impaired barrier function.10,11
GWAS have identified TSLP as a major candidate gene. TSLP is released by activated epithelial cells and has an important role in promoting Th2 differentiation by inducing the Th2-polarizing capacity of dendritic cells.12 Levels of TSLP are significantly higher in patients with atopic diseases, including EoE.13 CAPN14 encodes a proteolytic enzyme that is specific to the esophagus and is induced by IL-13.14 Unlike TSLP, which is associated with multiple allergic disorders, CAPN14 may account for the tissue specificity of esophageal disease in EoE, as CAPN14 invokes a pathway that alters basic epithelial cell functions including barrier integrity.14 STAT6 has been shown to be important for Th2 development and is a signaling intermediate for IL-4 and IL-13 post IL-4 receptor alpha (IL-4Ra) engagement. LRRC32 is a TGF-beta binding protein, and EMSY is involved in transcriptional regulation.
Taken together, this genetic profile supports a microenvironment predisposed to develop allergic and eosinophilic inflammation of the esophagus (Table 1). It is notable that the genes implicated in EoE are distinct from those involved in GERD and inflammatory bowel disease and are much closer to those involved in allergen sensitization and squamous epithelial cell dysfunction.
Allergic milieu predisposes to eosinophilic inflammation
Multiple lines of evidence support an allergic etiology as an underlying mechanism for EoE. First, patients with EoE have a high incidence of concurrent atopic disease (Figure 1).3 Recent evidence shows that EoE is correlated with higher rates of asthma and airway hyperresponsiveness.15 Sensitization to cutaneous, ingested, and/or inhaled allergens is likely necessary in the development of EoE, and in some patients, seasonal allergens may play a role.16–18 Second, the success of dietary antigen elimination has provided profound insights into the role of food allergens in EoE. Removing the 6 most common food allergens leads to clinicopathologic remission in 50–75% of children and adults, and further studies and clinical experiences reveal that adding some of these allergens back into the diet leads to reoccurrence of the mucosal eosinophilia.19,20 Third, murine models using sensitization and challenges with oral ovalbumin, peanut, or inhaled aspergillus or dust mite antigen lead to IL-5–, IL-13–, and eotaxin-dependent esophageal eosinophilia.16,21–24
The role of Th2 cytokines remains central to our understanding of EoE. Early mouse studies revealed the important nature of IL-5, a required interleukin for eosinophilopoiesis, in driving mucosal esophageal eosinophilia and in potentially mediating tissue remodeling. The clinical relevance of IL-5 is partially underscored by studies that revealed humanized anti–IL-5 antibodies significantly, but not completely, reduced esophageal eosinophilia compared to placebo; however, there was no difference in clinical symptoms between individuals treated with anti–IL-5 antibody or a placebo, suggesting an incomplete effect. Additional strategies to inhibit IL-5–induced eosinophilia include the use of the eosinophil-depleting IL-5Rα antibody benralizumab, though this has not yet been formally studied in EoE.25
Later studies determined the critical nature of IL-13 to EoE. IL-13 is well recognized for its role in other atopic diseases, such as asthma, atopic dermatitis, and chronic sinusitis; in both basic and clinical studies, IL-13 is shown to contribute to eosinophil chemotaxis, goblet cell hyperplasia, collagen deposition, and smooth muscle contractility. In patients with EoE, the esophagus expresses elevated levels of IL-13, and IL-13–overexpressing transgenic mice develop an EoE-like inflammatory response in the esophagus, which has an esophageal transcriptome that partially overlaps with the EoE transcriptome.26 IL-13 also promotes EoE-like changes by promoting eosinophil recruitment by inducing eosinophil-activating chemokines such as eotaxin 3; by inducing tissue remodeling, including collagen deposition and angiogenesis. IL-13 also disrupts the epithelial barrier via a mechanism involving downregulation of the desmosomal protein desmoglein 1 (DSG1); preliminary studies have shown that this process is CAPN14 dependent, as CAPN14 is markedly induced by IL-13 and dysregulated expression of CAPN14 impairs epithelial architecture and barrier formation, including promotion of DSG1 degradation.27 The first clinical trial of a monoclonal antibody against IL-13 showed improved markers of esophageal barrier function and tissue remodeling and decreased inflammation.28 The results of a phase 2 study of a different anti–IL-13 antibody showed a marked decrease in esophageal eosinophilia, including in cases that were previously non-responsive to steroids; in this study, endoscopic severity and symptoms also improved with active therapy.29 Along these lines, blocking IL-4Rα (dupilumab) may prove useful in EoE, as this strategy has shown benefit in prospective trials for atopic dermatitis; a phase 2 study of this agent in EoE is ongoing (clinicaltrials.gov NCT02379052) (Table 2).
Table 2. Emerging Therapies in EoE.
| Emerging Therapies | Type | Mechanism |
|---|---|---|
| RPC4046 | Anti–IL-13 antibody | IL-13 regulates multiple genes within the EoE transcriptome including eotaxin 3, desmoglein 1, periostin, and filaggrin |
| OC000459 | CRTH2 inhibitor | CRTH2 is important for chemotaxis of eosinophils |
| Reslizumab/Mepolizumab | Anti–IL-5 antibody | IL-5 specifically stimulates expansion of eosinophils |
| Dupilumab | Anti–IL-4Rα antibody | IL-4rα is a high-affinity receptor for IL-4, which induces Th2 cell differentiation |
| Benralizumab | Anti–IL-5Rα antibody | IL-5Rα is the high-affinity receptor for IL-5, which stimulates expansion of eosinophils |
Abbreviations: CRTH2, chemoattractant receptor-homologous molecule
Increasing evidence suggests that IgE does not have a prominent role in the pathogenesis of EoE. Though patients with EoE have increased levels of food-specific IgE compared to control individuals, the level of food-specific IgE is only relatively modestly increased in patients with EoE compared with patients with food anaphylaxis. In addition, elevated serum food-specific IgE does not necessarily predict EoE-triggering foods.30 Consistent with these findings, anti-IgE therapy (omalizumab) in humans was neither effective in reducing levels of esophageal eosinophilia nor clinical symptoms in clinical trials.31,32 Recent evidence suggests a potential role for tissue-resident IgG4, including total and food antigen–specific IgG4, in the pathogenesis of EoE. Immunohistochemical analysis of esophageal mucosal biopsies from adult subjects revealed IgG4 staining only in those with active EoE and not controls.32 Interesting, cases of EoE that respond to dietary treatment have elevated ratios of esophageal and plasma food-specific IgG4 and tissue values that decrease during disease remission.33 IgG4 is generally thought to be a neutralizing antibody as it only weakly binds to IgG receptors, does not fix complement or engage antibody-dependent cellular cytotoxicity, and undergoes Fab-arm exchange and hence has limited ability to cross-link receptors.34 It is interesting to speculate that EoE may be part of a spectrum of IgG4-related diseases, which often involve extensive, eosinophil-associated tissue-remodeling processes.35
Th2 cytokines are likely produced by the recently described pathogenic effector Th2 cells (peTH2 cells), which were identified at higher numbers in the blood of patients with EoE compared to control individuals.36–37 These peTH2 cells are chemoattractant receptor-homologous molecule–positive (CRTH2+), hematopoietic prostaglandin D synthase–positive (HPSD+), and CD161high CD4 T cells.36–37 CRTH2 is present on peTH2 cells, eosinophils, and basophils and is involved in the chemotaxis of these cells via its response to prostaglandin D2. A recent clinical trial in patients with severe EoE with a CRTH2 antagonist demonstrated a statistically significant decrease, but not complete resolution, in esophageal eosinophilic inflammation.38 In addition, group 2 innate lymphocytes that are capable of expressing IL-5, -9, and -13 have been shown to be elevated in active EoE and to correlate with the degree of esophageal eosinophilia.39 Another cell type that has been shown to be a source of Th2 cytokines in EoE, at least in murine models of EoE, is the invariant natural killer (iNKT) cell and their depletion attenuates experimental EoE, highlighting the potential importance of this cell as a contributor and therapeutic target.40–43 Mucosal mast cells influx and degranulate into the EoE esophageal epithelium and resolve following successful therapy.44,45 In contrast, it appears that subepithelial connective tissue mast cells are relatively static in the esophagus. Murine models demonstrate that mast cells increase smooth muscle mass.46 In addition, via producing pro-fibrotic factors such as TGF-β1, mast cells likely play a role in esophageal remodeling.47,48 A new IL-9–producing mucosal mast cell (MMC9s) described in immediate hypersensitivity may play a role in EoE, but this remains to be explored.49
A recent advance in treating IgE-mediated food allergy is oral immunotherapy (OIT). However, 10-20% of cases will fail OIT due to recurrent gastrointestinal symptoms.50 Recent meta-analysis has shown that EoE is observed in about 3% of esophageal biopsies from patients with OIT and abdominal symptoms were seen in 8-15%.51,52 In addition, recent examination of patients with IgE-mediated food allergy has shown that EoE occurs in 4.7% compared to 0.4% in the general population, indicating a link between atopic phenotypes and EoE.52 Since patients have a tendency to drop out of IgE desensitization trials due to abdominal pain and without undergoing esophageal biopsy, it is possible that these rates are underestimates. Conversely, because the patients are highly atopic, they might also have pre-existing undiagnosed or sub-clinical EoE that is exacerbated by the immunotherapy.53 The link between OIT and EoE provides insight about the underlying pathoetiology, which undoubtedly involves food antigen–driven adaptive immune responses that involve the interplay of IgE-mediated responses (e.g., IL-4), EoE-mediated responses (e.g., IL-5 and IL-13), and checkpoints such as IgG4 and likely T regulatory cells (Figure 2).
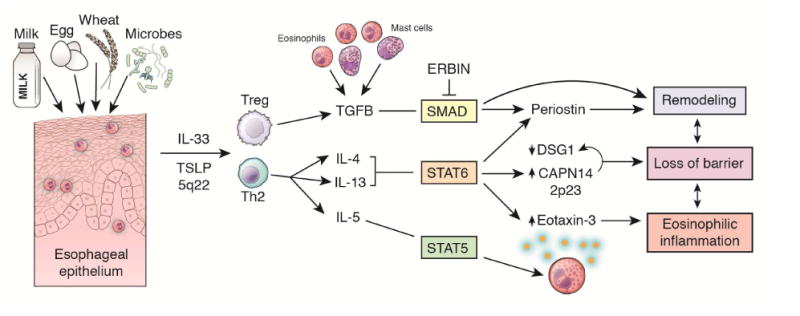
Figure 2. Pathophysiologic overview of EoE.
Environmental factors, including foods and the microbiome, interact with the esophageal epithelium to elicit production of the pro-atopy cytokines IL-33 and TSLP. Activated T regulatory and Th2 cells secrete bioactive cytokines including TGF-β, IL-4, IL-13, and IL-5, which elicit barrier disruption, tissue remodeling, and eosinophilic inflammation.
Microbial imbalance may contribute to esophagitis
Similar to patterns in other atopic diseases, an emerging body of evidence suggests a role for intestinal dysbiosis in the pathogenesis of EoE. Host commensal populations may be skewed toward a Th2 profile by early life events such as Caesarian section delivery and antibiotic exposure during infancy, which appear to increase the risk of EoE in both children and adults.54–57 Of note, similar risk factors have been identified in other atopic conditions and inflammatory bowel disease.58
Although early investigations revealed only a few bacterial populations in the esophagus, culture-independent techniques uncovered a microbial content with over 300 species. In an effort to define the role of bacteria in EoE, two recent investigations determined esophageal microbial patterns in children and adults with EoE. Collectively, the results revealed striking differences, as well as many similarities, between the oral and esophageal cavities. For instance, the Prevotella and Streptococcus genera were similar in the two sites, whereas the Firmicutes genus was increased in the esophagus.59 Comparisons between active EoE and normal controls revealed increased Proteobacteriae in subjects with active disease and Streptococus in controls.60 Ingesting EoE-triggering foods leads to changes in the esophageal microbiome, with the emergence of Granulicatella and Campylobacter genera on mucosal biopsies.59 An interesting observation relates to the association of herpes simplex viral infection with EoE. Case reports identify a preceding herpes simplex virus infection in some patients who go on to develop EoE; however, whether the viral infection and EoE development are related is presently unknown.61 The exact pathogenetic mechanisms that microbes contribute to the initiation, perpetuation, or even prevention of mucosal eosinophilia remain to be determined. It remains to be determined if esophageal eosinophilia leads to the changes in microbiome and/or if the dysbiosis influences EoE development.
Interestingly, commensal bacteria in a mouse model may limit food sensitization.62 In support of this is the observational finding of an inverse relationship between Helicobacter pylori infection and EoE, which has been demonstrated in several independent populations at different centers.63–66 This is intriguing as the rapid decrease in H. pylori prevalence over the past several decades matches the increase in EoE prevalence. From a mechanistic standpoint, H. pylori polarizes the immune system to a Th1 milieu, whereas lack of H. pylori results in a Th2 environment similar to what is seen in EoE.64,67 Despite this association, there have been no mechanistic studies that confirm a protective role of H. pylori. In addition, H. pylori infection is also protective against atopy but this effect wanes after childhood, indicating that the protective effect is complex, multifactorial and not EoE specific (Lionetti W J Gastro 2014 and Taye et al CEA and den Hollander ref).
Practical aspects of future human studies of the microbiome in EoE will need to focus on determining methods of collection of samples (e.g., mucosal biopsies or scrapings), specific host features (e.g., antibiotics, mouthwashes, PPIs), and the impact of EoE-related treatments. In addition, it is interesting to speculate that stool microbial content could have a diagnostic or monitoring role as a non-invasive tool for EoE.
Eosinophil transmigration and activation
The esophageal epithelium is composed of non-keratinized, stratified squamous epithelium that is bathed by a layer of mucus and covers the rete pegs with its vascular elements. Though the normal gastrointestinal tract contains eosinophils in varying density in times of good health, the normal esophageal mucosa does not contain any eosinophils. Thus, identifying eosinophils and their progenitor cells in the esophageal mucosa indicates a pathogenic role for these cells in an inflammatory response.68 In this realm, studies have elucidated pathogenic mechanisms related to eosinophil migration in EoE. Blanchard et al. identified that the CCL26 gene is the most upregulated gene in EoE human esophageal tissue.8 Eotaxin 3 acts through the G-protein coupled receptor CCR3, leading to eosinophil chemotaxis. Eotaxin-3 is upregulated by IL-13, a key EoE-related cytokine in vitro, and genetic deletion of the murine eotaxin receptor gene CCR3 leads to diminished esophageal eosinophilia in vivo. Periostin is also directly induced by IL-13 and promotes eosinophil adhesion and recruitment by direct and indirect mechanisms.69 It is notable that periostin is one of the top upregulated genes in the EoE transcriptome. Whether transmigration across the endothelial surface follows a different pathway than other diseases is not certain. Staining studies reveal increases in vascular cell adhesion molecule 1 and CD31 expression during active EoE. Immunostaining revealed decreased CD18 following topical corticosteroid treatment.70
Following transmigration across the epithelial space, eosinophil activation is evidenced by the intense pattern of eosinophil-derived granule protein deposition observed in active disease. Eosinophil-derived major basic protein (MBP), eosinophil-derived neurotoxin (EDN), eosinophil cationic protein (ECP), and eosinophil peroxidase (EPO) are not only deposited in the epithelium of patients with EoE but also may carry functional consequences relevant to EoE pathogenesis.71 For instance, EDN activates dendritic cells, which promote the Th2 cell response. ECP increases membrane permeability of target cells, EPO may perpetuate tissue-injuring peroxidase formation, and MBP can disrupt epithelial barrier.71–73 Translational studies have also shown that measurements of these proteins in gastrointestinal secretions may be indicative of EoE activity.74,75
Barrier dysfunction contributes to esophageal inflammation
The basal layer of esophageal tissues from patients with EoE becomes hyperplastic, and its integrity as a barrier structure is impaired as evidenced by dilated intercellular spaces and spongiosis.10 Using impedance monitoring, van Rhijn et al. demonstrated a significantly decreased esophageal barrier during active EoE compared to inactive EoE in adult subjects.76 Ussing chamber analyses identified similar findings with mucosal biopsies.10 Ultrastructural studies show not only increased intercellular spaces but also decreased junctional proteins. It is important to note, as discussed later, that inborn errors in barrier formation predispose to EoE, indicating that intrinsic defects are contributory, at least in rare cases. Barrier dysfunction is associated with impaired epithelial turnover as hyperplastic basal cells replace the normally differentiated epithelium, which exhibit a loss of tissue identity markers.77 This process is likely mediated by increased expression of follistatin, a natural inhibitor of bone morphogenetic protein (BMP) signaling, resulting in impaired basal progenitor cell differentiation.78 A dysfunctional barrier could facilitate passage of allergenic molecules to a genetically predisposed microenvironment.
Mechanistically, CAPN14 is overexpressed by the esophageal epithelia in patients with EoE, and IL-13 stimulation of esophageal epithelia results in impaired barrier function and overexpression of CAPN14. In vitro overexpression of CAPN14 in esophageal epithelial cells results in diminished barrier function and architectural changes indicative of barrier impairment, such as epidermal clefting and loss of the normal expression pattern of DSG1 and filaggrin,27 a cutaneous structural protein that is important to barrier integrity and downregulated by IL-13.79 Proof for the importance of this pathway in the pathoetiology of EoE and other atopic diseases is illustrated by the rare genetic disease, SAM (severe dermatitis, multiple allergies and metabolic wasting) syndrome, in which the desmosomal proteins DSG1 or desmoplankin (DSP) are disrupted due to homozygous loss-of-function mutations in either gene.80,81 Loss of DSG1 or DSP leads to weakened barrier function in squamous surfaces, providing an entry pathway for allergens. Further re-enforcing the importance of barrier function is the capability of IL-13 to downregulate formation of not only filaggrin, but also DSG1.80 Notably, DSG1 and filaggrin are downregulated in EoE, and gene silencing of DSG1 in esophageal epithelial cells is sufficient to induce many of the features of EoE including acantholysis and barrier impairment.10 In addition, patients with atopic dermatitis, which often is associated with loss of function mutations in filaggrin, exhibit this impaired barrier function, emphasizing the ability of barrier impairment to pre-dispose to atopic diseases of the squamous epithelium.79 Notably, disruptive mutations in filaggin are enriched in patients with EoE, independent of the presence of atopic dermatitis, indicating the direct involvement of filaggrin in EoE susceptibility.82
Esophageal fibrosis and strictures are a chronic feature of EoE
Emerging clinical evidence supports that the likely outcome of unbridled eosinophilia is esophageal fibrosis and stricture formation. Esophageal remodeling is defined by histologic parameters in the epithelium, including basal zone hyperplasia, dilated intercellular spaces, rete peg elongation, and desquamation, and by subepithelial lamina propria features such as increased vascularization and fibrosis.3,83–85 The confounding aspect of this pathogenic process is why remodeling sometimes directs mucosal healing without sequelae and other times goes on to develop clinically relevant long and short segment narrowing. In this regard, it is notable that CAPN14 has been implicated in eliciting and repairing the epithelial damage associated with EoE, suggesting that remodeling could involve a genetically controlled balance between these two processes.27,86 Epithelial products such as plasminogen activator inhibitor 1 (aka serpineE1) correlate with the severity of lamina propria fibrosis.85 Such epithelial markers of subepithelial fibrosis may be clinically valuable because the size and adequacy of esophageal tissue procurement for lamina propria evaluation is variable. In addition, features of epithelial mesenchymal transition with increased vimentin-positive epithelial cells have been documented as part of the remodeling process.87,88 In the lamina propria, potential markers of remodeling include the pro-fibrotic factor, transforming growth factor beta (TGF-β) and its signaling molecules pSMAD2 and pSMAD3.23,83
A validated endoscopic scoring system, EoE Endoscopic Reference Score (EREFS), characterizes the gross features thought to reflect remodeling, such as the presence and severity of strictures, rings, narrowing, and crepe paper esophagus.89 Additional features, such as the endoscopic “pull” sign, which occurs during biopsy procurement, has been reported as a sign of remodeling.90 Studies in children and adults have shown that that esophageal narrowing may be better captured by esophagram, whereas >50% of strictures may be missed by isolated endoscopic assessment.91 The ultimate consequence of esophageal remodeling and fibrosis is stiffening and dysmotility of a normally compliant tube capable of coordinated contractions that propel the food bolus distally.92,93 Dysmotility is alluded to by the fact that food impactions can occur in the absence of frank stricture formation. Using an endoscopic functional lumen imaging probe (EndoFLIP), strictured and non-strictured EoE esophagi have been shown to be more rigid than non-inflamed esophagi.94 In addition, the fibrostenotic esophagus is more prone to dysmotility.95 The natural history of untreated EoE in adults is to progressive fibrostenosis and preliminary pediatric studies show that esophageal rigidity begins in childhood.92,93 (Figure 1)
Using translational studies of pediatric esophageal biopsies and primary human esophageal cells, the presence of remodeling in both children and adults and the activation of the TGF-β pathway in EoE has been elucidated.83,87,96–98 TGF-β has a number of molecular consequences including increasing fibrotic gene expression, altering fibroblast phenotype to myofibroblast, and increasing esophageal smooth muscle cell contraction via the expression of contractile proteins, such as the sarcoendoplasmic reticulum protein phospholamban, and via the induction of periostin.47,69,84,85,98,99 Further, independent of inflammation, a rigid environment increases esophageal smooth muscle cell gene expression of phospholamban and collagen I and induces smooth muscle hypertrophy.100 Similar changes occur in EoE fibroblasts.98 These data suggest that the mechanical environment significantly alters structural cell function and document an inflammation-independent pathway for esophageal remodeling.
Whether remodeling and fibrosis can be reversed may depend on patient age and/or the duration of disease.93,101–106 It is clear that the subset of children who respond histologically to topical corticosteroid therapy can have improvements in histologic remodeling and that this can be sustained long term.107 In addition, epithelial mesenchymal transition can resolve following successful therapy.87 Adult data show that topical steroids can improve esophageal diameter and decrease food impactions, but whether histologic remodeling or the process of fibrostenosis can be uniformly reduced remains unclear.104,108 Indeed, the patient with the fibrostenotic esophagus is often the most challenging to treat.109 The fact that a rigid environment alone alters the function of esophageal structural cells coupled with the clinical observations of therapy-resistant disease underscores the importance of finding non-steroidal, remodeling-altering treatments.110
Association with other conditions provides insight into pathogenetic mechanisms
Notably, EoE has known associations with several genetic conditions (Table 3), particularly connective tissue disorders with hypermobility syndromes, such as Loeys-Dietz syndrome and Ehlers-Danlos syndrome, hypermobility type.111 A common denominator between these two conditions is the increased production and/or signaling of TGF-β, which is thought to lead to increased contractility of smooth muscle, tissue remodeling, and Th2 responses.47,84 Another condition associated with increased production of TGF-β is a loss-of-function mutation in ERBB2-interacting protein (ERBIN), a protein that negatively regulates TGF-β signaling.112 EoE is also associated with other syndromes including PTEN hamartoma tumor syndrome (PHTS), hyper-IgE syndromes, and SAM syndrome.80 EoE has also been associated with Netherton's syndrome, which is caused by autosomal dominant mutations in the protease inhibitor SPINK5, which are normally expressed in the skin.113 In addition, EoE has been associated with esophageal granular cell tumors; whether this is a disease association or a concerning consequence of EoE is not certain.114 Finally, EoE has been associated with a number of autoimmune conditions including Hashimoto's thyroiditis, rheumatoid arthritis, celiac disease, inflammatory bowel disease, combined variable immunodeficiency, multiple sclerosis, and Sjögren's syndrome.115 Table 3 summarizes the known Mendelian diseases associated with EoE and attempts to synthesize what we can learn from them.
Table 3. Mendelian diseases associated with EoE.
| Mendelian disease associated with EoE | Inheritance | Genetic mutation | Plausible etiologic mechanism |
|---|---|---|---|
| Hyper-IgE syndrome | Atopic dermatitis | Deleterious mutations in signal transducer and activator of transcription 3 (STAT3) | Dysregulated response to IL-6 and possibly IL-5 |
| Hyper-IgE syndrome | Allergic rhinitis | Loss-of-function mutations in dedicator of cytokinesis 8 (DOCK8) | Loss of T cell homeostasis; lack of durable secondary antibody response against specific antigens |
| Ehlers-Danlos syndrome, hypermobility type | Atopic dermatitis | Unknown – other subtypes of Ehlers-Danlos syndrome are caused by mutations in collagen genes | Disrupted joint and skin development; increased activity of transforming growth factor beta (TGF-β) due to altered binding by extracellular matrix |
| ERBIN Deficiency | Atopic dermatitis | Loss-of-function mutation in ERBB2-interacting protein (ERBIN) | Increased TGF-β pathway activation in T cells with increased Th2 responses |
| Loeys-Dietz syndrome (LDS) | Allergic rhinitis | Mutations in TGF-β receptors 1 and 2 (TGFBR1 and TGFBR2, respectively) | Enhanced TGF-β signaling |
| Netherton's syndrome | Allergic rhinitis | Loss-of-function mutations in skin protease inhibitor, kazal type 5 (SPINK5) | Unrestricted protease activity of kallikrein 5 and 7 (KLK5, KLK7) |
| PTEN hamartoma tumor syndrome (PHTS) | Atopic dermatitis | Mutations in phosphatase and tensin homolog (PTEN) | Inhibited regulation of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling pathway |
| Severe atopy syndrome associated with metabolic wasting (SAM) syndrome | Allergic rhinitis | Homozygous mutations in desmoglein 1 (DSG1) or desmoplankin (DSP) | Disrupted epithelial barrier |
Closing
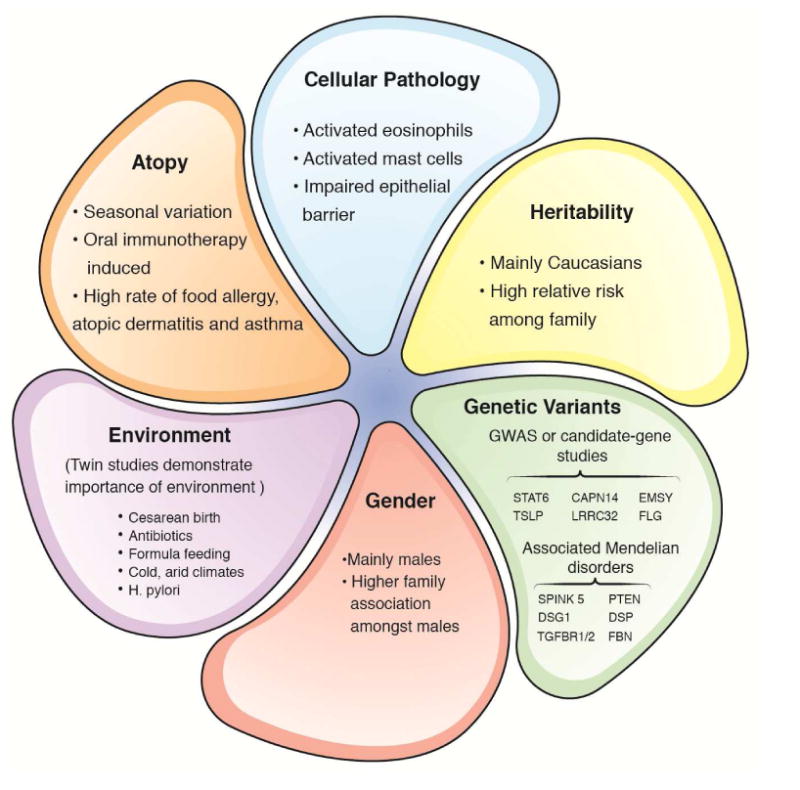
The recent formation of the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR), which is part of the Rare Diseases Clinical Research Network (RDCRN) of the NIH, will undoubtedly lead to better understanding and treatment of EoE and related rare eosinophil-associated gastrointestinal diseases. CEGIR is focused on defining the natural history of eosinophilic gastrointestinal disorders, developing disease criteria, identifying improved dietary intervention strategies, and training the next generation of clinical and research leaders in the field. In the short time since EoE has received attention (i.e., the last two decades), there has been much progress in understanding its pathogenic bases. We have furthered the perspective that EoE is unlike GERD and inflammatory bowel disease but rather closely aligned with an allergic etiology and thus involves the interplay of a different set of experts and clinical interventions than is typically encountered in the gastroenterology practice. Accordingly, this review article integrated the input of experts in gastroenterology and allergy, consistent with the composition of CEGIR. Collectively, we are establishing that EoE is a unique disease process, characterized by the expression of a marked esophageal transcriptome that provides deep insight into the effector phase of the disease. Transcriptome analysis from only a single biopsy has similar sensitivity and specificity as histologic characterization,116,117 and the routine usage of this test has the potential to transform clinical care.118,119 We have reviewed the likely importance of the interplay of early life environmental factors and distinct genetic susceptibility elements, with a focus on 5q22 (TSLP) and 2p23 (CAPN14), the two loci that have been genetically replicated and most studied in the context of EoE. We have shown that rare genetic syndromes can predispose to EoE and provide valuable insight into disease mechanisms that may not only be operational in the rare disease but also informative for the common patient. These studies have uncovered the contribution of barrier disruption, mediated in part by defective desmosomes and dysregulated TGF-β production and signaling. Experimental modeling has defined a cooperative role of activated eosinophils, mast cells, and the cytokines IL-5 and IL-13, likely mediated by allergic sensitization to multiple foods. Figure 3 synthesizes our understanding of the pathophysiology of EoE. Understanding these processes is opening the way to better treatment based on disrupting allergic inflammatory and Th2 cytokine– mediated responses including anti-cytokine therapeutics and dietary therapy.
Figure 3. Factors that contribute to the development of EoE.
Acknowledgments
This work was supported in part by NIH U19 AI070235, NIH R01 AI124355, R37 R37 A1045898, NIH K24DK100303, the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation, the Buckeye Foundation, and the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning. The study is also funded by U54 AI117804, which is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Disease Research (ORDR), NCATS, and is funded through collaboration between NCATS, NIAID and NIDDK, as well as the patient advocacy groups American Partnership for Eosinophilic Disorders (APFED), CURED and the Eosinophilic Family Coalition (EFC), which have collectively resulted in the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). We also thank Shawna Hottinger for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dellon ES, Gonsalves N, Hirano I, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE) Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 2.Mansoor E, Cooper GS. The 2010-2015 Prevalence of Eosinophilic Esophagitis in the USA: A Population-Based Study. Dig Dis Sci. 2016;61:2928–34. doi: 10.1007/s10620-016-4204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 4.Collins MH. Histopathologic features of eosinophilic esophagitis. Gastroenterology Clinics of North America. 2014;43:257–68. doi: 10.1016/j.gtc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Alexander ES, Martin LJ, Collins MH, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1084–92. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottyan LC, Davis BP, Sherrill JD, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286:13193–204. doi: 10.1074/jbc.M110.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard C, Mingler MK, Vicario M, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Sherrill JD, Kc K, Wu D, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–29. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017 Jan 17; doi: 10.1016/j.jaci.2016.11.042. pii: S0091-6749(17)30036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitajima M, Lee HC, Nakayama T, Ziegler SF. TSLP enhances the function of helper type 2 cells. Eur J Immunol. 2011;41:1862–71. doi: 10.1002/eji.201041195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui CC, Rusta-Sallehy S, Asher I, Heroux D, Denburg JA. The effects of thymic stromal lymphopoietin and IL-3 on human eosinophil-basophil lineage commitment: Relevance to atopic sensitization. Immun Inflamm Dis. 2014;2:44–55. doi: 10.1002/iid3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. The Journal of Allergy and Clinical Immunology. 2017 Jan 25; doi: 10.1016/j.jaci.2016.09.027. pii: S0091-6749(16)31215-5. doi:10 1016/j jaci 2016 09 027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krupp NL, Sehra S, Slaven JE, Kaplan MH, Gupta S, Tepper RS. Increased prevalence of airway reactivity in children with eosinophilic esophagitis. Pediatr Pulmonol. 2016;51:478–83. doi: 10.1002/ppul.23327. [DOI] [PubMed] [Google Scholar]
- 16.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–94. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Dellon ES, Gibbs WB, Fritchie KJ, et al. Clinical, endoscopic, and histologic findings distinguish eosinophilic esophagitis from gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2009;7:1305–13. doi: 10.1016/j.cgh.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen ET, Shah ND, Hoffman K, Sonnenberg A, Genta RM, Dellon ES. Seasonal variation in detection of oesophageal eosinophilia and eosinophilic oesophagitis. Aliment Pharmacol Ther. 2015;42:461–9. doi: 10.1111/apt.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arias A, Gonzalez-Cervera J, Tenias JM, Lucendo AJ. Efficacy of Dietary Interventions for Inducing Histologic Remission in Patients With Eosinophilic Esophagitis: A Systematic Review and Meta-analysis. Gastroenterology. 2014;146:1639–48. doi: 10.1053/j.gastro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Fogg MI, Ruchelli E, Spergel JM. Pollen and eosinophilic esophagitis. J Allergy Clin Immunol. 2003;112:796–7. doi: 10.1016/s0091-6749(03)01715-9. [DOI] [PubMed] [Google Scholar]
- 21.Rajavelu P, Rayapudi M, Moffitt M, Mishra A, Mishra A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G645–54. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayapudi M, Mavi P, Zhu X, et al. Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2010;88:337–46. doi: 10.1189/jlb.0110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho JY, Doshi A, Rosenthal P, et al. Smad3-deficient mice have reduced esophageal fibrosis and angiogenesis in a model of egg-induced eosinophilic esophagitis. Journal of pediatric gastroenterology and nutrition. 2014;59:10–6. doi: 10.1097/MPG.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan SP, Mishra A, Brandt EB, et al. A pathological function for eotaxin and eosinophils in eosinophilic gastrointestinal inflammation. Nat Immunol. 2001;2:353–60. doi: 10.1038/86365. [DOI] [PubMed] [Google Scholar]
- 25.Fulkerson PC, Rothenberg ME. Targeting eosinophils in allergy, inflammation and beyond. Nat Rev Drug Discov. 2013;12:117–29. doi: 10.1038/nrd3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo L, Fulkerson PC, Finkelman FD, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185:660–9. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis BP, Stucke EM, Khorki ME, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1:e86355. doi: 10.1172/jci.insight.86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135:500–7. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 29.Hirano ICM, Assouline-Dayan Y, Evans L, Gupta S, Schoepfer A, Straumann A, Grimm M, Smith H, Tomkins C, Woo A, Peach R, Frohna P, Gujrathi S, Aranda R, Dellon ES. A randomized, double-blind, placebo-controlled trial of a novel recombinant, humanized, anti-interleukin-13 monoclonal antibody (RPC4046) in patients with active eosinophilic esophagitis: Results of the HEROES study. United European Gastroenterology Journal. 2016;4 [Google Scholar]
- 30.Aceves SS. Food allergy testing in eosinophilic esophagitis: what the gastroenterologist needs to know. Clinical gastroenterology and hepatology: the official clinical practice Journal of the American Gastroenterological Association. 2014;12:1216–23. doi: 10.1016/j.cgh.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erwin EA, Tripathi A, Ogbogu PU, et al. IgE Antibody Detection and Component Analysis in Patients with Eosinophilic Esophagitis. J Allergy Clin Immunol Pract. 2015;3:896–904.e3. doi: 10.1016/j.jaip.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clayton F, Fang JC, Gleich GJ, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147:602–9. doi: 10.1053/j.gastro.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 33.Wright BL, Kulis M, Guo R, et al. Food-specific IgG4 is associated with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138:1190–2 e3. doi: 10.1016/j.jaci.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies AM, Sutton BJ. Human IgG4: a structural perspective. Immunol Rev. 2015;268:139–59. doi: 10.1111/imr.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao M, Liu M, Fan G, Yang X, Li J. Diagnostic Value of Serum IgG4 for IgG4-Related Disease: A PRISMA-compliant Systematic Review and Meta-analysis. Medicine (Baltimore) 2016;95:e3785. doi: 10.1097/MD.0000000000003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitson-Salazar A, Yin Y, Wansley DL, et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human T(H)2 cell subpopulation with enhanced function. J Allergy Clin Immunol. 2016;137:907–18.e9. doi: 10.1016/j.jaci.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Wen T, Rothenberg ME, Wang YH. Hematopoietic prostaglandin D synthase: Linking pathogenic effector CD4(+) T(H)2 cells to proeosinophilic inflammation in patients with gastrointestinal allergic disorders. J Allergy Clin Immunol. 2016;137:919–21. doi: 10.1016/j.jaci.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straumann A, Hoesli S, Bussmann C, et al. Anti-eosinophil activity and clinical efficacy of the CRTH2 antagonist OC000459 in eosinophilic esophagitis. Allergy. 2013;68:375–85. doi: 10.1111/all.12096. [DOI] [PubMed] [Google Scholar]
- 39.Doherty TA, Baum R, Newbury RO, et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J Allergy Clin Immunol. 2015;136:792–4. doi: 10.1016/j.jaci.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jyonouchi S, Smith CL, Saretta F, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014;44:58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayapudi M, Rajavelu P, Zhu X, et al. Invariant natural killer T-cell neutralization is a possible novel therapy for human eosinophilic esophagitis. Clin Transl Immunology. 2014;3:e9. doi: 10.1038/cti.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lexmond WS, Neves JF, Nurko S, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol. 2014;109:646–57. doi: 10.1038/ajg.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajavelu P, Rayapudi M, Moffitt M, Mishra A. Significance of para-esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302:G645–54. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 45.Dellon ES, Chen X, Miller CR, et al. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–71. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niranjan R, Mavi P, Rayapudi M, Dynda S, Mishra A. Pathogenic role of mast cells in experimental eosinophilic esophagitis. American journal of physiology Gastrointestinal and liver physiology. 2013;304:G1087–94. doi: 10.1152/ajpgi.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204 e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 48.Rawson R, Anilkumar A, Newbury RO, et al. The TGFbeta1 Promoter SNP C-509T and Food Sensitization Promote Esophageal Remodeling in Pediatric Eosinophilic Esophagitis. PloS one. 2015;10:e0144651. doi: 10.1371/journal.pone.0144651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CY, Lee JB, Liu B, et al. Induction of Interleukin-9-Producing Mucosal Mast Cells Promotes Susceptibility to IgE-Mediated Experimental Food Allergy. Immunity. 2015;43:788–802. doi: 10.1016/j.immuni.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldberg MR, Elizur A, Nachshon L, et al. Oral immunotherapy-induced gastrointestinal symptoms and peripheral blood eosinophil responses. J Allergy Clin Immunol. 2017;139:1388–90.e4. doi: 10.1016/j.jaci.2016.09.053. [DOI] [PubMed] [Google Scholar]
- 51.Lucendo AJ, Arias A, Tenias JM. Relation between eosinophilic esophagitis and oral immunotherapy for food allergy: a systematic review with meta-analysis. Ann Allergy Asthma Immunol. 2014;113:624–9. doi: 10.1016/j.anai.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Hill DA, Dudley JW, Spergel JM. The Prevalence of Eosinophilic Esophagitis in Pediatric Patients with IgE-Mediated Food Allergy. J Allergy Clin Immunol Pract. 2017;5:369–75. doi: 10.1016/j.jaip.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burk CM, Dellon ES, Steele PH, et al. Eosinophilic esophagitis during peanut oral immunotherapy with omalizumab. J Allergy Clin Immunol Pract. 2017;5:498–501. doi: 10.1016/j.jaip.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen ET, Kappelman MD, Kim HP, Ringel-Kulka T, Dellon ES. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;57:67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 55.Jensen ETSO, Koutlas NT, Chang A, Martin LJ, Rothenberg ME, Baron JA, Dellon ES. Early life factors are associated with risk for eosinophilic esophagitis diagnosed in adulthood. J Allergy Clinical Immunol. doi: 10.1093/dote/doaa074. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radano MC, Yuan Q, Katz A, et al. Cesarean section and antibiotic use found to be associated with eosinophilic esophagitis. J Allergy Clin Immunol Pract. 2014;2:475–7. doi: 10.1016/j.jaip.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 57.Slae M, Persad R, Leung AJ, Gabr R, Brocks D, Huynh HQ. Role of Environmental Factors in the Development of Pediatric Eosinophilic Esophagitis. Dig Dis Sci. 2015;60:3364–72. doi: 10.1007/s10620-015-3740-7. [DOI] [PubMed] [Google Scholar]
- 58.Jensen ET, Bertelsen RJ. Assessing Early Life Factors for Eosinophilic Esophagitis: Lessons From Other Allergic Diseases. Curr Treat Options Gastroenterol. 2016;14:39–50. doi: 10.1007/s11938-016-0083-1. [DOI] [PubMed] [Google Scholar]
- 59.Benitez AJ, Hoffmann C, Muir AB, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. doi: 10.1186/s40168-015-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dellon ES. The Esophageal Microbiome in Eosinophilic Esophagitis. Gastroenterology. 2016;151:364–5. doi: 10.1053/j.gastro.2016.06.026. [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann D, Criblez DH, Dellon ES, et al. Acute Herpes Simplex Viral Esophagitis Occurring in 5 Immunocompetent Individuals With Eosinophilic Esophagitis. ACG Case Rep J. 2016;3:165–8. doi: 10.14309/crj.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stefka AT, Feehley T, Tripathi P, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–50. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnenberg A, Dellon ES, Turner KO, Genta RM. The influence of Helicobacter pylori on the ethnic distribution of esophageal eosinophilia. Helicobacter. 2016 Dec 28; doi: 10.1111/hel.12370. doi:101111/hel12370 2016. [DOI] [PubMed] [Google Scholar]
- 64.Dellon ES, Peery AF, Shaheen NJ, et al. Inverse association of esophageal eosinophilia with Helicobacter pylori based on analysis of a US pathology database. Gastroenterology. 2011;141:1586–92. doi: 10.1053/j.gastro.2011.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuta K, Adachi K, Aimi M, et al. Case-control study of association of eosinophilic gastrointestinal disorders with Helicobacter pylori infection in Japan. J Clin Biochem Nutr. 2013;53:60–2. doi: 10.3164/jcbn.13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Arnim U, Wex T, Link A, et al. Helicobacter pylori infection is associated with a reduced risk of developing eosinophilic oesophagitis. Aliment Pharmacol Ther. 2016;43:825–30. doi: 10.1111/apt.13560. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167:821–7. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 68.Morris DW, Stucke EM, Martin LJ, et al. Eosinophil progenitor levels are increased in patients with active pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138:915–8. doi: 10.1016/j.jaci.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blanchard C, Mingler MK, McBride M, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lingblom C, Bergquist H, Johnsson M, et al. Topical corticosteroids do not revert the activated phenotype of eosinophils in eosinophilic esophagitis but decrease surface levels of CD18 resulting in diminished adherence to ICAM-1, ICAM-2, and endothelial cells. Inflammation. 2014;37:1932–44. doi: 10.1007/s10753-014-9926-x. [DOI] [PubMed] [Google Scholar]
- 71.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–74. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 72.Yang D, Chen Q, Su SB, et al. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J Exp Med. 2008;205:79–90. doi: 10.1084/jem.20062027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bystrom J, Amin K, Bishop-Bailey D. Analysing the eosinophil cationic protein--a clue to the function of the eosinophil granulocyte. Respir Res. 2011;12:10. doi: 10.1186/1465-9921-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subbarao G, Rosenman MB, Ohnuki L, et al. Exploring potential noninvasive biomarkers in eosinophilic esophagitis in children. J Pediatr Gastroenterol Nutr. 2011;53:651–8. doi: 10.1097/MPG.0b013e318228cee6. [DOI] [PubMed] [Google Scholar]
- 75.Masterson JC, McNamee EN, Fillon SA, et al. Eosinophil-mediated signalling attenuates inflammatory responses in experimental colitis. Gut. 2015;64:1236–47. doi: 10.1136/gutjnl-2014-306998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Rhijn BD, Weijenborg PW, Verheij J, et al. Proton pump inhibitors partially restore mucosal integrity in patients with proton pump inhibitor-responsive esophageal eosinophilia but not eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2014;12:1815–23.e2. doi: 10.1016/j.cgh.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 77.Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang M, Ku WY, Zhou Z, et al. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 2015;125:1557–68. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Regan GM, Sandilands A, McLean WH, Irvine AD. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2008;122:689–93. doi: 10.1016/j.jaci.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Samuelov L, Sarig O, Harmon RM, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–8. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McAleer MA, Pohler E, Smith FJ, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol. 2015;136:1268–76. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blanchard C, Stucke EM, Burwinkel K, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:206–12. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 84.Beppu LY, Anilkumar AA, Newbury RO, Dohil R, Broide DH, Aceves SS. TGF-beta1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1100–7. doi: 10.1016/j.jaci.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rawson R, Yang T, Newbury RO, et al. TGF-β1-induced PAI-1 contributes to a profibrotic network in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2016;138:791–800. doi: 10.1016/j.jaci.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. The Journal of allergy and clinical immunology. 2017 doi: 10.1016/j.jaci.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kagalwalla AF, Akhtar N, Woodruff SA, et al. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. J Allergy Clin Immunol. 2012;129:1387–96. doi: 10.1016/j.jaci.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muir AB, Dods K, Noah Y, et al. Esophageal epithelial cells acquire functional characteristics of activated myofibroblasts after undergoing an epithelial to mesenchymal transition. Exp Cell Res. 2015;330:102–10. doi: 10.1016/j.yexcr.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system. Gut. 2013;62:489–95. doi: 10.1136/gutjnl-2011-301817. [DOI] [PubMed] [Google Scholar]
- 90.Dellon ES, Gebhart JH, Higgins LL, Hathorn KE, Woosley JT, Shaheen NJ. The esophageal biopsy “pull” sign: a highly specific and treatment-responsive endoscopic finding in eosinophilic esophagitis (with video) Gastrointest Endosc. 2016;83:92–100. doi: 10.1016/j.gie.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Menard-Katcher C, Swerdlow MP, Mehta P, Furuta GT, Fenton LZ. Contribution of Esophagram to the Evaluation of Complicated Pediatric Eosinophilic Esophagitis. Journal of Pediatric Gastroenterology and Nutrition. 2015;61:541–6. doi: 10.1097/MPG.0000000000000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointestinal Endoscopy. 2014;79:577–85. doi: 10.1016/j.gie.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schoepfer AM, Safroneeva E, Bussmann C, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145:1230–6. doi: 10.1053/j.gastro.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 94.Kwiatek MA, Pandolfino JE, Hirano I, Kahrilas PJ. Esophagogastric junction distensibility assessed with an endoscopic functional luminal imaging probe (EndoFLIP) Gastrointest Endosc. 2010;72:272–8. doi: 10.1016/j.gie.2010.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Colizzo JM, Clayton SB, Richter JE. Intrabolus pressure on high-resolution manometry distinguishes fibrostenotic and inflammatory phenotypes of eosinophilic esophagitis. Dis Esophagus. 2016;29:551–7. doi: 10.1111/dote.12360. [DOI] [PubMed] [Google Scholar]
- 96.Mishra A, Wang M, Pemmaraju VR, et al. Esophageal remodeling develops as a consequence of tissue specific IL-5-induced eosinophilia. Gastroenterology. 2008;134:204–14. doi: 10.1053/j.gastro.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rieder F, Nonevski I, Ma J, et al. T-helper 2 cytokines, transforming growth factor β1, and eosinophil products induce fibrogenesis and alter muscle motility in patients with eosinophilic esophagitis. Gastroenterology. 2014;146:1266–77. doi: 10.1053/j.gastro.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muir AB, Dods K, Henry SJ, et al. Eosinophilic Esophagitis-Associated Chemical and Mechanical Microenvironment Shapes Esophageal Fibroblast Behavior. J Pediatr Gastroenterol Nutr. 2016;63:200–9. doi: 10.1097/MPG.0000000000001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beppu L, Yang T, Luk M, et al. MMPs-2 and -14 Are Elevated in Eosinophilic Esophagitis and Reduced Following Topical Corticosteroid Therapy. J Pediatr Gastroenterol Nutr. 2015;61:194–9. doi: 10.1097/MPG.0000000000000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tkachenko E, Rawson R, La E, et al. Rigid substrate induces esophageal smooth muscle hypertrophy and eosinophilic esophagitis fibrotic gene expression. J Allergy Clin Immunol. 2016;137:1270–2. doi: 10.1016/j.jaci.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aceves SS, Newbury RO, Chen D, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65:109–16. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abu-Sultaneh SM, Durst P, Maynard V, Elitsur Y. Fluticasone and food allergen elimination reverse sub-epithelial fibrosis in children with eosinophilic esophagitis. Dig Dis Sci. 2011;56:97–102. doi: 10.1007/s10620-010-1259-5. [DOI] [PubMed] [Google Scholar]
- 103.Lieberman JA, Morotti RA, Konstantinou GN, Yershov O, Chehade M. Dietary therapy can reverse esophageal subepithelial fibrosis in patients with eosinophilic esophagitis: a historical cohort. Allergy. 2012;67:1299–307. doi: 10.1111/j.1398-9995.2012.02881.x. [DOI] [PubMed] [Google Scholar]
- 104.Lucendo AJ, Arias A, De Rezende LC, et al. Subepithelial collagen deposition, profibrogenic cytokine gene expression, and changes after prolonged fluticasone propionate treatment in adult eosinophilic esophagitis: a prospective study. J Allergy Clin Immunol. 2011;128:1037–46. doi: 10.1016/j.jaci.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 105.Straumann A, Conus S, Degen L, et al. Long-term budesonide maintenance treatment is partially effective for patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2011;9:400–9. doi: 10.1016/j.cgh.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 106.Kuchen T, Straumann A, Safroneeva E, et al. Swallowed topical corticosteroids reduce the risk for long-lasting bolus impactions in eosinophilic esophagitis. Allergy. 2014;69:1248–54. doi: 10.1111/all.12455. [DOI] [PubMed] [Google Scholar]
- 107.Rajan J, Newbury RO, Anilkumar A, Dohil R, Broide DH, Aceves SS. Long-term assessment of esophageal remodeling in patients with pediatric eosinophilic esophagitis treated with topical corticosteroids. J Allergy Clin Immunol. 2016;137:147–56. doi: 10.1016/j.jaci.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee J, Huprich J, Kujath C, et al. Esophageal diameter is decreased in some patients with eosinophilic esophagitis and might increase with topical corticosteroid therapy. Clin Gastroenterol Hepatol. 2012;10:481–6. doi: 10.1016/j.cgh.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 109.Eluri S, Runge TM, Cotton CC, et al. The extremely narrow-caliber esophagus is a treatment-resistant subphenotype of eosinophilic esophagitis. Gastrointest Endosc. 2016;83:1142–8. doi: 10.1016/j.gie.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aceves SS. Unmet therapeutic needs in eosinophilic esophagitis. Digestive diseases. 2014;32:143–8. doi: 10.1159/000357131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abonia JP, Wen T, Stucke EM, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132:378–86. doi: 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lyons JJ, Liu Y, Ma CA, et al. ERBIN deficiency links STAT3 and TGF-β pathway defects with atopy in humans. J Exp Med. 2017;214:669–80. doi: 10.1084/jem.20161435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Paluel-Marmont C, Bellon N, Barbet P, et al. Eosinophilic esophagitis and colonic mucosal eosinophilia in Netherton syndrome. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 114.Riffle ME, Polydorides AD, Niakan J, Chehade M. Eosinophilic Esophagitis and Esophageal Granular Cell Tumor: An Unexpected Association. Am J Surg Pathol. 2017;41:616–21. doi: 10.1097/PAS.0000000000000832. [DOI] [PubMed] [Google Scholar]
- 115.Peterson K, Firszt R, Fang J, Wong J, Smith KR, Brady KA. Risk of Autoimmunity in EoE and Families: A Population-Based Cohort Study. Am J Gastroenterol. 2016;111:926–32. doi: 10.1038/ajg.2016.185. [DOI] [PubMed] [Google Scholar]
- 116.Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dellon ES, Yellore V, Andreatta M, Stover J. A single biopsy is valid for genetic diagnosis of eosinophilic esophagitis regardless of tissue preservation or location in the esophagus. J Gastrointestin Liver Dis. 2015;24:151–7. doi: 10.15403/jgld.2014.1121.242.bsy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–63. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 119.Dellon ES, Veerappan R, Selitsky SR, et al. A Gene Expression Panel is Accurate for Diagnosis and Monitoring Treatment of Eosinophilic Esophagitis in Adults. Clin Transl Gastroenterol. 2017;8 doi: 10.1038/ctg.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]


