Abstract
Because of its wide geographical adaptation and importance in human nutrition, wheat is one of the most important crops in the world. However, wheat yield has reduced due to drought stress posing threat to sustainability and world food security in agricultural production. The first stage of drought tolerant variety breeding occurs on the molecular and biochemical characterization and classification of wheat genotypes. The aim of the present study is characterization of widely grown bread wheat cultivars and breeding lines for drought tolerance so as to be adapted to different regions in Turkey. The genotypes were screened with molecular markers for the presence of QTLs mapped to different chromosomes. Results of the molecular studies identified and detected 15 polymorphic SSR markers which gave the clearest PCR bands among the control genotypes. At the end of the research, bread wheat genotypes which were classified for tolerance or sensitivity to drought and the genetic similarity within control varieties were determined by molecular markers. According to SSR based dendrogram, two main groups were obtained for drought tolerance. At end of the molecular screening with SSR primers, genetic similarity coefficients were obtained that ranged from 0.14 to 0.71. The ones numbered 8 and 11 were the closest genotypes to drought tolerant cultivar Gerek 79 and the furthest genotypes from this cultivar were number 16 and to drought sensitive cultivar Sultan 95. The genotypes as drought tolerance due to their SSR markers scores are expected to provide useful information for drought related molecular breeding studies.
Keywords: Bread wheat, Drought, Molecular marker, QTL, Triticum aestivum
Introduction
Global warming is a threat to world food security and lack of rain as a result of it has severly affected food security. The temperature increase has a direct impact on water resources and agricultural activities, leading to more severe drought. Plant productivity is declining because of various climatic events that have increased or changed, and they threaten global food security (Mickelbart et al. 2015). When plants are exposed to abiotic stress conditions such as drought, salinity, excessive rainfall and high temperature, this affects the development and growth of the plant negatively and it causes metabolic and physiological changes in the plant. Changing climate events are predicted to cause an increase in the frequency of floods, drought and high temperatures (Bita and Gerats 2013). In these events, drought is the major abiotic stress factor that adversely affects crop production and quality especially wheat. The wheat being affected negatively from drought and climate changes makes the situation worse (Shao et al. 2005; Kirigwi et al. 2007; Huseynova and Rustamova 2010). In order to minimize effects of drought caused by the weather changes, studies to produce drought tolerant plants should be continued. The effect of abiotic stress on plants has quite complex characteristics, and therefore many studies are being conducted to gain better understanding. Despite limitations caused by the environment in terms of polygenic properties, morphological markers have been used for breeding studies. Molecular markers overcome the morphological and biochemical markers limitations (Gupta et al. 1999; Dodig et al. 2010; Ateş Sönmezoğlu and Balkan 2014). In order to elucidate the genotypic mechanisms of drought-tolerant wild species; classical breeding approaches, genetic engineering studies and marker technologies must be used together.
Most crops, increased in yield by conventional breeding methods, grow under favorable conditions. For this reason most of the varieties of crops used for agricultural production are not tolerant to drought (Öztürk 2015). Therefore, it is very important that biotechnological studies continue at a rapid speed in order to develop drought-tolerant plants that can adapt to local conditions, since they have a large share in production. The first stage of drought-tolerant variety breeding begins with research studies focussing on various apects such as reducing the effects of drought, molecular and biochemical characterization and classification of wheat varieties and putting them in terms of tolerance to drought.
Because of its wide adaptability and nutrition, wheat is one of the most important plants in the world; however the decrease in yield based on the drought threatens the sustainability of agricultural production and world nutritional safety. Global wheat production in the main production areas has gradually diminished due to recurring drought which has come as a consequence of the increase in climate change (Li et al. 2009; Mwadzingeni et al. 2016). According to USDA (United States Department of Agriculture) data for 2015–2016 (https://apps.fas.usda.gov/psdonline/circulars/production), Turkey ranks ninth in the world with 19, 5 million tons of wheat produced in year. Despite of its importance in the world production, some regions in Turkey are at the risk of drought that is caused by the effects of global climate change.
Wheat has a large genome sizes (16.000 Mb for bread wheat), therefore, drought tolerance is a complex and quantitative trait controlled by multiple genes (Bernardo 2008; Huseynova and Rustamova 2010). In order to overcome these challenges, many studies have been carried out about the molecular mechanism of drought tolerance and molecular breeding for drought (Rampino et al. 2006; Zhao et al. 2008; Wei et al. 2009; Ashraf 2010; Huseynova and Rustamova 2010; Dvorak et al. 2011; Esmail et al. 2012; Ameen 2013; El-Rawy and Youssef 2014; Faheem et al. 2015; Dreisigacker et al. 2016). Recently, several molecular markers and quantitative trait loci (QTLs) have been found to be associated with genes responsible for the drought signaling mechanism (Budak et al. 2015). Significant progress has been made in molecular identification of genes of interest (Yıldırım et al. 2013). Recent advances in molecular and genomic technologies have enabled the development of descriptive molecular markers that can be used for many crops and for the identification of QTLs. These improvements have allowed for the development of crop that is tolerant to drought conditions in future (Salvi and Tuberosa 2015). To achieve this, numerous molecular markers are used. Among these markers, the most notable ones are the DNA markers which are based on the Polymerase Chain Reaction (PCR). In genetic characterization studies of wheat; amplified fragment length polymorphisms (AFLP) (Barrett and Kidwell 1998), Sequence tagged microsatellite site markers (STMSs) or generally simple sequence repeats (SSRs) (Prasad et al. 2000) and chloroplast-specific microsatellite markers (CPSSR) (Tomar et al. 2013) are all used as molecular markers based on PCR. SSR markers are frequently used for molecular studies of wheat because it has a large number in genomes, co-dominant type of inheritance, locus specificity, reproducibility, and high informational content (Röder et al. 1998; Yıldırım et al. 2009). Additionally, it has multiallelic nature, chromosome specificity, high polymorphism ratio and wide distribution throughout the wheat genome, all these make it a suitable molecular marker for genetic characterization studies in wheat (Huang et al. 2002; Prasad et al. 2009; Dodig et al. 2010; Ateş Sönmezoğlu et al. 2010; Yıldırım et al. 2011; Bousba et al. 2012; Ateş Sönmezoğlu et al. 2012). Tomar et al. (2016) made a correlation analysis of morphological and agronomic characters in drought stress conditions, and determined that the phylogenetic relationship between 31 wheat genotypes through SSR markers exists. The genetic diversity of winter and spring bread wheat varieties in terms of tolerance to drought was examined by phenotypic observations and simple sequence repeats (SSR) (Dodig et al. 2010). Golabadi et al. (2011) used microsatellite markers to identify QTLs with yield-trait competent such as thousand grain weight and harvest index. Faheem et al. (2015) studied D genome-based genetic diversity research in terms of tolerance to drought using SSR markers. In another study, Ramya et al. (2015) reported about physiological and genetic characterization of 24 modern wheat genotypes to use in breeding studies to examine the drought and temperature tolerance. Taking into account the results from previous studies, it can be concluded that SSR markers can be effectively used to determinate drought tolerance in wheat.
The aim of this study is to examine the genetic characterization of bread wheat conditions, developed in Eskisehir Parade Belt Agricultural Research Institute for drought tolerance in genotypes of bread wheat cultivated in Turkey and that has adapted to different local conditions. For this purpose, microsatellite (SSR), single nucleotide polymorphisms (SNP), randomly amplified fragment polymorphisms (RAPD) markers were used in molecular screening that were developed by different researchers and associated with tolerance to drought.
Materials and methods
Plant materials
In this study, 10 bread wheat cultivars (Triticum aestivum L.) and 9 breeding lines from different regions in Turkey were used and examined by using SSR microsatellite markers. Pedigree of the bread wheat genotypes are summarized in Table 1. The bread wheat breeding lines were provided by the Transitional Zone Agricultural Research Institute, Eskişehir, Turkey. Additionally, Gerek 79 (drought tolerant) and Sultan 95 (recommended for irrigated farming lands) were used as control cultivars for molecular characterization.
Table 1.
Bread wheat cultivars and breeding lines were used in this study
| No | Institute | Variety/breeding line |
|---|---|---|
| 1 | TZARI | SULTAN 95 |
| 2 | TZARI | GEREK 79 |
| 3 | BDIARI | KARAHAN |
| 4 | TZARI | SÖNMEZ 2001 |
| 5 | TARI | KATE-1 |
| 6 | TZARI | ALTAY 2000 |
| 7 | FCRI | BAYRAKTAR |
| 8 | TZARI | HARMANKAYA 99 |
| 9 | TZARI | İZGİ 2001 |
| 10 | TZARI | HAYMANA79/ALTAY2000 |
| 11 | TZARI | GRK/CTY//MESA/3/RL6043/4*NAC/4/MNCH |
| 12 | TZARI | T 98-9//VORONA/HD2402 |
| 13 | TZARI | ATTILA//AGRI/NAC/3/ESKINA-8 |
| 14 | TZARI | SMZ01/BEZ1 |
| 15 | TZARI | PASTOR/DEMIR2000//MUFITBEY |
| 16 | TZARI | TRK13 RESEL//TRAP#1/BOW/4/EKG15// TAST/SPRW/3/2*ID800994.W/VEE/5/SOYER02 |
| 17 | TZARI | CALIBASAN/MUFITBEY |
| 18 | TZARI | PM ME1 IRR_S-5/2*YAKAR99 |
| 19 | TZARI | PM ME1 IRR_S-32//TMP64/YY305/3/MUFITBEY |
TZARI transitional zone agricultural research institute, BDIARI Bahri Dagdas international agricultural research institute, TARI Trakya agricultural research institute, FCRI field crops central research institute
DNA isolation and PCR amplification
DNA was extracted from the young leaves of each wheat genotype according to the method by Doyle and Doyle (1990) with some modifications. A total of 45 SSR, SNP and RAPD primers were pre-screened and only 15 most polymorphic primers were applied for molecular characterization. The chromosomal location, annealing temperature, primer sequences and references to SSR markers are shown in Table 2.
Table 2.
Microsatellite markers used for molecular screening
| SSR primer | Chromosomal location | Annealing temp. (°C) | Primer sequences (5′ → 3′) | References |
|---|---|---|---|---|
| Xgwm 11 | 1B | 61 °C | F- GGATAGTCAGACAATTCTTGTG R-GTGAATTGTGTCTTGTATGCTTCC |
Röder et al. (1998) Dodig et al. (2010) |
| Xgwm 99 | 1A | 50 °C | F- AAGATGGACGTATGCATCACA R- GCCATATTTGATGACGCATA |
Röder et al. (1998) Dodig et al. (2010) |
| Xgwm 108 | 3B | 63 °C | F- CGACAATGGGGTCTTAGCAT R- TGCACACTTAAATTACATCCGC |
Galindo (2012) |
| Xgwm 186 | 5A | 55 °C | F- GCAGAGCCTGGTTCAAAAAG R- CGCCTCTAGCGAGAGCTATG |
Dodig et al. (2010) |
| Xgwm 337 | 1D | 55 °C | F- CCTCTTCCTCCCTCACTTAGC R- TGCTAACTGGCCTTTGCC |
Dodig et al. (2010) Faheem et al. (2015) |
| Xgwm 357 | 1A | 54 °C | F- TATGGTCAAAGTTGGACCTCG R- AGGCTGCAGCTCTTCTTCAG |
Röder et al. (1998) Dodig et al. (2010) |
| Xgwm 389 | 3B | 50 °C | F- ATCATGTCGATCTCCTTGACG R- TGCCATGCACATTAGCAGAT |
Röder et al. (1998) Dodig et al. (2010) |
| Xgwm 484 | 2D | 52 °C | F- ACATCGCTCTTCACAAACCC R- AGTTCCGGTCATGGCTAGG |
Röder et al. (1998) Faheem et al. (2015) |
| Xgwm 603 | 7A | 61 °C | F-ACAAACGGTGACAATGCAAGGA R-CGCCTCTCTCGTAAGCCTCAAC |
Röder et al. (1998) |
| Xgwm 626 | 6B | 50 °C | F- GATCTAAAATGTTATTTTCTCTC R- TGACTATCAGCTAAACGTGT |
Röder et al. (1998) Dodig et al. (2010) |
| Xpsp 3200 | 6D | 54 °C | F- GTTCTGAAGACATTACGGATG R- GAGAATAGCTGGTTTTGTGG |
Bryan et al. (1997) Dodig et al. (2010) |
| Xwmc 78 | 3B | 61 °C | F- AGTAAATCCTCCCTTCGGCTTC R- AGCTTCTTTGCTAGTCCGTTGC |
Somers et al. (2004) |
| Xwmc 89 | 4A | 55 °C | F- ATGTCCACGTGCTAGGGAGGTA R- TTGCCTCCCAAGACGAAATAAC |
Somers et al. (2004) Tomar et al. (2016) |
| Xwmc 118 | 5B | 58 °C | F- AGAATTAGCCCTTGAGTTGGTC R- CTCCCATCGCTAAAGATGGTAT |
Somers et al. (2004) |
| Xwmc 304 | 1A | 58 °C | F- CGATACAAGGAAGACCAGCC R- GGTTCGTCTGGTTCGCAAGT |
Somers et al. (2004) |
PCR reactions were performed using BIO-RAD C1000 Touch Thermal Cycler in total of 30 μl mix containing 50 ng wheat DNA, 0.25 μM each primer, 0.2 μM dNTP mix, 2 μM MgCl2, 10X PCR buffer, and 0.5 units of Taq DNA polymerase (Thermo Fisher USA). The details of PCR cycling reactions were as follows: 5 min at 94 °C initial denaturation, followed by 37 cycles of 94 °C for 30 s, 50–60 °C (different annealing temperatures of primers) for 30 s, 72 °C for 45 s with a final extension step of 5 min at 72 °C and storage at 4 °C. PCR products were separated using 2% agarose gels with ethidium bromide. Electrophoresis was applied at 100 V constant for 4–5 h, and a 0.5X TBE buffer was used during electrophoresis.
Statistical analysis
Polymorphisms in the amplified bands were determined by using the Biorad ChemiDoc MP. The SSR markers were scored as presence (1) or absence (0) of amplified bands (Nei and Li 1979). Comparisons of genotypes for drought tolerance were carried out using DendroUPGMA (D-UPGMA) Analysis System software (http://genomes.urv.es/UPGMA). The genetic similarity and dissimilarity coefficient among the wheat genotypes was calculated using Jaccard’s coefficient (Jaccard, 1908). SSR marker polymorphism rates were determined using polymorphism information content (PIC) values, which were estimated using PICCalc program (Botstein et al. 1980; Nagy et al. 2012), and calculated according to Liu (1997).
Results and discussion
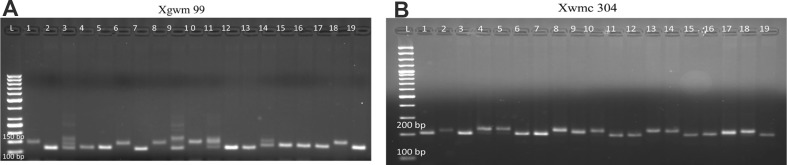
A total of 45 SSR, SNP and RAPD primers were tested and 15 polymorphic SSR primers were used in this study (Table 2; Fig. 1). The allele numbers and allele sizes of the primers are presented in Table 3. The number of alleles detected by the primers ranged from 4 to 8 among the bread wheat genotypes. The most polymorphic microsatellite marker was Xgwm 11 with 8 alleles, followed by Xwmc 78, Xgwm 626 and Xwmc 89 and they all had 7 alleles (Table 3). A total 88 polymorphic allele were obtained from screening 19 bread wheat cultivars using the 15 SSR markers with an average of 5.9 alleles per locus. The lowest number of alleles was found in Xwmc 118 with 4 alleles. Xgwm 11 had the highest number of alleles (8 alleles) per locus and the highest PIC value (0.82). The lowest number of alleles per locus and the PIC was calculated to be 5 and 0.51 respectively in Xgwm 389.
Fig. 1.
The SSR marker profiles of bread wheat genotypes using a Xgwm 99 and b Xwmc 304 SSR primers. List of 19 wheat accessions is present in Table 1
Table 3.
Allele frequency, number, size, heterozygosity and PIC value of 15 SSR primers used in the research
| SSR marker | Major allele frequency | Allele no | Allele size (bp) | Heterozygosity (He) | PIC value |
|---|---|---|---|---|---|
| Xgwm 11 | 0.23 | 8 | 180–209 | 0.84 | 0.82 |
| Xgwm 99 | 0.29 | 6 | 120–165 | 0.78 | 0.75 |
| Xgwm 108 | 0.47 | 5 | 150–170 | 0.68 | 0.64 |
| Xgwm 186 | 0.58 | 5 | 105–140 | 0.60 | 0.56 |
| Xgwm 337 | 0.30 | 6 | 180–210 | 0.80 | 0.78 |
| Xgwm 357 | 0.37 | 5 | 120–135 | 0.75 | 0.72 |
| Xgwm 389 | 0.56 | 5 | 130–145 | 0.58 | 0.51 |
| Xgwm 484 | 0.31 | 6 | 155–190 | 0.78 | 0.75 |
| Xgwm 603 | 0.31 | 6 | 100–130 | 0.78 | 0.75 |
| Xgwm 626 | 0.26 | 7 | 100–135 | 0.80 | 0.78 |
| Xpsp 3200 | 0.37 | 6 | 165–195 | 0.76 | 0.73 |
| Xwmc 78 | 0.31 | 7 | 130–175 | 0.81 | 0.79 |
| Xwmc 89 | 0.30 | 7 | 155–205 | 0.80 | 0.77 |
| Xwmc 118 | 0.37 | 4 | 110–130 | 0.73 | 0.68 |
| Xwmc 304 | 0.42 | 5 | 200–230 | 0.71 | 0.68 |
| Total | – | 88 | 100–230 | – | – |
| Mean | 0.34 | 5.9 | – | 0.75 | 0.71 |
The polymorphism information content (PIC) values of the analyzed microsatellite markers ranged from 51 to 82%, with average of 71%. Among the 15 SSR markers used in this study, Xgwm 11 primer had the highest PIC values, followed by primer Xwmc 78 at 0.79 PIC and Xgwm 337 and Xgwm 626 SSR primers with 0.78 PIC value. According to PIC values of each marker; the lowest PIC value was presenting Xgwm 389 primer with 0.51 PIC. Heterozygosity (He) rates were similar to the PIC values. It was found that the highest He value was found in Xgwm 11 primer with the value of 0.84 and Xwmc 78 with the value of 0.81 and the lowest He value was found in Xgwm 389 primer with 0.58 value. The most widely used criterion for testing genetic variation in a population is heterozygosity. The SSR markers used in this study indicates high heterozygosity with an average of 0.75 He value in the investigated wheat genotypes have wide genetic variation.
The polymorphism information content (PIC) value for the primers used in this study, had 15 loci that were considered to be informative, since they had a PIC value greater than 0.5. The PIC value can be used to evaluate the level of gene variation in a plant. When the PIC value is > 0.5 the locus is considered to be of high diversity, while if the PIC is < 0.25 the locus is considered to be of low diversity (Botstein et al. 1980; Nagy et al. 2012; Ramadugu et al. 2015). The mean PIC value used SSR markers in this study was 0.71 and the range was from 0.51 to 0.82. Therefore, all of the primers used in this study were found to be highly informative. The results that used SSRs are potential markers that could be used as marker to assist in selection for drought stress tolerance by molecular plant breeding. Moreover, the results are in agreement with those reported by Dodig et al. (2010), Brbaklic et al. (2015), Faheem et al. (2015), Ramya et al. (2015), and Tomar et al. (2016), who assigned SSR markers to drought tolerance in wheat genotypes using molecular markers. Hao et al. (2006) suggested that the allele’s number in each locus and the calculated PIC values of these alleles should be evaluated together as part of an objective assessment of genetic diversity in genotype collections. Since PIC values correlates positively with the number of alleles for all genotypes. The number of alleles in SSR markers ranged from 8 (Xgwm 11) to 4 (Xwmc 118) with an average being 5.9 in total 88 alleles were found (Table 3). The PIC ranged from 0.82 (Xgwm 11) to 0.64 (Xgwm 108) with a mean PIC value being 0.71. It was determined that the primers that had fewer alleles also had lower PIC values.
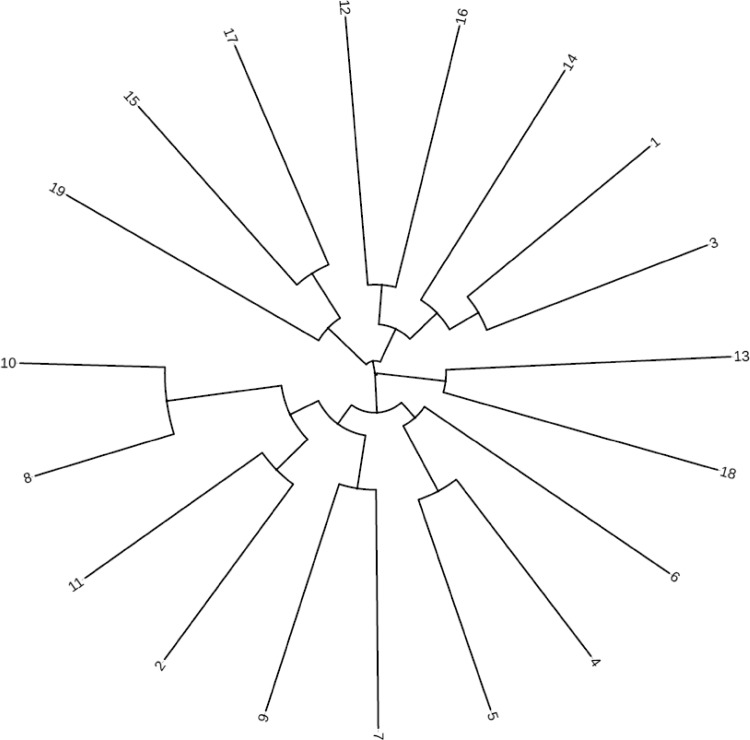
A dendrogram based on the genetic similarity values among the genotypes was prepared using the SSR marker information (Fig. 2). It is clearly seen from the dendrogram that the bread wheat genotypes were divided into 2 main groups, and these groups were then separated into several subgroups. Group 1 consisted of 4, 5, 6, 7, 8, 9, 10, 11, 13 and 18 number genotypes and drought tolerant cultivar Gerek 79, while group 2 included all the remaining genotypes. According to SSR based dendrogram, two main groups were obtained for drought tolerance (Fig. 2). The genotypes examined in the study showed a genetic diversity of 69% in terms of drought tolerance by drought-related DNA regions-specific SSR primers. At end of the molecular screening that was done with SSR primers, a genetic similarity coefficients were obtained that ranged from 0.14 (9 with 17) to 0.71 (8 with 10). Number 8 and 11 were the closest genotypes to drought tolerant cultivar Gerek 79 and the farthest genotypes of this cultivar was number 16 and drought sensitive cultivar Sultan 95. The closest genotypes to drought sensitive control cultivar Sultan 95 were number 3 and 14 genotypes, while the farthest genotypes of Sultan 95 were Gerek 79, number 8, 10 and 18 genotypes. Genotypes that were grouped in terms of tolerance to drought and SSR markers are expected to provide a useful information for drought related molecular breeding studies. When the data from the used primers was evaluated, it was determined that the genetic similarity coefficient between drought sensitive cultivar Sultan 95 and Gerek 79 which is known to be tolerant to drought, was 0.24. Therefore, marker assisted selection via the SSR markers could be used in identifying genotypes in terms of drought tolerant.
Fig. 2.
Dendrogram based on 15 SSR marker data of the bread wheat genotypes was generated using DendroUPGMA software. List of 19 wheat accessions is present in Table 1
Conclusion
Characterizing genetic diversity, especially with respect to important stress factors such as drought, has a great influence on increasing genetic variation for future wheat breeding programs. Wheat genotypes exhibit different level of drought tolerance which makes their molecular characterization and classification important to develop drought-tolerant wheat varieties. In this study, genetically characterization of some bread wheat genotypes was examined for drought stress and a wide genetic variation has been determined between observed genotypes. In this paper, we used 15 polymorphic SSR markers to determine drought tolerance of bread wheat genotypes. The most polymorphic microsatellite marker was Xgwm 11 with 8 alleles and the highest PIC value (0.82). According to SSR based dendrogram, two main groups were found when it came to drought tolerance. At end of the molecular screening with SSR primers genetic similarity coefficients were obtained that ranged from 0.14 to 0.71. Number 8 and 11 were the closest genotypes to the drought tolerant cultivar Gerek 79 and the furthest genotypes from this cultivar was number 16 and drought sensitive cultivar Sultan 95. It was determined that the genetic similarity coefficient between drought tolerant cultivar Gerek 79 and drought sensitive cultivar Sultan 95 was 0.24. Genotypes that were grouped in terms of their tolerance to drought and SSR markers are expected to provide useful information for drought related molecular breeding studies.
This study has shown that SSR markers are very useful, reliable and useful in genetic characterization of drought tolerance studies in bread wheat. It has been also determined that microsatellites can be successfully used for genetic characterization, genotype identification and drought-related genetic resources. Therefore, the SSR primers used in the study will be useful during MAS in search of more drought-resistant wheat varieties. In conclusion, both the genotypes investigated and the findings of the SSR markers used are thought to help in creating preliminary data for future breeding studies and genetic studies on drought.
Acknowledgements
The authors would like to express their thanks to Savaş BELEN from Transitional Zone Agricultural Research Institute, Eskişehir, Turkey for providing bread wheat breeding lines. Furthermore, this work was financially supported by the Scientific Research Commission of Karamanoglu Mehmetbey University under Project No. 18-M-15.
References
- Ameen ET. Molecular markers for drought tolerance in bread wheat. Afr J Biotechnol. 2013;12(21):3148–3152. [Google Scholar]
- Ashraf M. Inducing drought tolerance in plants: recent advances. Biotechnol Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Ateş Sönmezoğlu Ö, Balkan AS. Molecular and biochemical analysis of durum wheat genotypes to examine carotenoid pigment content and lipoxygenase enzyme activity. Cereal Res Commun. 2014;42(2):218–228. doi: 10.1556/CRC.2013.0070. [DOI] [Google Scholar]
- Ateş Sönmezoğlu Ö, Yıldırım A, Eserkaya Güleç T, Kandemir N, Sayaslan A, Koyuncu M. Molecular breeding of Selçuklu-97 durum wheat cultivar for some genes affecting pasta quality. J Appl Biol Sci. 2010;4(2):51–55. [Google Scholar]
- Ateş Sönmezoğlu Ö, Bozmaz B, Yıldırım A, Kandemir N, Aydın N. Genetic characterization of Turkish Bread wheat landraces based on microsatellite markers and morphological characters. Turk J Biol. 2012;36:589–597. [Google Scholar]
- Barrett BA, Kidwell KK. AFLP-based genetic diversity assessment among wheat cultivars from The Pacific Northwest. Crop Sci. 1998;38:1261–1271. doi: 10.2135/cropsci1998.0011183X003800050025x. [DOI] [Google Scholar]
- Bernardo R. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 2008;48:1649–1664. doi: 10.2135/cropsci2008.03.0131. [DOI] [Google Scholar]
- Bita C, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White R, Skolnick M, Davis R. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Bousba R, Baum M, Djekoune A, Labadidi S, Djighly A. Screening for drought tolerance using molecular markers and phenotypic diversity in durum wheat genotypes. World Appl Sci J. 2012;16:1219–1226. [Google Scholar]
- Brbaklic L, Trkulja D, Kondic-Spika A, Mikih S, Tomičić M, Kobiljski B. Determination of population structure of wheat core collection for association mapping. Cereal Res Commun. 2015;43(1):22–28. doi: 10.1556/CRC.2014.0027. [DOI] [Google Scholar]
- Bryan GJ, Collins AJ, Stephenson P, Orry A, Smith JB, Gale MD. Isolation and characterisation of microsatellites from hexaploid bread wheat. Theor Appl Genet. 1997;94:557–563. doi: 10.1007/s001220050451. [DOI] [Google Scholar]
- Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N. From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci. 2015;6:1012. doi: 10.3389/fpls.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodig D, Zori M, Kobiljski B, Momirovi GS, Quarrie SA. Assessing drought tolerance and regional patterns of genetic diversity among spring and winter bread wheat using simple sequence repeats and phenotypic data. Crop Pasture Sci. 2010;61:812–824. doi: 10.1071/CP10001. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dreisigacker S, Sukumaran S, Guzmán C, He X, Bonnett D, Crossa J. Molecular marker-based selection tools in spring bread wheat improvement: CIMMYT experience and prospects. Mol Breed Sustain Crop Improv. 2016;11:421–474. doi: 10.1007/978-3-319-27090-6_16. [DOI] [Google Scholar]
- Dvorak J, Luo MC, Akhunov E. NI Vavilov's theory of centres of diversity in the light of current understanding of wheat diversity, domestication and evolution. Czech J Genet Plant Breed. 2011;47:20–27. [Google Scholar]
- El-Rawy MAE, Youssef M. Evaluation of drought and heat tolerance in wheat based on seedling traits and molecular analysis. J Crop Sci Biotechnol. 2014;17(3):183–189. doi: 10.1007/s12892-014-0053-x. [DOI] [Google Scholar]
- Esmail RM, Sattar AAA, Sherin Mahfouze A, El-Enany Magda AM, Mostafa EAH, Abou-Ellail MA, Mahfouze HA, Fathallah FB. Genetic characterizations among drought tolerant wheat genotypes by biochemical and molecular markers. J Appl Sci Res. 2012;8(12):5886–5896. [Google Scholar]
- Faheem M, Mahmood T, Shabbir G, Akhtar N, ul Kazi AG, Kazi AM. Assessment of D-genome based genetic diversity in drought tolerant wheat germplasm. Int J Agric Biol. 2015;17:791–796. doi: 10.17957/IJAB/14.0018. [DOI] [Google Scholar]
- Galindo MAA (2012) Meta-analysis of wheat QTL regions associated with heat and drought stress. Master Theses and Dissertations, University of Arkansas 645
- Golabadi M, Arzani A, Mirmohammadi Maibody SAM, Sayed Tabatabaei BE, Mohammadi SA. Identification of microsatellite markers linked with yield components under drought stress at terminal growth stages in durum wheat. Euphytica. 2011;177:207–221. doi: 10.1007/s10681-010-0242-8. [DOI] [Google Scholar]
- Gupta PK, Varshney RK, Sharma PC, Ramesh B. Molecular markers and their applications in wheat breeding. Plant Breed. 1999;118:369–390. doi: 10.1046/j.1439-0523.1999.00401.x. [DOI] [Google Scholar]
- Hao CY, Zhang XY, Wang LF, Dong YS, Shang XW, Jia JZ. Genetic diversity and core collection evaluations in common wheat germplasm from the northwestern spring wheat region in China. Mol Breed. 2006;17(1):69–77. doi: 10.1007/s11032-005-2453-6. [DOI] [Google Scholar]
- Huang XQ, Börner A, Röder MS, ve Ganal MW. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet. 2002;105:699–707. doi: 10.1007/s00122-002-0959-4. [DOI] [PubMed] [Google Scholar]
- Huseynova IM, Rustamova SM. Screening for drought stress tolerance in wheat genotypes using molecular markers. Proc ANAS Biol Sci. 2010;65(5–6):132–139. [Google Scholar]
- Jaccard P. Nouvelles research sur la distribution florare. Bull De Lasociete Vaudoise Des Sciences Naturelles. 1908;44:223–270. [Google Scholar]
- Kirigwi FM, Van Ginkel M, Brown-Guedira G, Gill BS, Paulsen GM, Fritz AK. Markers associated with a QTL for grain yield in wheat under drought. Mol Breed. 2007;20:401–413. doi: 10.1007/s11032-007-9100-3. [DOI] [Google Scholar]
- Li Y, Ye W, Wang M, Yan X. Climate change and drought: a risk assessment of crop-yield impacts. Climate Res. 2009;39:31–46. doi: 10.3354/cr00797. [DOI] [Google Scholar]
- Liu BH. Statistical genomics: linkage, mapping and QTL analysis. Boca Raton: CRC Press; 1997. [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16(4):237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Mwadzingeni L, Shimelis H, Dube E, Laing MD, Tsilo TJ. Breeding wheat for drought tolerance: progress and technologies. J Integr Agric. 2016;15(5):935–943. doi: 10.1016/S2095-3119(15)61102-9. [DOI] [Google Scholar]
- Nagy S, Poczai P, Cernák I, Gorji AM, Hegedűs G, Taller J. PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem Genet. 2012;50(9–10):670–672. doi: 10.1007/s10528-012-9509-1. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk NZ. Literature review and new approaches on plant drought stress response. Turk J Agric Food Sci Technol. 2015;3(5):307–315. [Google Scholar]
- Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK. The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet. 2000;100:584–592. [Google Scholar]
- Prasad B, Babar MA, Xu XY, Bai GH, Klatt AR. Genetic diversity in U.S. hard red winter wheat cultivars as revealed by microsatellite markers. Crop Pasture Sci. 2009;60:16–24. doi: 10.1071/CP08052. [DOI] [Google Scholar]
- Ramadugu C, Keremane ML, Hu X, Karp D, Federici CT, Kahn T, Lee RF. Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms. Sci Hortic. 2015;195:124–137. doi: 10.1016/j.scienta.2015.09.004. [DOI] [Google Scholar]
- Rampino P, Pataleo S, Gerardi C, Perotta C. Drought stress responses in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006;29:2143–2152. doi: 10.1111/j.1365-3040.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- Ramya P, Jain N, Singh PK, Singh GP, Prabhu KV. Population structure, molecular and physiological characterisation of elite wheat varieties used as parents in drought and heat stress breeding in India. Indian J Genet Plant Breed. 2015;75(2):250–252. doi: 10.5958/0975-6906.2015.00038.3. [DOI] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149(4):2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S, Tuberosa R. The QTLome comes of age. Curr Opin Biotechnol. 2015;32:179–185. doi: 10.1016/j.copbio.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Shao HB, Liang ZS, Shao MA, Sun Q. Dynamic changes of anti-oxidative enzymes of 10 wheat genotypes at soil water deficits. Colloids Surf B. 2005;42:187–195. doi: 10.1016/j.colsurfb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Somers DJ, Isaac P, Edwards K (2004) A high-density wheat microsatellite consensus map for bread wheat (Triticum aestivum L.). Theory and Appl Genet 109(6):1105–1114 [DOI] [PubMed]
- Tomar RS, Deshmukh RK, Naik KB, Tomar SMS. Development of chloroplast-specific microsatellite markers for molecular characterization of alloplasmic lines and phylogenetic analysis in wheat. Plant Breed. 2013;133(1):12–18. doi: 10.1111/pbr.12116. [DOI] [Google Scholar]
- Tomar RSS, Tiwari S, Naik BK, Chand S, Deshmukh R, Mallick N, Tomar SMS. Molecular and morpho-agronomical characterization of root architecture at seedling and reproductive stages for drought tolerance in wheat. PLoS ONE. 2016;11(6):156–528. doi: 10.1371/journal.pone.0156528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Jing R, Wang Ch, Chen J, Mao X, Chang X, Jia J. Dreb1 Genes in Wheat (Triticum aestivum L.): development of functional markers and gene mapping based on SNPs. Mol Breed. 2009;23:13–22. doi: 10.1007/s11032-008-9209-z. [DOI] [Google Scholar]
- Yıldırım A, Kandemir N, Sönmezoğlu ÖA, ve Güleç TE. Transferability of microsatellite markers among cool season cereals. Biotechnol Biotechnol Equip. 2009;23(3):1299–1302. doi: 10.1080/13102818.2009.10817657. [DOI] [Google Scholar]
- Yıldırım A, Ateş Sönmezoğlu Ö, Gökmen S, Kandemir N, Aydın N. Determination of genetic diversity among Turkish durum wheat landraces by microsatellites. Afr J Biotechnol. 2011;10(19):3915–3920. [Google Scholar]
- Yıldırım A, Ateş Sönmezoğlu Ö, Sayaslan A, Koyuncu M, Güleç T, Kandemir N. Marker-assisted breeding of a durum wheat cultivar for gliadin and LMW-glutenin proteins affecting pasta quality. Turk J Agric For. 2013;37:527–533. doi: 10.3906/tar-1207-75. [DOI] [Google Scholar]
- Zhao CX, Guo LY, Cheruth AJ, Shao HB, Yang HB. Prospective for applying molecular and genetic methodology to improve wheat cultivars in drought environments. CR Biol. 2008;331:579–586. doi: 10.1016/j.crvi.2008.05.006. [DOI] [PubMed] [Google Scholar]