Abstract
Background
Neutrophil extracellular traps (NETs) are part of the innate immune response and are essential in local pathogen control, but are associated with pathological inflammation, organ damage, autoimmunity, and thrombosis. Immune‐mediated hemolytic anemia (IMHA) is a pro‐inflammatory, prothrombotic disease associated with high mortality.
Hypothesis/Objectives
Neutrophil extracellular traps (NETs) are a feature of the inflammatory process in dogs with IMHA. The objective of the study was to evaluate plasma from dogs with IMHA for the presence of 2 indirect markers and 1 direct marker of NETs.
Animals
Healthy client‐owned dogs (56) and hospitalized dogs with IMHA (n = 35).
Methods
Prospective study. Plasma samples for all dogs were evaluated for cell‐free DNA using a fluorescence assay, histone‐DNA (hisDNA) complex using an ELISA, and citrullinated histone H3 (specific for NETosis) using Western blot. Reference intervals were generated using plasma from healthy dogs.
Results
In dogs with IMHA, cell‐free DNA concentration was above the reference interval in 17% of samples with a median (range) of 1.0 μg/mL (0.1–17.3), and hisDNA concentration was above the reference interval in 94% of samples with a median (range) of 30.7 × pooled normal plasma (PNP; 0.6–372.1). Western blot for citrullinated histone H3 identified detectable bands in 84% samples from dogs with IMHA.
Conclusions and Clinical Importance
The assay for cell‐free DNA detected evidence of NETs in fewer dogs than did the other approaches. Excessive NETs appears to be a feature of IMHA in dogs and contributions to the prothrombotic state deserve further study.
Keywords: DNA, Histone, IMHA, Inflammation
Abbreviations
- CHAOS
canine hemolytic anemia score
- DNA
deoxyribonucleic acid
- hisDNA
histone‐DNA complexes
- IMHA
immune‐mediated hemolytic anemia
- NETs
neutrophils extracellular traps
- NETosis
the process of NET formation
- PAD4
peptidylarginine deiminase 4
- PNP
pooled normal plasma
- PVDF
polyvinylidene difluoride
- TBST
tris‐buffered saline with tween
Neutrophils, the most abundant leukocyte in mammals, are essential to host immunity. They are highly mobile and rapidly recruited to sites of injury and inflammation by chemokines, cytokines, and other inflammatory mediators.1, 2 Neutrophils target pathogens by phagocytosis, degranulation, and generation of reactive oxygen species. In 2004, a novel mechanism by which neutrophils kill bacteria was described.3 In response to pro‐inflammatory mediators (eg, IL‐8, endotoxin) or to pathogens themselves, neutrophils extrude a chromatin mesh originating from the nucleus (in some cases resulting in cell death)3 or from mitochondria (conserving the integrity and function of the neutrophil).4 This extracellular chromatin matrix (termed neutrophil extracellular trap or NET) creates a physical barrier to trap pathogens, and the attached histones and bactericidal enzymes kill those pathogens.1, 3 Eosinophils,5 basophils,6 mast cells,7 and macrophages8 also can produce extracellular chromatin traps.
The process of NET formation (NETosis) is active, and distinct from other mechanisms leading to cell death such as apoptosis. Neutrophil activation leads to migration of elastase to the nucleus where it promotes chromatin decondensation.9 Concurrently, in a process unique to NETosis, arginines in histones undergo citrullination by peptidylarginine deiminase 4 (PAD4).10, 11, 12 Current methods to quantify NETs in plasma include approaches that measure either cell‐free DNA or complexes between histones and DNA fragments (hisDNA), but neither of these approaches is specific for NET formation. The presence of citrullinated histones is a specific marker for NETosis, but there are no currently commercially available assays to identify citrullinated histones in plasma.
Although NETs are essential for innate immunity, they also have been implicated in the pathophysiology of a number of noninfectious inflammatory processes in humans and rodent models, including acute lung injury,13 ischemia‐reperfusion injury,14 systemic lupus erythematosus,15 vasculitis,16 rheumatoid arthritis,17 and the development of thrombosis.18, 19, 20, 21, 22, 23 Immunohistochemistry on venous thrombi has indicated the presence of citrullinated histone H3.18, 22 Furthermore, knockout of PAD4 has been shown to prevent deep vein thrombosis in mice,24 suggesting that NETosis may be a useful pharmacologic target for thromboprophylaxis.
Immune‐mediated hemolytic anemia (IMHA) in dogs is known to be associated with profound systemic inflammation often with marked neutrophilia,25 and thrombosis is a major contributor to the morbidity and mortality of IMHA in dogs.26, 27 We hypothesized that NETosis is a feature of the inflammatory process in dogs with IMHA. Our objective was to evaluate plasma from dogs with IMHA for the presence of markers that would indicate NETosis was occurring in this disease.
Materials and Methods
Ours was a prospective, observational study. Dogs admitted to the University of Illinois Veterinary Teaching Hospital that were identified to the clinical laboratory as having been diagnosed with IMHA were eligible for inclusion. A definitive diagnosis then was determined based on review of medical records. Diagnosis of IMHA was based on: (1) the presence of regenerative anemia (hematocrit <30% with >60,000 reticulocytes/μL or anisocytosis and polychromasia); (2) evidence of hemolysis (hyperbilirubinemia, hemoglobinemia, bilirubinuria, hemoglobinuria, or some combination of these); and (3) a positive saline agglutination test, spherocytosis, a positive Coombs test, or some combination of these.
Sample Collection and Processing
Whole blood EDTA or citrate samples that had been collected by the responsible clinician were obtained from the clinical laboratory after clinician‐requested assays had been completed. Plasma was obtained from 26 apparently healthy dogs, as previously described,28 to create reference intervals for the assays. The study was approved by the University of Illinois Institutional Animal Care and Use Committee, and informed owner consent was obtained for all apparently healthy dogs. Informed consent was not required for samples obtained from clinical patients that were submitted to the clinical pathology laboratory. For plasma production, blood was centrifuged at 1,500 × g for 10 minutes, and the upper 2/3 of the plasma removed. Plasma samples were divided into separate aliquots and then frozen at −80°C until analysis.
Citrated pooled normal plasma (PNP) was produced by pooling an equal volume of plasma from 30 apparently healthy dogs, as previously described.28 For assays using citrated PNP as a standard, the results obtained using EDTA plasma were corrected for the amount of plasma dilution by the citrate by dividing the EDTA result by 1.11.
Fluorescence Assay for DNA
Plasma cell‐free DNA was quantified using SYTOX Greena as previously described.28 Briefly, plasma was diluted 10‐fold to 1,280‐fold in phosphate‐buffered saline containing 0.1% bovine serum albumin (PBSA) then mixed 2 : 1 with 1 mM SYTOX Green in PBSA in a black 98‐well microplate.b Fluorescence (excitation 485 nm, emission 538 nm) was recorded on a Spectramax M2 fluorimeterc and corrected for background (by subtracting SYTOX emission in PBSA without plasma) and autofluorescence (measured in identically diluted samples without SYTOX Green added). The DNA concentrations were calculated based on a standard curve (0–1,000 ng/mL) of known concentrations of DNAd diluted in PBSA. Concentrations in citrated plasma were corrected for the dilution due to collection into the citrate volume.
ELISA for Histone‐associated DNA Fragments (hisDNA)
Plasma was evaluated for the presence of hisDNA using a commercially available ELISA kite as previously described.28 Briefly, plasma samples were diluted 3‐fold to 280‐fold into the supplied incubation buffer, and applied to the plate in duplicate, then handled according to the kit instructions. Rate of substrate cleavage in each well was evaluated over 20 minutes at 25°C on a Versamax Spectrophotometerf and compared to rates obtained from a standard curve consisting of serially diluted locally prepared mammalian hisDNA, and then normalized to the value obtained for canine PNP.
Western Blot for Citrullinated Histone H3
Plasma samples were thawed at 37°C for 5 minutes, then diluted 4‐fold to 100‐fold into 20 mM Hepes NaOH pH 7.4, 100 mM NaCl (HBS). Each gel included an internal reference sample of canine histones containing citrullinated H3. This reference material was produced by purifying histones from septic canine abdominal fluid (from a dog with a gastrointestinal tract perforation verified on cytology and at surgery) using EpiQuick Total Histone Extraction kitg according to the manufacturer's instructions. Samples were boiled in reducing Laemeli buffer, loaded (20 μL) onto a 4–20% Tris‐glycine mini gel,h electrophoresed for 1.25 hours at 100V, then transferred onto polyvinylidene difluoride (PVDF) membrane for 1 hour at 100 V. The membrane was blocked in 5% milk in tris‐buffered saline with Tween (TBST) (10 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.05% Tweens‐20) for 2 hours at 25°C, then exposed to 0.5 mg/mL primary antibody (antihistone H3, citrulline R2 + R8 + R17i) in 5% milk/TBST overnight at 4°C. Membranes were washed 5 times in TBST, and then exposed to 1,000‐fold diluted secondary antibody (donkey anti‐rabbit IgG‐horseradish peroxidasej) in 5% milk/TBST for 2 hours at 25°C, then again washed 5 times. The chemiluminescence detection was performed using Pierce ECL‐2 substrate.k Films were developed using a standard photographic procedure, and quantitative analysis of detected bands was carried out by densitometer scanning using GelDoc XR+ and ImageLab software.l
Statistical Analysis
Data sets were tested for normality by Shapiro‐Wilk test. Correlations between data sets were calculated using Pearson Product Moment Correlation. Comparisons between data for survivors and nonsurvivors at specific time points were determined using Mann‐Whitney Rank. Tests were performed by SigmaStat 4.0.m A P value of <0.05 was considered significant.
Results
Patient Characteristics
Samples initially were collected from 41 patients, but 6 samples were excluded because of failure to meet the defined criteria for a diagnosis of IMHA (5) or lack of adequate samples to perform the assays (1). The cohort included 25 spayed females, 4 intact females, and 6 neutered males. Represented breeds included Labrador Retriever (3), Miniature poodle (3), Shih tzu (2), Miniature Schnauzer (2), and 1 each of Bichon Frise, Bloodhound, Boston Terrier, Chesapeake Bay Retriever, Chihuahua, Chinese crested, Cocker spaniel, English springer spaniel, Mastiff, Pomeranian, and Standard poodle. The remainder (14) were of mixed breed. The median (range) body weight for included dogs was 13.1 (4.5–50.0) kg. A summary of the results of relevant initial laboratory analysis is presented in Table 1. Hemoglobinemia consistent with intravascular hemolysis was identified in 7 samples, and 28 samples had evidence of autoagglutination. Sufficient diagnostic tests to rule out underlying disease were not performed in 7 patients. In 30 patients, diagnostic tests (eg, CBC, serum biochemical profile, thoracic radiographs, abdominal ultrasound examination, and SNAP 4Dxn) failed to identify an underlying disease, suggesting a diagnosis of primary IMHA; 2 patients had concurrent hepatic disease, and 2 had previous drug or vaccine exposure that could have triggered secondary IMHA. Thirty patients had thoracic radiographs interpreted as normal, 31 had abdominal ultrasound findings with 4 recorded as abnormal but interpreted as not clinically relevant (no further details available). Of the 32 patients that had a SNAP 4Dx test performed for infectious disease, none were positive. Of the 31 patients that had sufficient data collected to calculate a Canine Hemolytic Anemia Score (CHAOS)29 score, the scores were 1 (n = 2), 2 (n = 7), 3 (n = 8), 4 (n = 6), 5 (n = 6), and 7 (n = 2). Spherocytosis was documented as low (n = 9), moderate (n = 4), many (n = 4), and present (n = 5). Outcomes during the initial hospitalization were survival to discharge (n = 26), euthanasia (n = 7), or death (n = 2). At 28 days, 5 additional patients had died or were euthanized, 1 patient was lost to follow‐up, and 20 patients were still alive. Of these 20, 13 patients were still alive at 180 days, and an additional 2 were lost to follow‐up.
Table 1.
Selected biochemical and hematological results from clinical samples of dogs with IMHA
| Parameter | Reference Interval | Median | Range | N | % Low | % High |
|---|---|---|---|---|---|---|
| Leukocytes (×103/μL) | 6–17 | 24.80 | 5.29–92.10 | 35 | 2.9 | 71.4 |
| Neutrophils (×103/μL) | 3–11.5 | 21.07 | 2.86–70.00 | 35 | 2.9 | 80.0 |
| Bands (×103/μL) | 0–0.3 | 1.34 | 0–11.05 | 35 | NA | 71.4 |
| Monocytes (×103/μL) | 0.2–1.4 | 2.22 | 0.44–9.09 | 35 | 0 | 65.7 |
| Lymphocytes (×103/μL) | 1–4.8 | 1.20 | 0–5.10 | 35 | 5.7 | 5.7 |
| Hematocrita (%) | 35–52 | 17.7a | 7.4a | 35 | 100.0 | 0 |
| Platelets (×103/μL) | 200–400 | 156.5 | 10–426 | 35 | 68.6 | 5.7 |
| BUN (mg/dL) | 6–30 | 20.5 | 9–55 | 34 | 0 | 22.9 |
| Creatinine (mg/dL) | 0.5–1.5 | 0.7 | 0.3–1.5 | 33 | 20.0 | 0 |
| Total bilirubin (mg/dL) | 0.1–0.3 | 1.9 | 0.2–29.7 | 35 | 0 | 91.4 |
| Albumina (g/dL) | 2.5–3.8 | 2.84a | 0.42a | 34 | 17.1 | 2.9 |
| ALT (U/L) | 8–65 | 67 | 21–3,129 | 34 | 0 | 48.6 |
Although the majority of the data sets were nonparametric, hematocrit and albumin sets were normally distributed. Consequently, mean (rather than median) and standard deviation (rather than range) are reported for these variables.
Cell‐free DNA
The results of the SYTOX Green assay for plasma cell‐free DNA are reported in Figure 1. For 9 samples, pigmentemia prevented acquisition of data because of severe autofluorescence. Of the 26 samples for which data could be obtained, 6 samples (17%) had cell‐free DNA concentrations above the established reference interval of <1.5 μg/mL, with a median (range) of 1.0 (0.1–17.3 μg/mL). Plasma concentration of cell‐free DNA did not correlate significantly with blood neutrophil concentration (P = 0.87).
Figure 1.
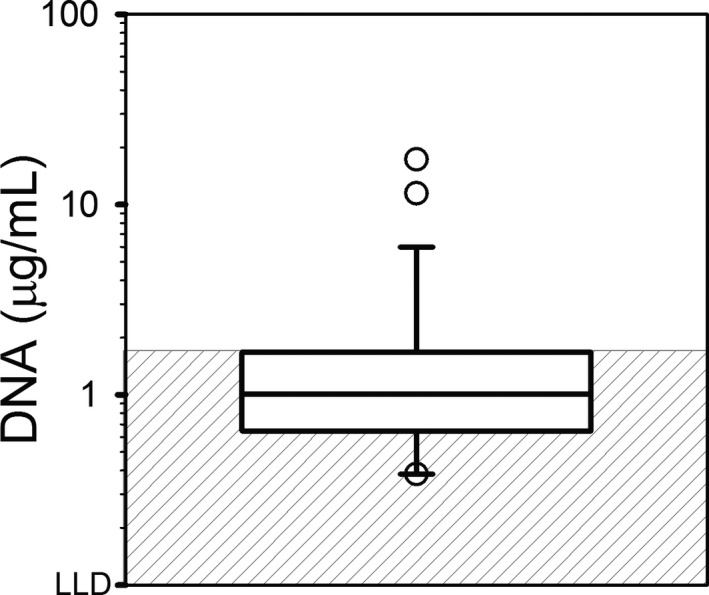
Cell‐free DNA in Plasma from Dogs with IMHA (n = 26). Note that results could not be obtained with 9 samples due to pigmentemia. Due to the wide distribution of results, for visualization purposes, the data are plotted on a log scale, but the data were not log‐transformed for analysis. The line of centrality denotes median, the box indicates 25th to 75th percentile, and the whiskers indicate 10th to 90th percentile. The gray‐striped box represents the established reference interval of <1.5 μg/mL. LLD, lower limit of detection.
HisDNA
The results of the ELISA for hisDNA are reported in Figure 2. Of the 35 samples evaluated, 33 samples (94%) contained concentrations of hisDNA above the established reference interval of <3.4×PNP, with a median (range) of 30.7 (0.6–372.1×). As observed in Figure 3, comparison of the results obtained with the ELISA for hisDNA to those obtained with the SYTOX Green assay for cell‐free DNA indicated a significant (P = 0.003) positive correlation (r = 0.59). Plasma concentration of hisDNA did not correlate significantly with blood neutrophil concentration (P = 0.97).
Figure 2.
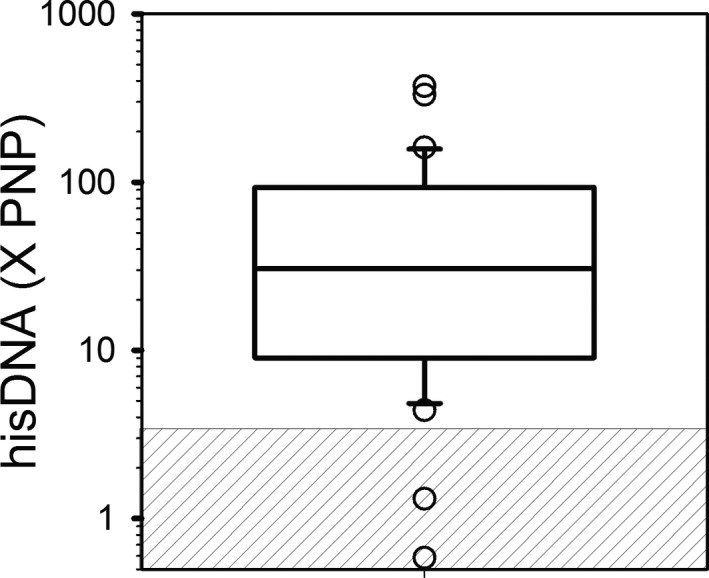
Histone‐DNA (hisDNA) Complexes in Plasma from Dogs with IMHA (n = 35). Plasma samples were evaluated for hisDNA using the cell death detection ELISA as described in the Methods section. Results were normalized to that obtained with pooled normal plasma (PNP) to allow for comparison between laboratories. For visualization purposes, the data are plotted on a log scale, but the data were not log‐transformed for analysis. The line of centrality denotes median, the box indicates 25th to 75th percentile, and the whiskers indicate 10th to 90th percentile. The gray‐striped box represents the established reference interval of <3.4 X PNP.
Figure 3.
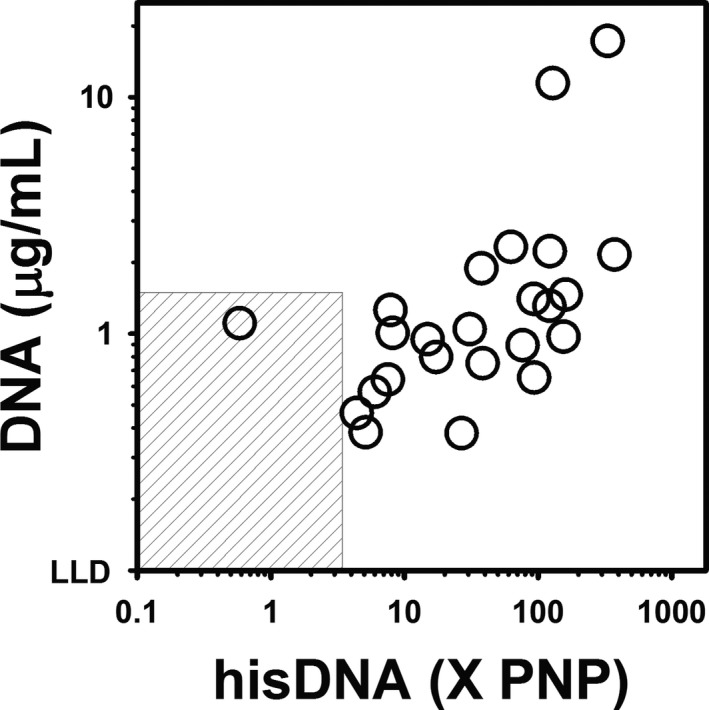
Comparison of cell‐free DNA and Histone‐DNA (hisDNA) Complexes in Plasma from Dogs with IMHA. Comparison of results obtained with the DNA fluorescence assay (y‐axis) and the ELISA for hisDNA complex (x‐axis). Results were significantly (P = 0.003) positively correlated (r = 0.59) via Pearson Product Moment Correlation. For visualization purposes, the data are plotted on a log‐log scale, but the data were not log‐transformed for the analysis. The gray‐striped box represents the reference interval for each assay. LLD, lower limit of detection. PNP, pooled normal plasma.
Citrullinated Histone H3
The results of the evaluation by Western blot for citrullinated histone H3 are reported in Figure 4A, and examples of gel images of samples from apparently healthy dogs and dogs with IMHA are presented in Figure 4B,C, respectively. Of the 32 samples from dogs with IMHA that were evaluated, 27 (84%) had detectable bands indicative of the presence of citrullinated histone H3. In comparison, of the 26 samples from apparently healthy dogs that were evaluated, 9 (35%) samples had detectable bands, and none were of marked intensity. Comparison of the results obtained with the ELISA for hisDNA to those obtained by densitometric assessment of the Western blots for citrullinated histone H3 did not indicate a significant correlation (P = 0.83). However, only 2 samples contained detectable citrullinated histone H3 but had hisDNA results within the reference range, and only 4 samples had abnormally high hisDNA results but no detectable band for citrullinated histone H3. There was no correlation between the citrullinated histone H3 and the SYTOX Green assay for plasma cell‐free DNA (Table S1).
Figure 4.
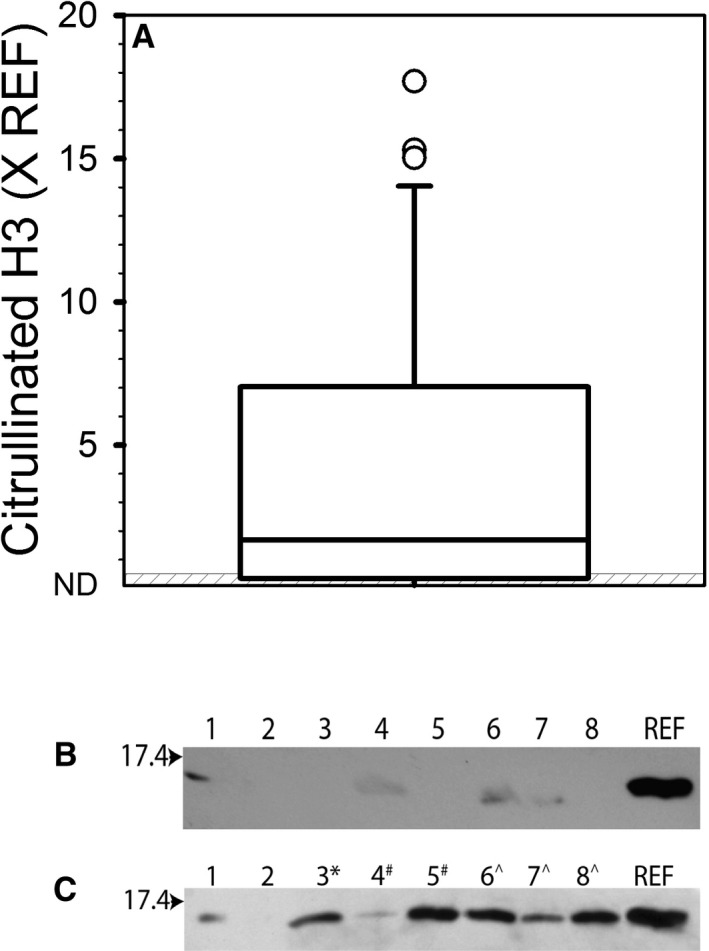
Citrullinated Histone H3 in Plasma. Plasma samples were evaluated for the presence of citrullinated histone H3 via Western blot as described in the Methods section, and quantified with respect to an internal reference sample. (A) Distribution of results obtained for plasma from dogs with IMHA (n = 32). The line of centrality denotes median, the box indicates 25th to 75th percentile, and the whiskers indicate 10th to 90th percentile. The gray‐striped box represents the reference interval of <0.48 × REF based on 95% confidence interval using samples from 26 apparently healthy dogs. (B) Example of Western blot image with samples from apparently healthy dogs. (C) Example of Western blot image with samples from dogs with IMHA. For B and C, the arrow denotes the location of the 17.4 KDa protein ladder band. Numbers at the top refer to the lane numbers. REF, the band lane containing the purified histone reference sample. Most plasma samples were diluted 4‐fold before loading, except for those denoted with *(8‐fold), #(16‐fold), and ^(32‐fold).
Comparison of Outcome to Assay results
Each data set was divided into 2 groups based on survival to discharge (Fig 5), survival to 28 days, and survival to 6 months. For all analyses, the median value for the nonsurvivor group was higher than that of the survivor group, but the difference was not significant.
Figure 5.
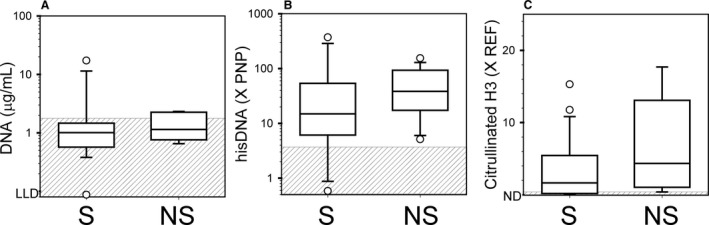
Comparison of Survivors to Nonsurvivors. Data for dogs with IMHA were separated into 2 groups as regards to whether or not the patient survived to be discharged from the hospital. (A) Results for cell‐free DNA. (B) Results for the histone‐DNA complex. (C) Results for citrullinated histone H3. LLD, lower limit of detection. PNP, pooled normal plasma. REF, reference band. ND, no band detected. S, survived. NS, did not survive.
Discussion
Several recent studies have evaluated human plasma for the presence of nucleosomal material in specific disease conditions,17, 30, 31, 32, 33, 34, 35 but published studies describing NETosis in the veterinary literature are limited. Canine neutrophils undergo NETosis in vitro in response to stimulation.36, 37 Recently, higher than normal concentrations of hisDNA were identified in the plasma of 7 dogs with IMHA.36 The presence of hisDNA, however, indicates only that nucleosomal release has occurred but does not determine the process that resulted in the presence of nucleosomal material in the plasma. Cell‐free DNA and hisDNA can be released from infective organisms, or be present as a result of cell necrosis, apoptosis or NETosis. Our results confirm and extend previous observations36 by demonstrating both excessive nucleosomal material in plasma and excessive citrullination of histones in most of the dogs with IMHA. Because citrullination may be a specific component of NETosis,38 our data suggest that NETosis is indeed excessive in dogs with IMHA. The lack of correlation between the Cell‐free DNA assay and the citrullinated Histone H3 assay could mean that the assays are, in part, detecting cell‐free DNA from different sources (eg, infective organisms, necrosis, and apoptosis) separate from or in addition to NETosis.
Immune‐mediated hemolytic anemia is a devastating disease associated with high mortality,29, 39, 40 and the marked inflammatory nature of IMHA may be an important contributor to mortality. Dogs with IMHA often have a marked leukemoid response, and may develop systemic inflammatory response syndrome along with its devastating sequelae.25 It is therefore not surprising that a disease associated with neutrophilia and hypercytokinemia41, 42, 43 results in excessive NET formation. Interestingly, of the 3 types of assays performed, none of the results correlated significantly with the severity of neutrophilia in the patient. This finding suggests that excessive NETosis in the IMHA population is not merely a product of mobilization of neutrophils to the peripheral circulation, but rather may be a specific feature of the inappropriate immune response in this particular disease process. Alternatively, only specific subsets of the neutrophil population might be undergoing NETosis in these patients.
NETosis is reported to be a feature of various autoimmune inflammatory diseases in humans (eg, systemic lupus erythematosus,15 vasculitis,16 and rheumatoid arthritis17) and may be an important contributor to morbidity and mortality in these conditions. Furthermore, recent evidence suggests that NETosis also is related to thrombosis. Citrullinated histone H3 has been identified in thrombi,18, 22 and knockout of PAD4 (the enzyme necessary for citrullination) has been shown to prevent deep vein thrombosis in a mouse model.24
Although it is widely accepted that thrombosis (or thromboembolism) is an important feature of the clinical presentation of IMHA in dogs,44 the exact mechanisms predisposing these patients to thrombosis are not clear. A relationship between systemic inflammation and increased risk for thrombosis is well described. Potential contributors also include procoagulant microparticles,45 increased intravascular tissue factor exposure,46 platelet hyper‐reactivity,47, 48 acquired antithrombin deficiency,25, 49 and antiphospholipid antibodies. Based on our results, excessive NET formation should be considered as a potential contributor to or cause of thrombogenicity in dogs with IMHA. If NETosis is indeed an important mechanism underlying thrombus development in these patients, future therapeutic agents targeting this process may be of benefit in dogs with IMHA. Approaches under investigation include enzymes that degrade DNA and inhibitors of either PAD4 or the complement cascade.50
We previously reported that the assay that detects cell‐free DNA in plasma, although rapid, inexpensive, and easy to perform, is relatively insensitive.28 The information presented here using plasma from dogs with IMHA confirms this previous finding, because the test only identified increased amounts of plasma cell‐free DNA in 17% of samples (compared to much higher percentages using other approaches). Because pigmentemia can interfere with the assay, we were not able to obtain a result for 26% of the samples. We suspect that the concentration of cell‐free DNA may also have been increased in some (or all) of the samples that could not be assayed, which means that had results been obtained for these samples the percentage of abnormal results might have been higher. Unfortunately, this is a major limitation of the cell‐free DNA assay, because the majority of samples from dogs with IMHA will have pigmentemia. This assay, however, still was relatively insensitive as compared to the hisDNA ELISA (see Fig 3). Interestingly, all of the samples with increased cell‐free DNA also were abnormal on the ELISA for hisDNA, suggesting high specificity for the cell‐free DNA assay. It may therefore be useful as a screening tool but is less likely to be of clinical utility as a prognosticator. Extracting the DNA from pigmented plasma may be an option to improve the utility of this method.
Our study had several limitations. Because of the observational nature of our study, we were not able to mandate the diagnostic tests that were performed on these patients. Our criteria for confirming a diagnosis of IMHA were broad and did not distinguish between primary and secondary causes of IMHA. Consequently, some of our patients may have had clinically relevant concurrent conditions (eg, neoplasia, infectious diseases) that could have contributed to increases in NET markers. Furthermore, our population was quite variable in nature, including both patients with evidence of intra‐ and extravascular hemolysis, as well as a wide range and severity of illness as suggested by the distribution of CHAOS scores. This heterogeneity, in addition to relatively small patient numbers, resulted in low power to detect statistically significant associations that might be present between outcome and markers for nucleosomal release that we evaluated. A comparison between healthy individual dogs and those with IMHA would have been beneficial and may be helpful in future studies. A definitive evaluation of the potential prognostic value of measuring indicators of NETosis awaits further study in a larger population.
Supporting information
Table S1. Correlation data between SYTOX Green cell‐free DNA and Citrullinated Histone H3 assays.
Acknowledgments
The authors thank Katrina Jung and Ron Achiel for their help with sample collection and Danika Rahn for her help with data acquisition.
Conflict of Interest Declaration
Authors declare no conflict of interest.
Off‐label Antimicrobial Declaration
Authors declare no off‐label use of antimicrobials.
This study was performed at University of Illinois, Urbana‐Champaign.
This study was not supported by a grant.
A portion of this data was presented in abstract form at the 2015 ACVIM Forum, Indianapolis, IN.
Footnotes
aSYTOX Green1, Life Technologies, Grand Isle, NY
bMicroplate (98 well), Corning, Tewksbury, MA
cSpectramax M2 fluorimeter, Molecular Devices, Sunnyvale, CA
dDNA, ThermoFisher, Waltham, MA
eCell Death Detection ELISA, Roche Diagnostics GmbH, Mannheim, Germany
fVersamax Spectrophotometer, Molecular Devices, Sunnyvale, CA
gEpiQuick Total Histone Extraction kit, EpiGenTek Group, Inc, Farmingdale, NY
hTris‐glycine mini gel, Bio‐Rad, Hercules, CA
iAntihistone H3, citrulline, Abcam, Cambridge, MA
jDonkey anti‐rabbit IgG‐horseradish peroxidase, ThermoFisher, Waltham, MA
kPierce ECL‐2 substrate, ThermoFisher, Waltham, MA
lGelDoc XR+ and ImageLab, Bio‐Rad, Hercules, CA
mSigmaStat 4.0., Systat Software, San Jose, CA
nSNAP 4Dx test, IDEXX, Westbrook, ME
References
- 1. Pruchniak MP, Arazna M, Demkow U. Life of neutrophil: From stem cell to neutrophil extracellular trap. Respir Physiol Neurobiol 2013;187:68–73. [DOI] [PubMed] [Google Scholar]
- 2. Hostetter SJ. Neutrophil function in small animals. Vet Clin North Am Small Anim Pract 2012;42:157–171. [DOI] [PubMed] [Google Scholar]
- 3. Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532–1535. [DOI] [PubMed] [Google Scholar]
- 4. Yousefi S, Mihalache C, Kozlowski E, et al. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ 2009;16:1438–1444. [DOI] [PubMed] [Google Scholar]
- 5. Yousefi S, Gold JA, Andina N, et al. Catapult‐like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med 2008;14:949–953. [DOI] [PubMed] [Google Scholar]
- 6. Morshed M, Hlushchuk R, Simon D, et al. NADPH oxidase‐independent formation of extracellular DNA traps by basophils. J Immunol 2014;192:5314–5323. [DOI] [PubMed] [Google Scholar]
- 7. von Kockritz‐Blickwede M, Goldmann O, Thulin P, et al. Phagocytosis‐independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008;111:3070–3080. [DOI] [PubMed] [Google Scholar]
- 8. Mohanan S, Horibata S, McElwee JL, et al. Identification of macrophage extracellular trap‐like structures in mammary gland adipose tissue: A preliminary study. Front Immunol 2013;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papayannopoulos V, Metzler KD, Hakkim A, et al. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol 2010;191:677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neeli I, Dwivedi N, Khan S, et al. Regulation of extracellular chromatin release from neutrophils. J Innate Immun 2009;1:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Li M, Stadler S, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol 2009;184:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li P, Li M, Lindberg MR, et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med 2010;207:1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas GM, Carbo C, Curtis BR, et al. Extracellular DNA traps are associated with the pathogenesis of TRALI in humans and mice. Blood 2012;119:6335–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu FC, Chuang YH, Tsai YF, et al. Role of neutrophil extracellular traps following injury. Shock 2014;41:491–498. [DOI] [PubMed] [Google Scholar]
- 15. Yu Y, Su K. Neutrophil extracellular traps and systemic lupus erythematosus. J Clin Cell Immunol 2013;4:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konstantinov KN, Ulff‐Moller CJ, Tzamaloukas AH. Infections and antineutrophil cytoplasmic antibodies: Triggering mechanisms. Autoimmun Rev 2015;14:201–203. [DOI] [PubMed] [Google Scholar]
- 17. Sur Chowdhury C, Giaglis S, Walker UA, et al. Enhanced neutrophil extracellular trap generation in rheumatoid arthritis: Analysis of underlying signal transduction pathways and potential diagnostic utility. Arthritis Res Ther 2014;16:R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brill A, Fuchs TA, Savchenko AS, et al. Neutrophil extracellular traps promote deep vein thrombosis in mice. J Thromb Haemost 2012;10:136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diaz JA, Fuchs TA, Jackson TO, et al. Plasma DNA is elevated in patients with deep vein thrombosis. J Vasc Surg Venous Lymphat Disord 2013;1:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Savchenko AS, Borissoff JI, Martinod K, et al. VWF‐mediated leukocyte recruitment with chromatin decondensation by PAD4 increases myocardial ischemia/reperfusion injury in mice. Blood 2014;123:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. von Bruhl ML, Stark K, Steinhart A, et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med 2012;209:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuchs TA, Brill A, Duerschmied D, et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci USA 2010;107:15880–15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Montfoort ML, Stephan F, Lauw MN, et al. Circulating nucleosomes and neutrophil activation as risk factors for deep vein thrombosis. Arterioscler Thromb Vasc Biol 2013;33:147–151. [DOI] [PubMed] [Google Scholar]
- 24. Martinod K, Demers M, Fuchs TA, et al. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc Natl Acad Sci USA 2013;110:8674–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott‐Moncrieff JC, Treadwell NG, McCullough SM, et al. Hemostatic abnormalities in dogs with primary immune‐mediated hemolytic anemia. J Am Anim Hosp Assoc 2001;37:220–227. [DOI] [PubMed] [Google Scholar]
- 26. Carr AP, Panciera DL, Kidd L. Prognostic factors for mortality and thromboembolism in canine immune‐mediated hemolytic anemia: A retrospective study of 72 dogs. J Vet Intern Med 2002;16:504–509. [DOI] [PubMed] [Google Scholar]
- 27. Piek CJ, Junius G, Dekker A, et al. Idiopathic immune‐mediated hemolytic anemia: Treatment outcome and prognostic factors in 149 dogs. J Vet Intern Med 2008;22:366–373. [DOI] [PubMed] [Google Scholar]
- 28. Smith SA, Lawson C, McMichael MA, et al. Evaluation of assays for quantification of DNA in canine plasma as an indirect marker of NETosis. Vet Clin Pathol 2017;46:278–286. [DOI] [PubMed] [Google Scholar]
- 29. Goggs R, Dennis SG, Di Bella A, et al. Predicting Outcome in dogs with Primary Immune‐Mediated Hemolytic Anemia: Results of a Multicenter Case Registry. J Vet Intern Med 2015;29:1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen JA, Meister S, Urbonaviciute V, et al. Sensitive detection of plasma/serum DNA in patients with systemic lupus erythematosus. Autoimmunity 2007;40:307–310. [DOI] [PubMed] [Google Scholar]
- 31. Geiger S, Holdenrieder S, Stieber P, et al. Nucleosomes in serum of patients with early cerebral stroke. Cerebrovasc Dis 2006;21:32–37. [DOI] [PubMed] [Google Scholar]
- 32. Moore DJ, Greystoke A, Butt F, et al. A pilot study assessing the prognostic value of CK18 and nDNA biomarkers in severe sepsis patients. Clin Drug Invest 2012;32:179–187. [DOI] [PubMed] [Google Scholar]
- 33. Craig DG, Lee P, Pryde EA, et al. Circulating apoptotic and necrotic cell death markers in patients with acute liver injury. Liver Int 2011;31:1127–1136. [DOI] [PubMed] [Google Scholar]
- 34. Schimmel M, Nur E, Biemond BJ, et al. Nucleosomes and neutrophil activation in sickle cell disease painful crisis. Haematologica 2013;98:1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeerleder S, Stephan F, Emonts M, et al. Circulating nucleosomes and severity of illness in children suffering from meningococcal sepsis treated with protein C. Crit Care Med 2012;40:3224–3229. [DOI] [PubMed] [Google Scholar]
- 36. Jeffery U, Kimura K, Gray R, et al. Dogs cast NETs too: Canine neutrophil extracellular traps in health and immune‐mediated hemolytic anemia. Vet Immunol Immunopathol 2015;168:262–268. [DOI] [PubMed] [Google Scholar]
- 37. Wei Z, Hermosilla C, Taubert A, et al. Canine Neutrophil Extracellular Traps Release Induced by the Apicomplexan Parasite Neospora caninum In Vitro. Front Immunol 2016;7:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirose T, Hamaguchi S, Matsumoto N, et al. Presence of neutrophil extracellular traps and citrullinated histone h3 in the bloodstream of critically ill patients. PLoS One 2014;9:e111755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson ML, Kruth SA. Immune‐mediated Hemolytic Anemia and Thrombocytopenia in the Dog: A retrospective study of 55 cases diagnosed from 1979 through 1983 at the Western College of Veterinary Medicine. Can Vet J 1985;26:245–250. [PMC free article] [PubMed] [Google Scholar]
- 40. Reimer ME, Troy GC, Warnick LD. Immune‐mediated hemolytic anemia: 70 cases (1988‐1996). J Am Anim Hosp Assoc 1999;35:384–391. [DOI] [PubMed] [Google Scholar]
- 41. Johnson V, Burgess B, Morley P, et al. Comparison of cytokine responses between dogs with sepsis and dogs with immune‐mediated hemolytic anemia. Vet Immunol Immunopathol 2016;180:15–20. [DOI] [PubMed] [Google Scholar]
- 42. Duffy AL, Olea‐Popelka FJ, Eucher J, et al. Serum concentrations of monocyte chemoattractant protein‐1 in healthy and critically ill dogs. Vet Clin Pathol 2010;39:302–305. [DOI] [PubMed] [Google Scholar]
- 43. Kjelgaard‐Hansen M, Goggs R, Wiinberg B, et al. Use of serum concentrations of interleukin‐18 and monocyte chemoattractant protein‐1 as prognostic indicators in primary immune‐mediated hemolytic anemia in dogs. J Vet Intern Med 2011;25:76–82. [DOI] [PubMed] [Google Scholar]
- 44. Kidd L, Mackman N. Prothrombotic mechanisms and anticoagulant therapy in dogs with immune‐mediated hemolytic anemia. J Vet Emerg Crit Care 2013;23:3–13. [DOI] [PubMed] [Google Scholar]
- 45. Kidd L, Geddings J, Hisada Y, et al. Procoagulant microparticles in dogs with immune‐mediated hemolytic anemia. J Vet Intern Med 2015;29:908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piek CJ, Brinkhof B, Teske E, et al. High intravascular tissue factor expression in dogs with idiopathic immune‐mediated haemolytic anaemia. Vet Immunol Immunopathol 2011;144:346–354. [DOI] [PubMed] [Google Scholar]
- 47. Hamzianpour N, Chan DL. Thromboelastographic assessment of the contribution of platelets and clotting proteases to the hypercoagulable state of dogs with immune‐mediated hemolytic anemia. J Vet Emerg Crit Care 2016;26:295–299. [DOI] [PubMed] [Google Scholar]
- 48. Weiss DJ, Brazzell JL. Detection of activated platelets in dogs with primary immune‐mediated hemolytic anemia. J Vet Intern Med 2006;20:682–686. [DOI] [PubMed] [Google Scholar]
- 49. Kuzi S, Segev G, Haruvi E, et al. Plasma antithrombin activity as a diagnostic and prognostic indicator in dogs: A retrospective study of 149 dogs. J Vet Intern Med 2010;24:587–596. [DOI] [PubMed] [Google Scholar]
- 50. Barnado A, Crofford LJ, Oates JC. At the Bedside: Neutrophil extracellular traps (NETs) as targets for biomarkers and therapies in autoimmune diseases. J Leukoc Biol 2016;99:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Correlation data between SYTOX Green cell‐free DNA and Citrullinated Histone H3 assays.


